Abstract
Free full text

SARS-CoV-2 infects and replicates in photoreceptor and retinal ganglion cells of human retinal organoids
Summary
Several studies have pointed to retinal involvement in COVID-19, yet many questions remain regarding the ability of SARS-CoV-2 to infect and replicate in retinal cells and its effects on the retina. Here, we have used human pluripotent stem cell-derived retinal organoids to study retinal infection by SARS-CoV-2. Indeed, SARS-CoV-2 can infect and replicate in retinal organoids, as it is shown to infect different retinal lineages, such as retinal ganglion cells and photoreceptors. SARS-CoV-2 infection of retinal organoids also induces the expression of several inflammatory genes, such as interleukin 33, a gene associated with acute COVID-19 and retinal degeneration. Finally, we show that the use of antibodies to block ACE2 significantly reduces SARS-CoV-2 infection of retinal organoids, indicating that SARS-CoV-2 infects retinal cells in an ACE2-dependent manner. These results suggest a retinal involvement in COVID-19 and emphasize the need to monitor retinal pathologies as potential sequelae of “long COVID.”
Introduction
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), is a global pandemic that is responsible for millions of fatalities. While mild-to-severe respiratory symptoms are most commonly associated with the disease (Huang et al., 2020), neurological (Asadi-Pooya and Simani, 2020; Mao et al., 2020) and ocular (Wu et al., 2020) symptoms have also been described in patients.
Growing evidence suggests retinal involvement in some cases of COVID-19. Different reports identified SARS-CoV-2 RNA in retinal biopsies taken from deceased COVID-19 patients (Casagrande et al., 2020, 2021), while different retinal anomalies were also identified in patients (Burgos-Blasco et al., 2020; Conrady et al., 2021; Marinho et al., 2020; Pereira et al., 2020; Rodriguez-Rodriguez et al., 2021; Virgo and Mohamed, 2020). However, retinal involvement in COVID-19 remains controversial, as other studies were unable to detect retinal pathologies in patients (Pirraglia et al., 2020). Furthermore, a study reported failure to isolate virus from SARS-CoV-2 RNA-positive retinal biopsies and an inability to detect any SARS-CoV-2 Spike protein from those biopsies by immunohistochemical analysis, leading investigators to suggest that SARS-CoV-2 can infect but not actively replicate in retinal cells (Casagrande et al., 2021). It also remains unclear which retinal structures are infected by SARS-CoV-2 and whether the retinal anomalies reported are due to retinal infection or systemic organ dysfunction (Casagrande et al., 2020; de Figueiredo et al., 2020).
Organoids are 3-dimensional tissue cultures that resemble specific organs in their morphology, cell-type composition, or function. Different research groups have used organoids generated from human pluripotent stem cells to study the infection of SARS-CoV-2 in the context of different organs (Lamers et al., 2020; Monteil et al., 2020; Pellegrini et al., 2020; Ramani et al., 2020; Zhang et al., 2020). Such studies have demonstrated that SARS-CoV-2 can infect neurons and neural progenitors (Ramani et al., 2020; Zhang et al., 2020). Retinal organoids are among the most physiologically accurate neural organoid models, as they contain all of the major retinal cell types, follow a developmental trajectory similar to that of the human retina, and are correctly organized in a typical layered structure (Achberger et al., 2019; Nakano et al., 2012; Zhong et al., 2014).
Here, we use retinal organoids to show that SARS-CoV-2 can infect retinal cells, mainly retinal ganglion cells (RGCs) but also photoreceptors, as well as replicate in them. Moreover, retinal SARS-CoV-2 infection induces the expression of several inflammatory genes and is reduced by treatment with an anti-ACE-2 antibody.
Results
SARS-CoV-2 infects retinal organoids and replicates in retinal cells
To generate retinal organoids, we have combined different widely used protocols (Capowski et al., 2019; Kuwahara et al., 2015; Zhong et al., 2014). The organoids generated followed correct retinal development and contained a thick layer of visual system homeobox 2-positive (VSX2+) retinal progenitors and a basal layer of differentiated paired box 6-positive (PAX6+)/VSX2-RGC and amacrine cells by day 50 of the protocol (Figure 1A). At later stages, the organoids had gained a layered retinal structure with an inner nuclear layer (INL), containing the different INL cell types such as orthodenticle homeobox 2 (OTX2+) bipolar cells, transcription factor AP2-Alpha (TFAP2A, AP2A+) amacrine and horizontal cells, and cellular retinaldehyde-binding protein-positive (CRALBP+) Müller glia (MG), separated from an outer nuclear layer (ONL), containing OTX2+ photoreceptors (Figure 1B). The photoreceptors expressed rod and cone opsins concentrated in outer segment-like structures (Figure 1C), indicating highly mature organoids.
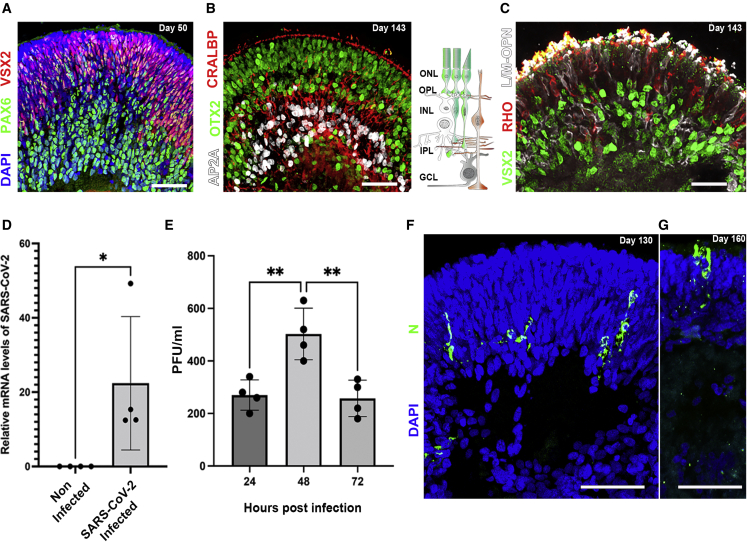
Retinal organoids can be infected by SARS-CoV-2
(A) Retinal organoids on day 50 of differentiation are shown to contain VSX2+ (green) retinal progenitors and PAX6+ (white)/VSX2-amacrine and retinal ganglion cells (RGCs).
(B and C) On day 143 of differentiation, the organoids contain AP2a+ (white) amacrine and horizontal cells, CRALBP+ (red) Müller glia, and OTX2+ (green) photoreceptors (B) and express long or medium wave length opsin (L/M-OPN white) and rhodopsin (RHO, red) (C). The sketch in (B) depicts the cell types and structure of the vertebrate retina. GCL: ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer.
(D) qPCR identified SARS-CoV-2 genomic RNA within retinal organoids treated with SARS-CoV-2 but not in noninfected controls at day 118. N = 4,  p = 0.0467.
p = 0.0467.
(E) A viral plaque assay performed in VeroE6 using the supernatant of SARS-CoV-2-infected organoids was used to assess viral titers in organoids infected on day 118. N = 4, ANOVA p = 0.0023, F = 12.85; 
 p = 0.0053 and 0.0038, respectively.
p = 0.0053 and 0.0038, respectively.
(F and G) IF was used to detect SARS-CoV-2 nucleocapsid (N)-positive cells in infected organoids at day 130 (F) or 160 (G) of differentiation.
Each repeat (N = 4) in (D) and (E) is a separate group of 5 organoids infected or not infected in a separate well. Scale bars: A, B, F, and G, 50 μm; C, 20 μm. Error bars represent standard deviation.
