Abstract
Free full text

Frequent somatic gene conversion as a mechanism for loss of heterozygosity in tumor suppressor genes
Abstract
The major processes in carcinogenesis include the inactivation of tumor-suppressor genes (TSGs). Although Knudson's two-hit model requires two independent inactivating mutations, perhaps more frequently, a TSG inactivation can occur through a loss of heterozygosity (LOH) of an inactivating mutation. Deletion and uniparental disomy (UPD) have been well documented as LOH mechanisms, but the role of gene conversion is poorly understood. Here, we developed a simple algorithm to detect somatic gene conversion from short-read sequencing data. We applied it to 6285 cancer patient samples, from which 4978 somatic mutations that underwent gene conversion to achieve LOH were found. This number accounted for 14.8% of the total LOH mutations. We further showed that LOH by gene conversion was enriched in TSGs compared with non-TSG genes, showing a significant contribution of gene conversion to carcinogenesis.
Knudson's two-hit theory (Knudson 1971) describes the major process for carcinogenesis through the inactivation of tumor-suppressor genes (TSGs). For TSG function to be lost, both the paternal and maternal alleles must be inactivated: The first hit (mutation) inactivates one allele (Fig. 1A), and then the other allele is inactivated by another independent mutation, or the second hit (Fig. 1B). Perhaps more frequently, the TSG function could be lost through a loss of heterozygosity (LOH) after the first hit (Michor et al. 2004, 2005; Nowak et al. 2004). Figure 1, C through E, illustrates three mechanisms for LOH. First, deletion: LOH is achieved if the active allele is lost by a loss of the entire chromosome or a large deletion of the chromosome (Fig. 1C). In cancer cells, the copy number of a gene frequently changes through duplication and deletion of chromosome (or chromosomal region), some of which are recognized as an abnormal karyotype. Second, uniparental disomy (UPD): UPD could arise through a somatic cell division such that a daughter cell receives two copies of one chromosome from its parental cell, resulting in a homozygote for the entire chromosome (Andersen et al. 2007; Tuna et al. 2009). Note that UPD does not change the karyotype, which is different from the case of deletion. LOH occurs if the functional allele is lost and the inactivated allele is doubled (Fig. 1D). Third, gene conversion: Somatic gene conversion has a similar UPD outcome, except that gene conversion only affects the chromosome locally; generally, the gene conversion tract length may be ~200–1000 bp (Chen et al. 2007). If the inactivated allele is unidirectionally transferred by a double-strand break (DSB)–induced gene conversion in a somatic cell division, gene conversion works as a mechanism for LOH (Fig. 1E).
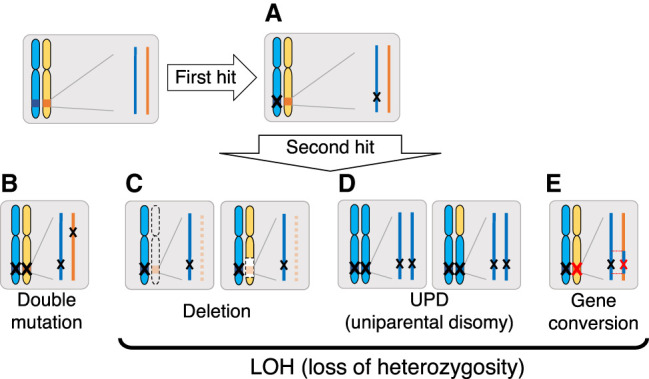
Possible scenarios for losing gene function. Start with two functional genes (deep blue and orange boxes) on the paternal and maternal chromosomes. The genic region is enlarged on the right. (A) A first mutation (black X) inactivates one of the active alleles, the paternal allele in this figure. (B) Double mutation: If a second mutation (black X) inactivates the maternal allele independently, the gene's function is completely lost. Additionally, loss of function can be achieved through LOH, and three major mechanisms for LOH are illustrated. (C) Deletion: The active allele is deleted by a chromosome loss (left) or a deletion of a chromosomal region (right). (D) Uniparental disomy (UPD): UPD creates a homozygote of the inactivated allele when only a chromosome (either the whole chromosome [left] or chromosome arm [right]) is inherited and doubled. (E) Gene conversion: Gene conversion transfers the first inactivating mutation to the active copy (red X), resulting in a homozygote for the inactivating mutation.
The roles of deletion and UPD have been well investigated (Rajagopalan et al. 2003; Sieber et al. 2003; Raghavan et al. 2005; Stark and Hayward 2007; Tuna et al. 2009; Zack et al. 2013), whereas there is very little documentation of gene conversion, except for some cases in which somatic gene conversion of pathogenic germline variants causes LOH (Zhang et al. 2006; Auclair et al. 2007). To our best knowledge, there is no genome-wide documentation of gene conversion to quantify its relative contribution to LOH. We here show a comprehensive survey of somatic mutations that achieved LOH (hereafter, referred to as SMLOH) in thousands of cancer genomes and discuss its potential role in carcinogenesis.
Results
Detecting somatic gene conversion
This work aims to identify somatic mutation in cancer genomes that underwent somatic gene conversion to achieve LOH (SMLOH,Conv). This work defined somatic mutations to include single-nucleotide variants (SNVs) and small indels (i.e., SNVs exclusively mean base substitutions). We used the whole-exome sequences (WXSs) data of 3,349,768 somatic mutations in 32 cancer types in 9482 patient samples in The Cancer Genome Atlas (TCGA; see Methods) (for details, see Supplemental Table S1). The coverage exceeded 10 reads for >99% of these mutations (average, 79.5). This fairly high somatic mutation quality allowed us to perform the following statistical analyses.
We developed a simple algorithm to detect gene conversion, as described in Figure 2. α1 and α2 represent the copy numbers of the paternal and maternal alleles. Figure 2 illustrates hypothetical short-read data aligned on the reference sequence, where both germline variants and somatic mutations were observed. In a region of (α1, α2) = (1, 1) (Fig. 2A), most germline variants (black bars) are heterozygote and so are somatic mutations (black X). Exceptions include one germline variant and one somatic mutation that are located adjacent to each other around the center of the illustrated region in Figure 2A. At both sites, all reads have these mutations, indicating that the two mutations on the paternal chromosome (black bar and X) were transferred to the maternal chromosome (red bar and X) by gene conversion (boxed by the red dotted line). In this work, we focused on regions of (α1, α2) = (1, 1) to detect gene conversion (see Fig. 1E), whereas regions of (α1, α2) = (1, 0) and (2, 0) were used as a comparison, representing the other two mechanisms for LOH: deletion and UPD (Fig. 2B,C).
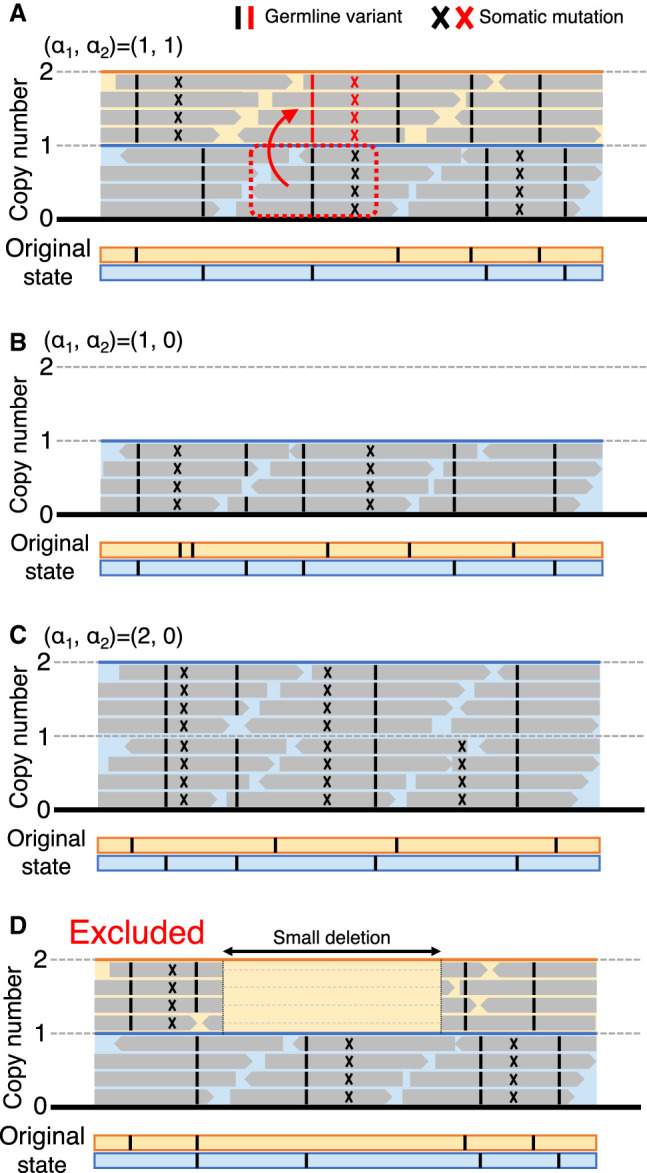