To test the ability of SARS-CoV-2 to infect retinal cells, relatively mature human retinal organoids (day of differentiation ≥118) were treated with SARS-CoV-2 and incubated for different time periods. The infected organoids were then analyzed by different methods to assess SARS-CoV-2 infection and replication. SARS-CoV-2 RNA was detected by qPCR in SARS-CoV-2-treated organoids but not in the controls (Figure 1D), suggesting that these organoids were indeed infected with SARS-CoV-2. Furthermore, a viral plaque assay was used to measure the active viral concentrations produced by the infected organoids after different incubation times. The appearance of viral plaques in the cell cultures treated with supernatant that was incubated for 24 h with the infected organoids indicated that new virus progeny was being generated, providing the first evidence that SARS-CoV-2 replicates within human retinal tissue. Accordingly, virus titers were increased after 48 h of incubation, but surprisingly were reduced back to levels that are similar to the 24-h time point after 72 h (Figure 1E). A similar phenomenon was reported in SARS-CoV-2-infected brain organoids (Zhang et al., 2020).
Finally, an immunofluorescence (IF) analysis identified a small number of SARS-CoV-2 nucleocapsid protein (N)-positive cells in SARS-CoV-2-treated organoids on days 130 and 160 of differentiation (Figures 1F and 1G) but not in controls (Figure 2F). Interestingly, many of the N+ cells were found in the ONL of the organoid and some exhibited a typical photoreceptor morphology (Figure 1G). While most of the work presented here was made using human induced pluripotent stem cells (iPSCs), organoids derived from human embryonic stem cells (hESCs) were also found to be open to infection (Figure S1).
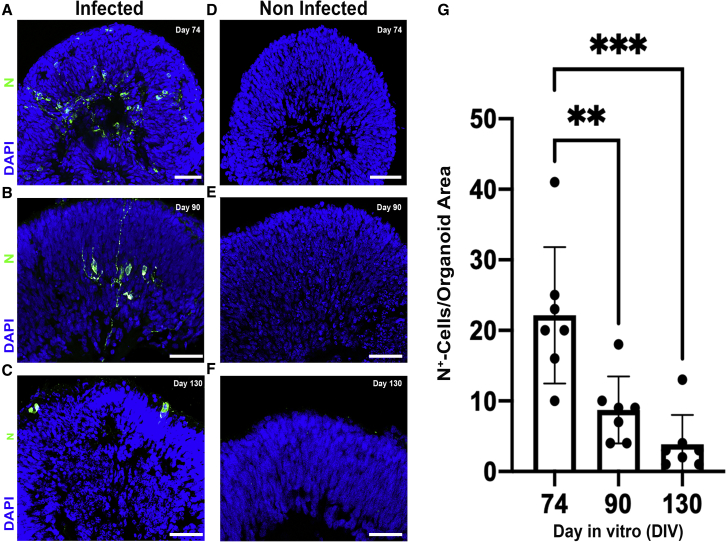
SARS-CoV-2 nucleocapsid-positive cells are more abundant in retinal organoids infected at an earlier stage of differentiation
(A–C) SARS-CoV-2 nucleocapsid (N) immunostaining was used to quantify the number of N+ (green) cells in retinal organoids that were infected at 3 different time points: day 74 (A), day 90 (B), and day 130 (C).
(D–F) An age-matched noninfected control for each stage: day 74 (D), day 90 (E), and day 130 (F) is also presented. All of the organoids were incubated for 96 h after infection.
(G) The number of N+ cells in the different stages was compared (G). N = 7 organoids, taken from 2 different infected or control wells, ANOVA p = 0.0002, F = 14.15; 
 p = 0.0038;
p = 0.0038; 

 p = 0.0002. Error bars represent standard deviation.
p = 0.0002. Error bars represent standard deviation.
Scale bars: 50 μm.
The number of N+ cells identified in retinal organoids at these late stages was relatively low (3.8 per organoid, on average, on day 130; Figure 2C). To investigate whether retinal organoids at earlier developmental stages are more susceptible to SARS-CoV-2 infection, we infected organoids at either day 74 or day 90, and compared the number of N+ cells to that of the older organoids (Figures 2A–2C and 2G). The number of N+ cells on day 74 infected retinal organoids was significantly higher than in either day 90 infected or day 130 infected organoids (an average of 22.14 per organoid, in contrast to 8.7 for day 90 and 3.8 for day 130). Strikingly, SARS-CoV-2-infected cells appeared to localize to the inner retina in younger retinal organoids, whereas they predominantly affected the outer retina in older organoids. Similarly, a plaque assay identified significantly higher virus titers in day 74 organoids compared to day 125 organoids (Figure S2A).
RGCs are particularly susceptible to SARS-CoV-2 infection
To identify the retinal cells that are infected, day 74 infected organoids were co-stained with antibodies directed against SARS-CoV-2 N and retinal cell-type-specific markers (Figures 3A–3D). Interestingly, RGCs appeared to be the most commonly infected cell type, as approximately 40% of the N+ cells also stained positive for the RGC marker SNCG (Figures 3A and 3D). A smaller number of OTX2+ photoreceptors (and perhaps a few bipolar cells) and AP2A+ amacrine cells (Figures 3B and 3D) also stained positive for N, indicating the permissibility of these cells. An even smaller number of VSX2+ progenitors (and perhaps a few bipolar cells) also stained positive for N (Figures 3C and 3D), suggesting that progenitor cells are less susceptible to infection than neurons but may still be infected. As MGs are still rare at this stage of retinal organoid development, we did not attempt to stain the organoids with an MG marker.

SARS-CoV-2 infects retinal ganglion cells at a higher rate than other retinal cell types
(A–C) Retinal organoids infected with SARS-CoV-2 on day 74 and incubated for 96 h before fixation were stained with antibodies against SARS-CoV-2 nucleocapsid (N, green) and the retinal ganglion marker SNCG (red) (A), the photoreceptor marker OTX2 (white) (B), the amacrine and horizontal marker AP2α (red) (B), and the retinal progenitor marker VSX2 (red) (C).
(D) The percentage of cells co-stained with each of the markers was compared. N = 6 organoids taken from 2 different wells, ANOVA p < 0.0001, F = 21.69;  p = 0.032;
p = 0.032; 

 p = 0.0008;
p = 0.0008; 


 p < 0.0001. Error bars represent standard deviation.
p < 0.0001. Error bars represent standard deviation.
(E–H) Day 74 control organoids were stained against OTX2 (white), HUC/D (red), and ACE2 (green) (E and G) or rabbit polyclonal isotype control (green) (F and H). Scale bars: 10 μm.
Angiotensin-converting enzyme 2 (ACE2) is the receptor most commonly associated with the SARS-CoV-2 infection pathway. While ACE2 mRNA expression in the human retina is thought to be low, ACE2 protein is present in the adult human and rat retinas (Senanayake et al., 2007; Tikellis et al., 2004). Accordingly, an IF analysis identified an ACE2 signal concentrated in OTX2+ photoreceptors and HUC/D+ RGC and amacrine cells in our retinal organoids (Figures 3E and 3G). These results are consistent with the retinal infection profile of SARS-CoV-2 shown here.
Day 90 infected organoids seemed to show (although reduced) infection patterns similar to that of day 74 (Figure S2B). Corresponding with the known depletion of RGCs in late-stage retinal organoids, the majority of N+ cells in day 130 infected organoids are co-stained with the photoreceptor/bipolar marker OTX2 (Figures S2C and S2D).
A transcriptomic analysis of SARS-CoV-2-infected retinal organoids
To study the effect of SARS-CoV-2 infection on retinal organoids, we conducted an RNA sequencing (RNA-seq) analysis that compared the transcriptome of noninfected retinal organoids to that of retinal organoids that were infected with SARS-CoV-2 on day 80 of differentiation and incubated for either 24 or 96 h after infection. Unsurprisingly, 10 SARS-CoV-2 transcripts were highly enriched within the infected samples, although no difference was seen between 24 h post-infection incubation and 96 h post-infection incubation (Figure 4A). This is consistent with the reduction in plaque-forming units (PFUs) observed in the plaque assay after 72 h (Figure 1E).