Illustrations of short-read sequencing data mapped on the reference sequence. The original state before somatic mutations occur is presented at the bottom using the blue and orange lines, representing the paternal and maternal chromosomes, where only germline variants are present (black bars). Somatic mutations can be detected in the short-read data, presented by X. (A) A hypothetical case of (α1, α2) = (1, 1), where a gene conversion event from the paternal to the maternal chromosomes occurred (boxed by the red dotted line). The mutated sites transferred by gene conversion are shown in red. (B,C) Cases of (α1, α2) = (1, 0) and (2, 0). (D) Cases with a small deletion within a region of (1, 1) that potentially causes false evidence for gene conversion (excluded from the analyses).
For estimating (α1, α2), the ASCAT software (Van Loo et al. 2010) was run to identify copy number changes using Affymetrix SNP6 genotyping arrays (downloaded from https://portal.gdc.cancer.gov/legacy-archive/) for each patient. ASCAT provides copy number estimates in integers for both paternal and maternal alleles (α1 and α2) at each marker SNP on the genotyping array. Note that ASCAT does not specify which is maternal or paternal; rather α1 and α2 are given such that α1 ≥ α2. We first screened for patient samples with estimated purity > 0.7 to secure the quality of our analysis, resulting in 6861 patient samples (see Methods) (Supplemental Table S2). Screening was then conducted based on the copy numbers of alleles. For all somatic mutations, (α1, α2) were obtained using ASCAT. We first screened out patient samples for which the estimates of α1 and α2 for the entire genome might be unreliable. We suspected such patients would show significant inconsistency between the ASCAT result (α1, α2) and sequence coverage (β, an estimate of the copy number based on coverage data). Based on this inconsistency, 564 patients were excluded, and 6296 patient samples remained (see Methods) (for details, see Supplemental Table S2). In each patient sample, local regions with extensive copy number changes were further excluded by focusing on the inconsistency between α and β (Methods). This screening excluded regions with presumably incorrect copy number estimates. Furthermore, our careful inspection successfully detected fairly short indels that could cause a serious problem in our analyses. As illustrated in Figure 2D, such a small deletion in a region of (1, 1), if ignored, could produce false evidence for gene conversion; therefore, such regions were excluded. After these screening processes, 1,875,968 somatic mutations (6285 patient samples) remained. We then classified them into three categories, (α1, α2) = (1, 0), (2, 0), and (1, 1), and others with α1 + α2 > 2 were excluded in the following analyses. The average lengths of the (1, 0), (2, 0), and (1, 1) regions per patient were 205, 224, and 1439 Mbp, respectively (Table 1). We obtained approximately 1.33 million somatic mutations (1,134,589 SNVs and 191,738 indels) in regions of (1, 1), and 34,479 and 66,612 somatic mutations in regions of (1, 0) and (2, 0), respectively (Supplemental Table S2).
Table 1.
Summary of the number of SMLOH
Using these data, we searched for gene conversions in regions with no copy number alternations (i.e., SMLOH,Conv). In a region of (1, 1), we typically observe germline variants in heterozygote (Fig. 2A); that is, VAF′ is ~50%, where VAF′ represents the variant allele fraction considering purity (see Methods). Somatic mutations usually arise in heterozygote (Fig. 2A, black X), and only when it undergoes gene conversion, VAF′ could increase up to around one (Fig. 2A, red X). To detect SMLOH,Conv, we searched for somatic mutations with two conditions: (1) VAF′ was larger than 0.8 because we were uninterested in somatic mutations in low frequencies; (2) The possibility of a heterozygote state was statistically ruled out (P < 0.001, one-tailed binomial test). (Because condition 1 about VAF′ alone increased the number of false positives for low coverages, condition 2 was used to eliminate the low coverage mutations [for details, see Supplemental Note]). We found that 4978 (0.38%) of the 1.33 million somatic mutations satisfied these two conditions, showing strong evidence for gene conversion (Table 1; Supplemental Table S3). This rate (i.e., 0.38%) can be considered an estimate of the rate at which a site experiences somatic gene conversion throughout life, which is hereafter called the per-site gene conversion rate. This estimate is conservative because our strategy cannot detect a very recent gene conversion in a low frequency. One might think we erroneously identified SMLOH,Conv in rearrangement-hot chromosomes (e.g., those that underwent chromothripsis). We confirmed that SMLOH,Conv was not enriched in such chromosomes (see Supplemental Note).
More convincing evidence for gene conversion could be obtained by looking at the linkage to germline variants in the surrounding region. As illustrated in Figure 3, A through C, we consider a two-locus system with sites-S and -G, the former corresponds to the focal site of somatic mutation and the latter is a linked site at which a germline variant is present in the heterozygote. Originally (before the somatic mutation arises), there are two haplotypes, 0-0 and 0-1, where the left and right numbers represent the alleles at sites-S and -G, respectively (Fig. 3A). Suppose allele 1 (somatic mutation) arises at site-S on haplotype 0-0, resulting in haplotype 1-0. At this moment, the somatic mutation is linked to only one allele at site-G (Fig. 3B, allele 0). Then, if this somatic mutation is transferred to the other chromosome by gene conversion, a new haplotype 1-1 arises (Fig. 3C), resulting in a situation in which the somatic mutation links to both alleles at site-G. Conversely, the presence of two haplotypes, 1-0 and 1-1, can be considered strong evidence for gene conversion. This line of evidence is very convincing but can only be obtained when these two conditions are satisfied. First, there must be a closely linked germline variant in the heterozygote. Second, the break-point of the gene conversion tract must be located between the two sites; a gene conversion involving the two sites cannot create a new haplotype (Fig. 3D). Our strategy also misses gene conversion from zero to one at the site-S (a transfer in the opposite direction to that in Fig. 3C). A caveat is that haplotype 1-1 that can also arise by somatic gene conversion at site-G (Fig. 3E), potentially resulting in false-positive evidence for gene conversion (if cells with or without gene conversion coexist in the tumor sample, we could observe both 1-0 and 1-1 haplotypes). However, the proportion of such a false positive should be very low if we apply our estimated per-site gene conversion rate (0.38%). The idea of this simple test for gene conversion is similar to Hudson's four-gamete test to detect recombination in polymorphism data (Hudson and Kaplan 1985).