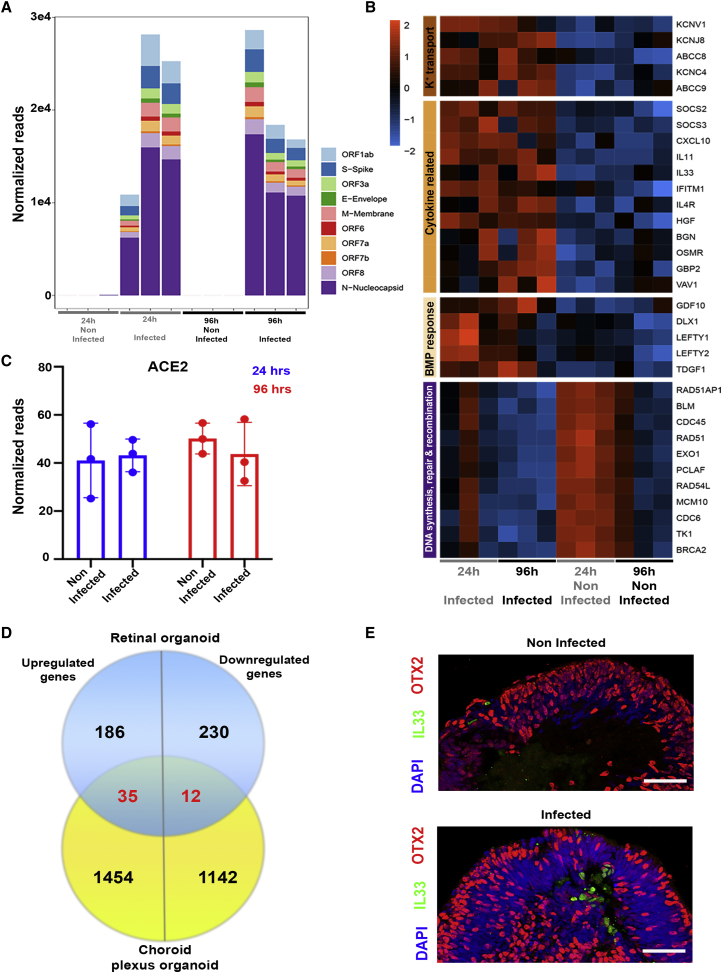
A transcriptomic analysis of SARS-CoV-2-infected retinal organoids
(A) A transcriptomic analysis was conducted using infected retinal organoids, incubated for 24 or 96 h following infection, and their respective controls. N = 3 per group. Each repeat included 5 separately treated organoids. SARS-CoV-2 transcripts were highly expressed in the infected samples.
(B) Genes that were significantly differentially expressed (DE) with a fold change of at least 2 (or −2) among all of the samples together were used for Gene Ontology (GO) analysis. A heatmap of genes related to the top enriched categories identified using GO is presented.
(C) The number of normalized reads of the ACE2 gene in noninfected and infected organoids was extracted from the analysis. A Student’s t test did not identify significant differences between the groups (p = 0.83 [24 h] and 0.48 [96 h]). Error bars represent standard deviation.
(D)The DE genes identified in the retinal organoids 24 h post-infection were compared to those identified in SARS-CoV-2-infected choroid plexus after 24 h. The comparison is presented as a Venn diagram.
(E) Control and retinal organoids infected on day 74 of differentiation and incubated for 24 h before fixation were stained against IL-33 (green) and OTX2 (red). Scale bars: 50 μm.
ACE2 transcripts were identified in all of the samples, with no change in expression due to virus infection at either incubation time (Figure 4C), suggesting that the infection of retinal cells does not result in ACE2 upregulation. This result contradicts studies suggesting ACE2 upregulation upon SARS-CoV-2 infection of other types of host cells (Butler et al., 2021).
Next, we compared the transcriptome of all of the infected samples to that of the control samples to identify genes that are differentially expressed (DE) upon infection. This analysis identified 759 DE genes (false discovery rate [FDR] adjusted p- ≤ 0.05), 366 of which were upregulated and 393 were downregulated (Figure S3B).
To gain further information regarding the processes occurring in the retinal organoids upon infection, a Gene Ontology (GO) analysis of the most highly upregulated genes (fold change [FC] ≥ 2) was performed. The top 5 most enriched biological process categories in this analysis were “potassium ion transport,” “cellular response to cytokine stimulus,” “cellular response to bone morphogenetic protein (BMP) stimulus,” “metal ion transport,” and “cytokine-mediated signaling pathway” (Figure 4B; Table 1).
Table 1
Enriched GO biological process categories identified in the top upregulated (adjusted p ≤ 0.05, fold change ≥2) and downregulated (adjusted p ≤ 0.05, fold change ≤−2) genes
| Enriched GO category | p | Adjusted p | |
|---|---|---|---|
| Enriched among top upregulated genes | potassium ion transport (GO: 0006813) | 0.00002763 | 0.01486 |
| cellular response to cytokine stimulus (GO: 0071345) | 0.00007025 | 0.01890 | |
| cellular response to BMP stimulus (GO: 0071773) | 0.0001522 | 0.02730 | |
| metal ion transport (GO: 0030001) | 0.0003060 | 0.02938 | |
| cytokine-mediated signaling pathway (GO: 0019221) | 0.0003120 | 0.02938 | |
| Enriched among top downregulated genes | DNA synthesis involved in DNA repair (GO: 0000731) | 3.688 × 10E−7 | 0.0002703 |
| strand displacement (GO: 0000732) | 9.469 × 10E−7 | 0.0003470 | |
| DNA recombination (GO: 0006310) | 0.000002408 | 0.0005882 | |
| DNA biosynthetic process (GO: 0071897) | 0.000007700 | 0.001411 | |
| DNA metabolic process (GO: 0006259) | 0.00001441 | 0.002113 |
The enrichment of cytokine-related genes indicates the mounting of an immune response upon SARS-CoV-2 infection. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis performed using the same genes found that SARS-CoV-2 infection also led to the enrichment of genes related to the Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway, a signaling pathway involved in inflammation and the innate antiviral interferon response (data not shown). The enrichment of potassium ion transport-related genes in the data may correspond with evidence suggesting that hypokalemia is relatively common in COVID-19 (Mabillard and Sayer, 2020). While data from other tissues and organoids suggested a strong chemokine response upon SARS-CoV-2 infection (Blanco-Melo et al., 2020; Yang et al., 2020), our analysis identified only 2 chemokines, C-X-C motif chemokine ligand 2 (CXCL2) (FC = 1.83, FDR adjusted p value = 7.94 × 10E−5) and CXCL10 (FC = 3.82, FDR adjusted p value = 0.0003), to be significantly upregulated in retinal tissue.
Comparison of the transcriptome changes of SARS-CoV-2-infected retinal organoids to those of other organoid systems may allow for a better understanding of the unique features of the retinal response to SARS-CoV-2 infection. To this end, we have used a dataset of genes altered in SARS-CoV-2-infected choroid plexus organoids (Jacob et al., 2020). While the choroid plexus is not a neuronal tissue like the retina, it does develop from neural progenitors (Liddelow, 2015) and may share some of the response to SARS-CoV-2 infection with the retina.