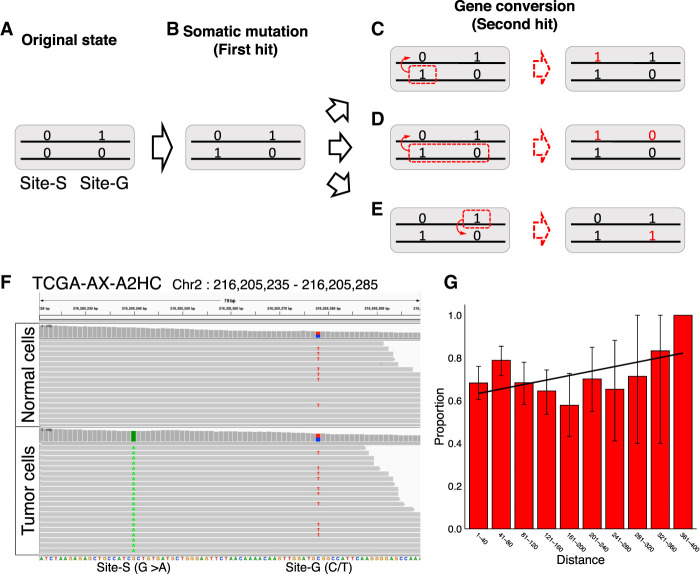
Evidence for gene conversion from a linked germline variant. (A–E) The process of producing clear evidence for gene conversion under a model with two sites, site-S and site-G. Zero and one represent the original and derived alleles. See text for details. (F) An example case (Patient ID, TCGA-AX-A2HC) with strong evidence for gene conversion. Short-reads of normal cells and tumor cells in regions Chr 2: 216,205,235–216,205,285 regions are shown. The figure was made using the Integrative Genomics Viewer (IGV) (Robinson et al. 2011). (G) The proportion of sites with evidence for gene conversion was confirmed by a linked germline variant, as a function of the distance to the germline variant site. The error bars represent the 95% confidence intervals.
Figure 3F shows an example case in which we found a somatic mutation (G > A) that arose in the tumor with VAF′ = 1, but not in the normal cells (Fig. 3F, site-S). Only 38 bp downstream from site-S, there is a site at which C and T segregate with a frequency of ~50% both in the tumor and normal cells (site-G), indicating that this is a germline variant inherited as heterozygote. This case clearly shows that the focal somatic mutation (A at site-S) links to both alleles C and T at site-G (i.e., both haplotypes 1-0 and 1-1 are observed).
As mentioned above, the number of SMLOH,Convs with such convincing evidence may be small. First, the number of somatic mutations with at least five shared reads with an adjacent germline variant was small, only 625 out of the 4978 somatic mutations (Supplemental Table S3). If our simple test was applied to these 625 somatic mutations, it was discovered that 462 (73.9%) mutations showed evidence for gene conversion (i.e., the case illustrated in Figure 3D applies; see Methods). A reason for the remaining 163 sites (26.1%) would be that gene conversion transferred the linked sites simultaneously (see Fig. 3D). To confirm this, we attempted to move away to another germline variant and found a secondary germline variant was available for only 30 of the 163 SMLOH,Convs. For these 30, we examined whether the pattern of Figure 3C holds between SMLOH,Conv and the secondary germline variants, and this was the case for eight of them (26.7%). Altogether, we found strong evidence for gene conversion for 462 + 8 = 470 somatic mutations (470/625 = 75.2%) (Supplemental Table S3). Another line of evidence for the case of Figure 3D was obtained by testing the prediction that the proportion of sites with positive evidence for gene conversion would increase with increasing distance between the two sites. As expected, we found that the proportion of somatic mutations with this evidence for gene conversion correlated positively with the distance (Fig. 3G; Supplemental Table S4), although insignificant (P = 0.112, permutation test).
The rate of somatic gene conversion
The number of SMLOH,Convs substantially varied among patients, from zero to about 226 with average = 0.79 (4978/6285) (Fig. 4A). The majority of the patients (92%) have no SMLOH,Conv, and the distribution has a long tail. We tested whether this variation is owing to the heterogeneity in the gene conversion rate among patients. If the rate is constant across different patients, the number of detected gene conversions should correlate highly with the number of detected somatic mutations. As expected, a strong linear correlation between them was found (Fig. 4B), indicating that the large variation may be well explained by the difference in the number of detected somatic mutations between patients and that the proportion of converted somatic mutations may not vary much between individuals.
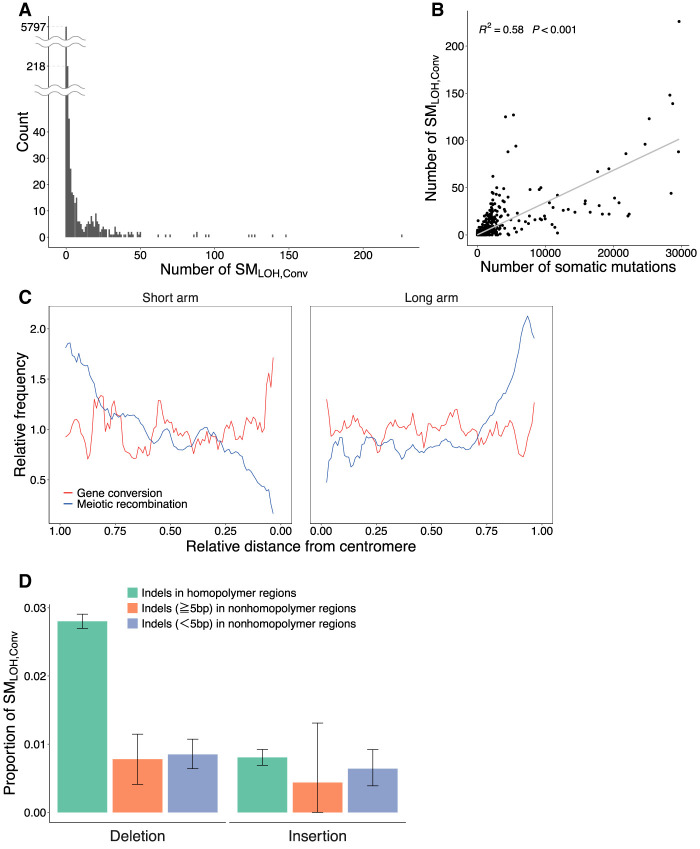
Summary of the observed somatic gene conversion events. (A) Distribution of the number of SMLOH,Convs per patient. (B) Correlation between the total number of somatic mutations and the number of SMLOH,Convs. (C) Distributions of the rates of meiotic recombination and somatic gene conversion along chromosomes. The estimated gene conversion (red line) and the meiotic recombination rates according to the estimates from the HapMap data (blue line) (The International HapMap Consortium 2007) are plotted against the relative distance from the centromere when each chromosome arm is assigned in the interval (0,1). The error bars represent the 95% confidence intervals. The two rates were standardized such that the genome average = 1. (D) Deletion versus insertion bias in SMLOH,Convs. Indels were classified into three categories: indels (≥1 bp) in homopolymer regions, indels with length ≥5 bp in nonhomopolymer regions, and indels with <5 bp in nonhomopolymer regions. Homopolymer regions were defined as those consisting of ≥3 bp of the same nucleotides in a row.
No striking heterogeneity in the gene conversion rate was found across the genome if the spatial distribution was investigated using a 1-Mbp window (Supplemental Fig. S1; Supplemental Table S5). We found 49 windows that had significantly high estimates of the gene conversion rate (FDR < 0.05, one-tailed exact test). We also investigated the effect of gene conversion rate on cancer disease status (i.e., tumor stage, overall survival, and progression free interval) but found no significant results (Supplemental Fig. S2).
A more important factor that might affect the somatic gene conversion rate may be the meiotic recombination (crossing-over) rate in germline cells, which highly varies between local genomic regions in the genome. The rates of meiotic recombination and meiotic gene conversion are highly correlated with each other because they occur through the same mechanism, that is, homologous recombination (Szostak et al. 1983; Ceccaldi et al. 2016). Suppose this also applies to somatic gene conversion. In that case, local heterogeneity in the somatic gene conversion rate along chromosomes is expected because the meiotic recombination rate generally increases with the distance from the centromere in the human genome (Kong et al. 2002; Nachman 2002). To explore this possibility, each chromosome arm (from the centromere to telomere) was assigned to the interval (0,1), and a sliding window analysis was performed using a 0.05 window size. In each window, the local per-site somatic gene conversion rates were computed (as defined above). The average overall chromosomes were plotted along the chromosome (Fig. 4C), with the average meiotic recombination rate (according to the estimates from the HapMap data) (The International HapMap Consortium 2007). The two rates were standardized such that the genome average = 1. We found that the somatic gene conversion rate distribution is almost flat over the chromosomal region, which is quite different from that of the meiotic recombination rate. This difference can be explained as follows. Homologous recombination causes somatic gene conversion and meiotic recombination (Chen et al. 2007; Hunter 2015; Ceccaldi et al. 2016), and DSB initiates homologous recombination. SPO11 mainly induces DSBs in meiosis (Chen et al. 2007; Hunter 2015), whereas extrinsic stress or replication error in mitosis induces somatic DSBs (Tubbs and Nussenzweig 2017). Therefore, the difference in what induces DSB could explain the observation of unclear correlation between the rates of somatic gene conversion and meiotic recombination.