Comparison of the DE genes of infected retinal organoids after incubation for 24 h to those of infected choroid plexus organoids after a similar incubation identified 35 genes that are upregulated and 12 genes that are downregulated in both organoid systems (Figure 4D). Of the genes upregulated in both systems, 17 were highly upregulated in the retinal organoids (FC ≥ 2). GO analysis identified enriched categories related to megakaryocyte differentiation, negative regulation of SMAD (suppressor of mothers against decapentaplegic) signaling and serine-threonine phosphorylation, peptidyl-tyrosine phosphorylation, and cell proliferation (Table 2). Interestingly, GO of genes that are specifically highly upregulated in the infected retinal organoids (and are not upregulated in choroid plexus organoids) identified several enriched categories related to transforming growth factor β (TGF-β) and BMP signaling (Table 2). Such a result suggests a difference in TGF-β response to SARS-CoV-2 infection between the retina and the choroid plexus.
Table 2
Enriched GO biological process categories identified in the top upregulated (adjusted p ≤0.05, fold change ≥2) genes in the retinal organoids 24 h post-infection that were also upregulated in choroid plexus organoids 24 h post-infection or that were not upregulated at 24 h post-infection
| Enriched GO category | p | Adjusted p | |
|---|---|---|---|
| A. Enriched among top upregulated genes shared between retinal and choroid plexus organoids | megakaryocyte differentiation (GO: 0030219) | 0.00002439 | 0.003050 |
| negative regulation of pathway-restricted SMAD protein phosphorylation (GO: 0060394) | 0.00003048 | 0.003050 | |
| negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway (GO: 0090101) | 0.00003589 | 0.003050 | |
| positive regulation of peptidyl-tyrosine phosphorylation (GO: 0050731) | 0.0001218 | 0.007767 | |
| regulation of cell proliferation (GO: 0042127) | 0.0002921 | 0.01490 | |
| B. Enriched among top upregulated genes not shared between retinal and choroid plexus organoids | cellular response to transforming growth factor β stimulus (GO: 0071560) | 0.00001530 | 0.009257 |
| transforming growth factor β receptor signaling pathway (GO: 0007179) | 0.00005211 | 0.01576 | |
| cellular response to BMP stimulus (GO: 0071773) | 0.00009952 | 0.01793 | |
| transmembrane receptor protein serine/threonine kinase signaling pathway (GO: 0007178) | 0.0001185 | 0.01793 | |
| potassium ion transport (GO: 0006813) | 0.0003084 | 0.03731 |
Unlike the choroid plexus organoids, retinal organoids show upregulation of the cytokine interleukin-33 (IL-33), the most upregulated (FC = 8.11, FDR adjusted p value = 4.27 × 10E−7) cytokine in these organoids. Accordingly, an IF analysis identified several IL-33+ cells in infected organoids, but hardly any in the controls (Figure 4E). Thus, SARS-CoV-2 retinal infection appears to induce IL-33 expression. IL-33 signaling is thought to play a role in COVID-19 pathology (Zizzo and Cohen, 2020), as well as in retinal inflammation and photoreceptor degradation following injury (Xi et al., 2016).
Another interesting gene found to be upregulated in SARS-CoV-2-infected retinal organoids is the inflammasome gene NLR family pyrin domain containing 1 (NLRP1) (FC = 2.2, FDR adjusted p value = 0.0346). NLRP1 inflammasomes are thought to be involved in RGC death in acute glaucoma (Yerramothu et al., 2018).
Interestingly, the top 5 categories enriched in the genes most significantly downregulated in infected retinal organoids (FDR adjusted p-value ≤ 0.05, FC ≤ −2) are related to DNA metabolism, repair, and recombination (Figure 4B; Table 1). This may suggest an effect of SARS-CoV-2 on proliferation or DNA damage repair mechanisms. Other coronaviruses have been shown to interact with the cell cycle and DNA damage response in different ways (Surjit et al., 2006; Xu et al., 2011; Zhou et al., 2008).
ACE2 blocking successfully reduces SARS-CoV-2 infection of retinal organoids
To assess the extent to which retinal SARS-CoV-2 infection is mediated by ACE2, retinal organoids were treated with an anti-ACE2 antibody that is documented to reduce SARS-CoV-2 infection (Hoffmann et al., 2020; Song et al., 2021). Interestingly, retinal organoids that were treated with the anti-ACE2 antibody contained a significantly lower number of N+ cells compared with organoids from the same batch that were treated with a normal goat immunoglobulin G (IgG) antibody isotype control (1.35 ± 2.02 versus 6.78 ± 7.78, on average; Figures 5A–5C). Thus, in contrast to recent suggestions that SARS-CoV-2 infection of the retina is not mediated by ACE2 (de Figueiredo et al., 2020), our results suggest that SARS-CoV-2 infection of retinal cells is dependent upon functional ACE2 receptors.
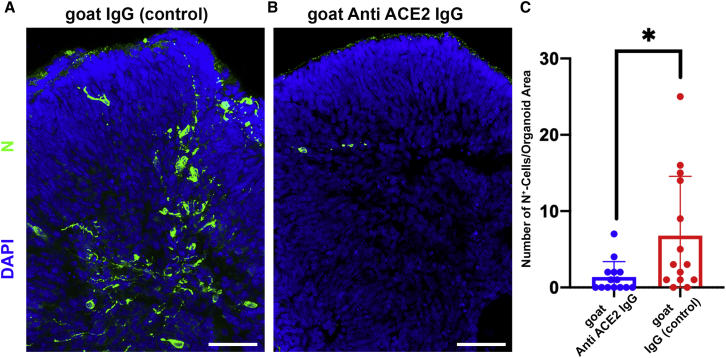
SARS-CoV-2 infection of retinal organoids is dependent upon ACE2
(A and B) SARS-CoV-2 nucleocapsid (N) staining (green) was used to detect SARS-CoV-2-infected cells in retinal organoids treated with either a goat IgG anti-ACE2 antibody (A) or a goat IgG isotype control (B) before being infected with SARS-CoV-2 and incubated for 96 h.
(C) The number of N+ cells was quantified and compared between organoids that went through the different treatments. N = 14. Organoids for each treatment were taken from 3 different infected wells;  p = 0.018. Scale bar: 50 μm. Error bars represent standard deviation.
p = 0.018. Scale bar: 50 μm. Error bars represent standard deviation.
Discussion
SARS-CoV-2 can actively infect retinal neurons
Retinal organoids show high similarity to the developing human retina in both morphology and transcriptome (Sridhar et al., 2020), rendering them a potent model for studying the biology of the human retina. Our data establish retinal organoids as a potent model to study retinal involvement in COVID-19. A recently published article showed that a recombinant lentiviral vector carrying the SARS-CoV-2 Spike protein can infect retinal organoids (Ahmad Mulyadi Lai et al., 2021). Our data go beyond that, showing that SARS-CoV-2 itself infects retinal organoids, and more important, we provide the first evidence that SARS-CoV-2 can actively replicate in retinal tissue.
Interestingly, younger retinal organoids appeared to be more open to infection than older ones. TMPRSS2 (transmembrane protease, serine 2) expression, which is known to be important in SARS-CoV-2 infection, was shown to decrease in retinal organoids over time (Ahmad Mulyadi Lai et al., 2021). This may suggest that more mature retinal cells are less likely to be infected by SARS-CoV-2 because of changes in TMPRSS2 expression. However, the expression of TMPRSS2 in our retinal organoids was extremely low on day 80, suggesting that its involvement in retinal SARS-CoV-2 infection is minimal.
In older retinal organoids, cells were also infected, albeit at a lower number, indicating that mature retinal cells are still susceptible to SARS-CoV-2 infection. In fact, progenitor cells were less likely to be infected than differentiated neurons, emphasizing the susceptibility of retinal neurons to infection. As retinal organoids differentiate, the number of cells contained therein increases, potentially increasing the organoid density. An increase in organoid density and size may decrease the permeability of the inner layers to SARS-CoV-2 and decrease the likelihood of the viral infection of inner retinal cells.
Moreover, changes in cell-type composition may affect the infectability of the organoids. Loss of RGCs is known to occur during the maturation of human retinal organoids, with nearly complete depletion of RGCs at the late stages (Sridhar et al., 2020; Zhong et al., 2014). The observed decrease in SARS-CoV-2-infected cells during retinal organoid maturation may be related to changes in the number of RGCs over time. Indeed, while our analysis identified SARS-CoV-2-infected cells from different retinal lineages, RGCs were significantly more likely to be infected.
Interestingly, many of the retinal symptoms associated with COVID-19, such as ganglion cell layer (GCL) lesions (Marinho et al., 2020) and swelling of the optic nerve (Burgos-Blasco et al., 2020), are related to RGCs. Pathology of the GCL could be the result of several issues, such as vascular dysfunction or increase in ocular pressure, and thus may be a secondary effect to other symptoms of the disease. However, as SARS-CoV-2 targets RGCs in the organoids, the infection of RGCs may have direct pathological consequences. It should be noted that because of their location in the inner layers of the retinal organoid sphere, RGCs may be exposed to a lower viral titer during the initial infection. If that is the case, then the high infectability of RGCs, despite their inner location, is a further testament to their high infectability.
RGCs are the cells that generate the optic nerve and connect the retina to the rest of the central nervous system (CNS). Viruses, such as the herpes simplex virus type 1, can be anterogradely transferred from the retina through the optic nerve into its targets in the brain (Sun et al., 1996). Considering this, the possibility that the retina represents a potential entry route for SARS-CoV-2 into the rest of the CNS should be considered. A recent publication suggested that SARS-CoV-2 can infect many nonretinal ocular tissues such as the cornea, the sclera, the choroid, the limbus, and the retinal pigmented epithelium (RPE), indicating the possibility of SARS-CoV-2 infection through the eye (Eriksen et al., 2021). Taken together, these results may suggest a direct route for SARS-CoV-2 infection from exposed ocular surface tissues such as the cornea to the CNS through the retina.
SARS-CoV-2 infection of retinal organoids induces inflammatory genes
The pathology of COVID-19 is highly attributed to uncontrolled hyperinflammation, referred to as a cytokine storm (Burke et al., 2020; Mehta et al., 2020). Our data suggest that the infection of retinal organoids results in the upregulation of several inflammatory genes. Among these are several genes involved in the immune response to viral infection, such as CXCL10 (Trifilo et al., 2004) and interferon-induced transmembrane protein 1 (IFTIM1) (Brass et al., 2009).
The most significantly upregulated cytokine in infected retinal organoids was IL-33. Several studies have identified a strong correlation between COVID-19 severity and the levels of IL-33 in the blood serum of patients (Burke et al., 2020; Munitz et al., 2021). In the mature retina, IL-33 is expressed mainly by MG and RPE cells (Xi et al., 2016). The elevated expression and secretion of IL-33 in the rodent retina were shown to be the result of different harmful stimuli such as infection with Toxoplasma gondii, retinal detachment, and light-induced damage (Augustine et al., 2019; Tong and Lu, 2015; Xi et al., 2016). While suggested to have a protective role in a mouse model of retinal detachment (Augustine et al., 2019), IL-33 was shown to pathologically amplify the innate immune response in the rat retina following light-induced damage or RPE disruption, promoting the expression of inflammatory chemokines and cytokines from MGs, the accumulation of myeloid cells in the ONL, and RGC and photoreceptor cell death (Xi et al., 2016).
Thus, our data suggest that SARS-CoV-2 infection of retinal cells results in an inflammatory response of immune system factors such as IL-33 and NLRP1, which are involved in retinal degenerative diseases that cause irreversible blindness. Cytokines that were upregulated in the infected organoids may prove to be worthy candidates for further studies regarding COVID-19 retinal pathology.
Comparison of the DE genes in retinal organoids and choroid plexus organoids identified the enrichment of TGF-β response genes among genes that were induced in the retinal organoids but not in the choroid plexus organoids. TGF-β signaling plays an important role in maintaining the immune-privileged status of the eye (Masli and Vega, 2011; Zhou and Caspi, 2010). For example, both RPE cells and RGCs suppress the activation of T cells via the secretion of TGF-β molecules (Edo et al., 2020; Sugita et al., 2011). Unlike most of the CNS, the choroid plexus is not an immune-privileged structure (Galea et al., 2007). Thus, differences in the TGF-β response of retinal and choroid plexus organoids to SARS-CoV-2 infection may be related to the immunosuppressive role of TGF-β.
SARS-CoV-2 retinal infection is dependent on ACE2
The relatively low expression of ACE2 in the retina has led researchers to speculate that SARS-CoV-2 retinal infection is dependent upon alternate pathways (de Figueiredo et al., 2020). However, ACE2 protein was identified in human retinas and retinal organoids (Ahmad Mulyadi Lai et al., 2021; Senanayake et al., 2007). Our data indicate that the infection of retinal cells can be significantly reduced by using anti-ACE2 antibodies. Thus, the infection of retinal cells most likely is primarily dependent upon functional ACE2 receptors. The possibility of reducing retinal infection by using ACE2 antibodies may also be helpful for the development of drugs in the treatment of SARS-CoV-2 retinal infection.
To conclude, our data indicate that SARS-CoV-2 can actively infect retinal cells in an ACE2-dependent manner. Further studies should consider the possibility that neuroretinal infection leads to retinal symptoms in patients with COVID-19 and that potential treatment options should perhaps be examined. SARS-CoV-2-dependent photoreceptor and/or RGC degeneration can cause permanent visual impairment or even blindness. While data suggesting vision impairment in patients with COVID-19 are scarce, it should be noted that visual impairment and subsequent blindness from retinal degenerative diseases may become evident only after a long course of progression. Long COVID-related vision impairment or even blindness may occur at a much later time point after an acute SARS-CoV-2 infection. The induction of inflammatory genes that are related to retinal degeneration in the organoids should prompt further investigation of the association between SARS-CoV-2 infection and retinal degenerative diseases.
While our study indicates that SARS-CoV-2 can infect and replicate in retinal cells in organoids, our experimental system has some limitations. Retinal organoids contain some RPE together with neuroretinal tissue but are devoid of other important tissues such as the cornea and the retinal vasculature present in the physiological environment (Achberger et al., 2019). This limits the ability to use the retinal organoid model to study the entry and route of SARS-CoV-2 infection in the retina and the interaction between the retina and other tissues during infection. Moreover, retinal organoids lack specialized immune cells such as microglia, which play an important role in retinal inflammation (Rashid et al., 2019), and cannot be used to fully characterize the retinal inflammatory response to SARS-CoV-2. Retinal organoids also represent the embryonic retina rather than a mature retina, and thus some age-related differences may affect retinal SARS-CoV-2 infection and pathology. However, the fact that SARS-CoV-2 also does infect relatively mature neurons and cause the upregulation of inflammatory genes in our organoids, combined with the growing evidence of retinal involvement in patients with COVID-19, indicates the relevance of the data produced from our retinal organoids and the need for further investigation into COVID-19-related retinal pathologies.
Experimental procedures
Organoid production
H9 hESCs, Gibco episomal iPSCs (line A18945), and IMR(90)-4 iPSCs were grown in feeder-free conditions in Matrigel (Corning)-coated plates in StemFlex medium (Thermo Fisher). To produce organoids, we combined different retinal organoid protocols (Capowski et al., 2019; Kuwahara et al., 2015; Zhong et al., 2014). In short, hESCs or iPSCs were grown until they reached ~80% confluence. At that stage, cell colonies were detached using 0.5 mM EDTA and placed in a costar ultra low attachment 6-well plate (Corning) well in 1 mL mTeSR1 medium (Stem Cell Technologies) with 10 μM blebbistatin (Sigma-Aldrich). Over the next 3 days, the medium was replaced gradually to neural induction medium (NIM) comprising DMEM/F12 (Thermo Fisher) 1:1, 1% N2 (Thermo Fisher), 1× MEM nonessential amino acids (Thermo Fisher), 1× GlutaMAX (Thermo Fisher), and 2 μg/mL heparin (Sigma-Aldrich). On day 6 of differentiation, NIM was replaced and supplemented with 1.5 nM BMP4 (R&D Systems). Half of the media was refreshed every 3 days. On day 16, NIM was replaced with retinal differentiation medium (RDM) comprising DMEM:F12 3:1, 2% B27 (Thermo Fisher), 1× MEM nonessential amino acids, 1× penicillin-streptomycin (Thermo Fisher), and 1× GlutaMAX. RDM was changed twice per week. On day 30 of differentiation, RDM was supplemented with 10% fetal bovine serum (FBS) and 100 μM taurine (Sigma-Aldrich). Between days 40 and 100 of differentiation, RDM was supplemented with 1 μM of all-trans-retinoic acid (Sigma-Aldrich).
Virus production and organoid infection
SARS-CoV-2 was isolated from a patient in South Tyrol (hCoV19/Germany/FI1103201/2020) and identified as type “Ischgl.” The virus sequence is available in GSAID: EPI-ISL_463008.
All SARS-CoV-2-containing experiments were conducted under biosafety level (BSL) 3 conditions. The virus was propagated on VeroE6-TMPRSS2 cells using DMEM (Sigma-Aldrich) supplemented with 2% FBS (Capricorn Scientific), 1% penicillin/streptomycin (Sigma-Aldrich), 1% sodium pyruvate solution (Sigma-Aldrich), 1% nonessential amino acid (NEAA) solution (Sigma-Aldrich), and 1% HEPES solution (Sigma-Aldrich) using an MOI of 0.01. Two days post-infection, the virus titer was determined.
SARS-CoV-2 was diluted to the desired amounts in RDM supplemented with 100 μM taurine (Sigma-Aldrich) without FBS. The organoids were washed twice with PBS and incubated for 1 h at 37 °C with the virus dilutions. Afterward, organoids were washed once with PBS and incubated in RDM with 100 μM taurine for specific time points.
Plaque assay
VeroE6 cells were grown to a confluent monolayer in 6-well plates. A 10-fold dilution series of viral solutions was prepared in PBS (Sigma-Aldrich) containing 1% (v/v) penicillin/streptomycin, 0.6% (v/v) BSA (35%) (Sigma-Aldrich), 0.01% (w/v) CaCl2 (1%), and 0.01% (w/v) MgCl2 (1%) from the medium in which the organoids were incubated. The VeroE6 cells were incubated with the dilution series for 1 h at 37 °C. Afterward, the inoculum was replaced by plaque medium: 63% (v/v) 2× MEM 20% (v/v), 10× MEM (Gibco), 3.2% (v/v) NaHCO3 (Lonza), 2% (v/v) HEPES (1 M; pH 7.2) (Sigma-Aldrich), 1.2% (v/v) BSA (35%), 1% (v/v) 100× penicillin/streptomycin/l-glutamine solution (10,000 U/mL penicillin; 10,000 μg/mL streptomycin, 29.2 mg/mL l-glutamine) (Gibco), 2% (v/v) FBS, and 35% (v/v) Agar (2%) (Oxoid). After 24–96 h, the plaques were counted.
Immunohistochemistry
Organoids were fixed in 4% paraformaldehyde (PFA) for 30 min (organoids used for ACE2 IF were fixed for 2 h), washed 3 times in PBS, and incubated overnight at 4°C in 30% sucrose in PBS. The organoids were then cryopreserved in optimum cutting temperature (O.C.T.; Tissue-Tek) and sectioned to a thickness of 20 μm. The sections were blocked and permeabilized with 10% normal donkey serum (Abcam) and 0.3% Triton X-100 in PBS for 1 h at room temperature (RT), and then incubated with primary antibodies diluted in a blocking medium overnight at 4°C. The primary antibodies used for the study can be found in Table S1. Sections were further washed and incubated with a species-specific secondary fluorescent antibody, with DAPI, for 1 h in the dark at RT and then mounted. Imaging was done using an LSM 780 scanning confocal microscope (Zeiss). All of the images presented are maximal projection images. Cell counting was performed manually on random sections from different organoids, with a similarly sized area.
RNA extraction and cDNA library preparation and RNA-seq
RNA from untreated and infected organoids was extracted with TRIzol reagent according to the manufacturer’s protocol, with minor modifications. Briefly, washed retina organoids were resuspended in 700 μL TRIzol reagent. The tissue was disrupted mechanically by vortexing for 30 s and pipetting 20 times. Chloroform 140 μL was added and samples were mixed vigorously for 15 s. Following centrifugation at 4°C, the aqueous phase was carefully separated and RNA was further precipitated by 350 μL 2-propanol. RNA pellets were washed twice with 75% ethanol and resuspended in diethyl pyrocarbonate (DEPC)-treated water for further library preparation.
Total RNA 600 ng was used for RNA isolation, fragmentation, and cDNA synthesis using the NEBNExt poly(A) mRNA magnetic isolation module (E7490). cDNA libraries were amplified and index labeled using the NEBNext Ultra TM II RNA Library Prep kit for Illumina (E7770, E7775). DNA library quality and concentration were analyzed on an Agilent Bioanalyzer DNA Chip. cDNA samples were loaded on a NextSeq 2000 system (Illumina) at a concentration of 800 pmol, and the sequencing was performed with single-end 100-bp reads.
Bioinformatic analysis
The sequencing data were first demultiplexed using the Illumina software bcl2fastq version 2.20.0. The quality of the resulting FASTQ files was then evaluated with the program FastQC version 0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
To perform the alignment, a customized combination of the human genome and the SARS-CoV-2 viral genome was created. The reference sequence and gene annotation for SARS-CoV-2 were obtained from the NCBI repository: NC_045512.2 “Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome.” The human genome sequence and gene annotation were also obtained from NCBI: GRCh38.p13. Both viral and human genomes were concatenated using a customized bash script to create the alignment reference sequence and annotation files. Finally, the alignment was performed using STAR version 2.7.7a (Dobin et al., 2013), with default parameters and the extra instruction “--quantMode GeneCounts” to create additional gene count tables.
The following secondary analysis was performed in the statistical environment R version 4.0.3 (https://www.r-project.org/). After importing the gene count tables, only genes that had a minimum of 5 reads in at least 3 samples were kept as a quality filter. Then, a dispersion trend among the samples was assessed with a principal-component analysis (PCA) applied over the normalized read counts, transformed through the regularized log transformation (Figure S3A).
Differential gene expression was assessed based on a negative binomial distribution through the package DEseq2 version 1.30.1 (Love et al., 2014). The Benjamini-Hochberg FDR method was used for multiple comparisons correction. Genes were considered DE if they presented an absolute value of FC of >1 and a Benjamini-Hochberg FDR of ≤5%. A very small number of SARS-CoV-2 transcripts was identified in 1 of the 24-h control samples; however, considering the vast difference in scale between the levels of SARS-CoV-2 transcripts in this sample and the levels found in the infected samples (Figure 4A, FC > 200), these transcripts most likely do not represent a SARS-CoV-2 infection. In addition, this sample clustered together with the control samples in a PCA analysis (Figure S3A). Thus, we decided to include this sample in the analysis.
Heatmap representation of the expression of selected genes was created with the package pheatmap version 1.0.12 using the values obtained after applying the regularized log transformation over the raw counts. GO and KEGG pathway analyses were performed by separately collecting all of the upregulated genes with an FC of ≥2 and all of the downregulated genes with an FC of ≤−2 and uploading the gene lists to the Enrichr software (Chen et al., 2013; Kuleshov et al., 2016).
Quantitative PCR (qPCR)
cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using iTaq SYBRGreen Supermix with ROX (Bio-Rad) and a QuantStudio3 machine (Applied Biosystems). Gene expression was normalized according to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated using the delta Ct method. qPCR primers can be found in Table S2.
ACE2 blocking
Retinal organoids on day 90 of differentiation were incubated at RT for 1 h in PBS with either a goat anti-ACE2 antibody (AF933, R&D Systems) or a normal goat IgG control (AB-108-C, R&D Systems), at a concentration of 100 μg/mL before being infected with SARS-CoV-2.
Statistical analysis
Statistical analysis was done using GraphPad Prism 9. An analysis of experiments with multiple groups was done by a 1-way ANOVA with Tukey’s multiple comparison test. Statistical analysis of experiments comparing 2 groups was done using an unpaired, 2-tailed Student’s t test. No statistical tests were used to predetermine the sample size.
Data availability
Raw and processed RNA-seq data were uploaded to GEO and can be found in GEO: GSE174843.
Author contributions
Y.M.-L., A.S., S.L., T.R., and H.R.S. conceived the study. Y.M.-L. and T.R. wrote the manuscript and prepared the figures. Y.M.-L., A.S., A.M.-Z., L.B., and O.E.P. performed the experiments. A.L. analyzed the data. All of the authors edited the manuscript.
Acknowledgments
This research was supported by the Max Planck Society’s White Paper-Project: Brain Organoids: Alternatives to Animal Testing in Neuroscience. Further financial support was received from the German Research Foundation (DFG), grants SFB1009 B13 and CRU342 P6; the German Ministry of Education and Research (BMBF); project OrganoStrat (01KX2021); SFB944 (Z-Project); iBiOs (PI 405/14-1); and the IZKF, grant Bru2/015/19. We thank Ingrid Gelker and Manuela Haustein for their technical help. We also thank the Core Facility Genomik in the medical faculty in the Münster team for RNA-seq and Areti Malapetsas for editing the manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.02.015.
Supplemental information
References
- Achberger K., Haderspeck J.C., Kleger A., Liebau S. Stem cell-based retina models. Adv. Drug Deliver Rev. 2019;140:33–50. [Abstract] [Google Scholar]
- Ahmad Mulyadi Lai H.I., Chou S.J., Chien Y., Tsai P.H., Chien C.S., Hsu C.C., Jheng Y.C., Wang M.L., Chiou S.H., Chou Y.B., et al. Expression of endogenous angiotensin-converting enzyme 2 in human induced pluripotent stem cell-derived retinal organoids. Int. J. Mol. Sci. 2021;22:1320. [Europe PMC free article] [Abstract] [Google Scholar]
- Asadi-Pooya A.A., Simani L. Central nervous system manifestations of COVID-19: a systematic review. J. Neurol. Sci. 2020;413:116832. [Europe PMC free article] [Abstract] [Google Scholar]
- Augustine J., Pavlou S., Ali I., Harkin K., Ozaki E., Campbell M., Stitt A.W., Xu H., Chen M. IL-33 deficiency causes persistent inflammation and severe neurodegeneration in retinal detachment. J. Neuroinflammation. 2019;16:251. [Europe PMC free article] [Abstract] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e39. [Europe PMC free article] [Abstract] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. [Europe PMC free article] [Abstract] [Google Scholar]
- Burgos-Blasco B., Guemes-Villahoz N., Donate-Lopez J., Vidal-Villegas B., Garcia-Feijoo J. Optic nerve analysis in COVID-19 patients. J. Med. Virol. 2020;93:190–191. [Europe PMC free article] [Abstract] [Google Scholar]
- Burke H., Freeman A., Cellura D.C., Stuart B.L., Brendish N.J., Poole S., Borca F., Phan H.T.T., Sheard N., Williams S., et al. Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020;21:245. [Europe PMC free article] [Abstract] [Google Scholar]
- Butler D., Mozsary C., Meydan C., Foox J., Rosiene J., Shaiber A., Danko D., Afshinnekoo E., MacKay M., Sedlazeck F.J., et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat. Commun. 2021;12:1660. [Europe PMC free article] [Abstract] [Google Scholar]
- Capowski E.E., Samimi K., Mayerl S.J., Phillips M.J., Pinilla I., Howden S.E., Saha J., Jansen A.D., Edwards K.L., Jager L.D., et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146:dev171686. [Europe PMC free article] [Abstract] [Google Scholar]
- Casagrande M., Fitzek A., Puschel K., Aleshcheva G., Schultheiss H.P., Berneking L., Spitzer M.S., Schultheiss M. Detection of SARS-CoV-2 in human retinal biopsies of deceased COVID-19 patients. Ocul. Immunol. Inflamm. 2020;28:721–725. [Abstract] [Google Scholar]
- Casagrande M., Fitzek A., Spitzer M., Puschel K., Glatzel M., Krasemann S., Aepfelbacher M., Norz D., Lutgehetmann M., Pfefferle S., et al. Detection of SARS-CoV-2 genomic and subgenomic RNA in retina and optic nerve of patients with COVID-19. Br. J. Ophthalmol. 2021 10.1136/bjophthalmol-2020-318618. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [Europe PMC free article] [Abstract] [Google Scholar]
- Conrady C.D., Faia L.J., Gregg K.S., Rao R.C. Coronavirus-19-Associated retinopathy. Ocul. Immunol. Inflamm. 2021;29:675–676. [Europe PMC free article] [Abstract] [Google Scholar]
- de Figueiredo C.S., Raony I., Giestal-de-Araujo E. SARS-CoV-2 targeting the retina: host-virus interaction and possible mechanisms of viral tropism. Ocul. Immunol. Inflamm. 2020;28:1301–1304. [Abstract] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [Europe PMC free article] [Abstract] [Google Scholar]
- Edo A., Sugita S., Futatsugi Y., Sho J., Onishi A., Kiuchi Y., Takahashi M. Capacity of retinal ganglion cells derived from human induced pluripotent stem cells to suppress T-cells. Int. J. Mol. Sci. 2020;21:7831. [Europe PMC free article] [Abstract] [Google Scholar]
- Eriksen A.Z., Moller R., Makovoz B., Uhl S.A., tenOever B.R., Blenkinsop T.A. SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium. Cell Stem Cell. 2021;28:1205–1220.e7. [Europe PMC free article] [Abstract] [Google Scholar]
- Galea I., Bechmann I., Perry V.H. What is immune privilege (not)? Trends Immunol. 2007;28:12–18. [Abstract] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [Europe PMC free article] [Abstract] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M., et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27:937–950.e39. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015;6:6286. [Abstract] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Liddelow S.A. Development of the choroid plexus and blood-CSF barrier. Front. Neurosci. 2015;9:32. [Europe PMC free article] [Abstract] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [Europe PMC free article] [Abstract] [Google Scholar]
- Mabillard H., Sayer J.A. Electrolyte disturbances in SARS-CoV-2 infection. F1000Res. 2020;9:587. [Europe PMC free article] [Abstract] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. [Europe PMC free article] [Abstract] [Google Scholar]
- Marinho P.M., Marcos A.A.A., Romano A.C., Nascimento H., Belfort R., Jr. Retinal findings in patients with COVID-19. Lancet. 2020;395:1610. [Europe PMC free article] [Abstract] [Google Scholar]
- Masli S., Vega J.L. Ocular immune privilege sites. Methods Mol. Biol. 2011;677:449–458. [Abstract] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. [Europe PMC free article] [Abstract] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. [Europe PMC free article] [Abstract] [Google Scholar]
- Munitz A., Edry-Botzer L., Itan M., Tur-Kaspa R., Dicker D., Marcoviciu D., Goren M.G., Mor M., Lev S., Gottesman T., et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci. Rep. 2021;11:3461. [Europe PMC free article] [Abstract] [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. [Abstract] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27:951–961.e5. [Europe PMC free article] [Abstract] [Google Scholar]
- Pereira L.A., Soares L.C.M., Nascimento P.A., Cirillo L.R.N., Sakuma H.T., Veiga G.L.D., Fonseca F.L.A., Lima V.L., Abucham-Neto J.Z. Retinal findings in hospitalised patients with severe COVID-19. Br. J. Ophthalmol. 2020;106:102–105. [Abstract] [Google Scholar]
- Pirraglia M.P., Ceccarelli G., Cerini A., Visioli G., d'Ettorre G., Mastroianni C.M., Pugliese F., Lambiase A., Gharbiya M. Retinal involvement and ocular findings in COVID-19 pneumonia patients. Sci. Rep. 2020;10:17419. [Europe PMC free article] [Abstract] [Google Scholar]
- Ramani A., Muller L., Niklas Ostermann P., Gabriel E., Abida-Islam P., Muller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A., et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39 [Europe PMC free article] [Abstract] [Google Scholar]
- Rashid K., Akhtar-Schaefer I., Langmann T. Microglia in retinal degeneration. Front. Immunol. 2019;10:1975. [Europe PMC free article] [Abstract] [Google Scholar]
- Rodriguez-Rodriguez M.S., Romero-Castro R.M., Alvarado-de la Barrera C., Gonzalez-Cannata M.G., Garcia-Morales A.K., Avila-Rios S. Optic neuritis following SARS-CoV-2 infection. J. Neurovirol. 2021;27:359–363. [Europe PMC free article] [Abstract] [Google Scholar]
- Senanayake P., Drazba J., Shadrach K., Milsted A., Rungger-Brandle E., Nishiyama K., Miura S., Karnik S., Sears J.E., Hollyfield J.G. Angiotensin II and its receptor subtypes in the human retina. Invest. Ophthalmol. Vis. Sci. 2007;48:3301–3311. [Abstract] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.E., Liu F., Dai Y., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218 [Europe PMC free article] [Abstract] [Google Scholar]
- Sridhar A., Hoshino A., Finkbeiner C.R., Chitsazan A., Dai L., Haugan A.K., Eschenbacher K.M., Jackson D.L., Trapnell C., Bermingham-McDonogh O., et al. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 2020;30:1644–1659.e44. [Europe PMC free article] [Abstract] [Google Scholar]
- Sugita S., Horie S., Yamada Y., Kawazoe Y., Takase H., Mochizuki M. Suppression of interleukin-17-producing T-helper 17 cells by retinal pigment epithelial cells. Jpn. J. Ophthalmol. 2011;55:565–575. [Abstract] [Google Scholar]
- Sun N., Cassell M.D., Perlman S. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J. Virol. 1996;70:5405–5413. [Europe PMC free article] [Abstract] [Google Scholar]
- Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. [Europe PMC free article] [Abstract] [Google Scholar]
- Tikellis C., Johnston C.I., Forbes J.M., Burns W.C., Thomas M.C., Lew R.A., Yarski M., Smith A.I., Cooper M.E. Identification of angiotensin converting enzyme 2 in the rodent retina. Curr. Eye Res. 2004;29:419–427. [Abstract] [Google Scholar]
- Tong X., Lu F. IL-33/ST2 involves the immunopathology of ocular toxoplasmosis in murine model. Parasitol. Res. 2015;114:1897–1905. [Abstract] [Google Scholar]
- Trifilo M.J., Montalto-Morrison C., Stiles L.N., Hurst K.R., Hardison J.L., Manning J.E., Masters P.S., Lane T.E. CXC chemokine ligand 10 controls viral infection in the central nervous system: evidence for a role in innate immune response through recruitment and activation of natural killer cells. J. Virol. 2004;78:585–594. [Europe PMC free article] [Abstract] [Google Scholar]
- Virgo J., Mohamed M. Paracentral acute middle maculopathy and acute macular neuroretinopathy following SARS-CoV-2 infection. Eye (Lond) 2020;34:2352–2353. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu P., Duan F., Luo C., Liu Q., Qu X., Liang L., Wu K. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in hubei province, China. JAMA Ophthalmol. 2020;138:575–578. [Europe PMC free article] [Abstract] [Google Scholar]
- Xi H., Katschke K.J., Jr., Li Y., Truong T., Lee W.P., Diehl L., Rangell L., Tao J., Arceo R., Eastham-Anderson J., et al. IL-33 amplifies an innate immune response in the degenerating retina. J. Exp. Med. 2016;213:189–207. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu L.H., Huang M., Fang S.G., Liu D.X. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase delta. J. Biol. Chem. 2011;286:39546–39559. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e27. [Europe PMC free article] [Abstract] [Google Scholar]
- Yerramothu P., Vijay A.K., Willcox M.D.P. Inflammasomes, the eye and anti-inflammasome therapy. Eye (Lond) 2018;32:491–505. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T., et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S., et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou B., Liu J., Wang Q., Liu X., Li X., Li P., Ma Q., Cao C. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1alpha. J. Virol. 2008;82:6962–6971. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou R., Caspi R.R. Ocular immune privilege. F1000 Biol. Rep. 2010;2:3. [Europe PMC free article] [Abstract] [Google Scholar]
- Zizzo G., Cohen P.L. Imperfect storm: is interleukin-33 the Achilles heel of COVID-19? Lancet Rheumatol. 2020;2:e779–e790. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Stem Cell Reports are provided here courtesy of Elsevier
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.stemcr.2022.02.015
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S2213671122001047/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/125311498
Article citations
Ocular A-to-I RNA editing signatures associated with SARS-CoV-2 infection.
BMC Genomics, 25(1):431, 01 May 2024
Cited by: 1 article | PMID: 38693480 | PMCID: PMC11061923
Optical Coherence Tomography Angiography Assessment of the Optic Nerve Head in Patients Hospitalized Due to COVID-19 Bilateral Pneumonia.
Medicina (Kaunas), 60(3):502, 19 Mar 2024
Cited by: 1 article | PMID: 38541228 | PMCID: PMC10972053
SARS-CoV-2 infects cells lining the blood-retinal barrier and induces a hyperinflammatory immune response in the retina via systemic exposure.
PLoS Pathog, 20(4):e1012156, 10 Apr 2024
Cited by: 1 article | PMID: 38598560 | PMCID: PMC11034659
Looking at the Data on Smoking and Post-COVID-19 Syndrome-A Literature Review.
J Pers Med, 14(1):97, 16 Jan 2024
Cited by: 1 article | PMID: 38248798 | PMCID: PMC10821354
Review Free full text in Europe PMC
SARS-CoV-2 neurovascular invasion supported by Mendelian randomization.
J Transl Med, 22(1):101, 24 Jan 2024
Cited by: 0 articles | PMID: 38268029 | PMCID: PMC10809787
Go to all (16) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE174843
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NC_045512.2
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids.
Int J Mol Sci, 22(3):1320, 28 Jan 2021
Cited by: 22 articles | PMID: 33525682 | PMCID: PMC7865454
SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids.
Cell Stem Cell, 29(2):217-231.e8, 25 Dec 2021
Cited by: 127 articles | PMID: 35032430 | PMCID: PMC8709832
Single-cell RNA sequencing analysis of SARS-CoV-2 entry receptors in human organoids.
J Cell Physiol, 236(4):2950-2958, 17 Sep 2020
Cited by: 16 articles | PMID: 32944935 | PMCID: PMC7537521
SARS-CoV-2 research using human pluripotent stem cells and organoids.
Stem Cells Transl Med, 10(11):1491-1499, 24 Jul 2021
Cited by: 12 articles | PMID: 34302450 | PMCID: PMC8550698
Review Free full text in Europe PMC