The relative contribution of somatic gene conversion to LOH
We ask “what is the relative contribution of gene conversion to archiving LOH compared with the other two mechanisms, deletion and UPD?” (see Fig. 1C,D). To do so, we compared the number of SMLOH,Convs with SMLOHs owing to deletion and UPD (denoted as SMLOH,Dels and SMLOH,UPDs, respectively). SMLOH,Dels were searched in regions of (1, 0), where LOH is achieved at most somatic mutations except for those present only in local subclones, as illustrated in Figure 2B. We defined SMLOH,Dels as those with VAF′ > 0.8. We searched for SMLOH,UPDs in regions of (2, 0) (Fig. 2C). To exclude SNVs that arose after UPD and appear as heterozygotes, we screened for LOH SNVs as those with VAF′ > 0.8, and the possibility of the heterozygous state was statistically ruled out (P < 0.001, one-tailed binomial test). We found 13,965 SMLOH,Dels (12,835 SNVs, 1130 indels) and 14,682 SMLOH,UPDs (13,453 SNVs, 1229 indels), indicating that 14.8% of the detected SMLOHs is owing to gene conversion (Table 1). Our results show a significant contribution of gene conversion to cause LOH of somatic mutations. It was confirmed that this proportion is robust to the cutoff values of purity and VAF′ in our screening process (for details, see Supplemental Note).
Table 1 shows that indels’ proportions in SMLOH,Conv (2679/4978, 53.8%) are markedly high compared with SNVs (P < 2.2 × 10−16, one-tailed exact test), indicating that indels are more likely involved in gene conversion. This association may be explained by considering the DSB repair system as follows. DSB repair mechanisms, as classical nonhomologous end-joining, single-strand annealing, and alternative end-joining, could be “error prone” and likely induce indels (Ceccaldi et al. 2016; Tubbs and Nussenzweig 2017), although homologous recombination, including gene conversion, is an accurate (“error-free”) repair mechanism (Ceccaldi et al. 2016). If this is true, a hotspot of DSBs can also be considered a hotspot of both indels and gene conversion. The observed association would be predicted if there are several DSB hotspots in the human genome (Durkin and Glover 2007; Tubbs and Nussenzweig 2017).
It was further discovered that the rate of the deletion type of SMLOH,Conv was particularly high when it occurred in homopolymer regions (Fig. 4D). This should not be an artifact owing to mutation call errors that likely occur in homopolymer regions because the insertion type proportion of SMLOH,Conv was not enriched (Fig. 4D). The molecular mechanism behind this observation is unknown.
Contribution of somatic gene conversion to TSG inactivation
We thus showed that many SMLOH,Convs were detected in regions of (1,1), thereby contributing to carcinogenesis, and then how the detected gene conversion potentially contributed to TSG inactivation. We focused on 242 TSGs according to the COSMIC definition (see Methods) (Futreal et al. 2004; Sondka et al. 2018) compared with all remaining genes defined as non-TSGs. If gene conversion-driven LOH contributed much to carcinogenesis, it was predicted that the detected gene conversion involving truncating mutations (STMLOH,Conv) should be enriched in TSGs compared with other non-TSG genes. As expected, we found that the proportion of STMLOH,Convs was significantly larger in TSGs compared with non-TSG genes (P < 2.2 × 10−16, one-tailed exact test) (Fig. 5A; Supplemental Table S6), whereas there were no significant differences in missense and silent mutations. The enrichment of STMLOH was specific to gene conversion in TSGs (Supplemental Fig. S3A). The result indicates that somatic gene conversion of truncating mutation plays a significant role in TSG inactivation, potentially contributing to carcinogenesis.
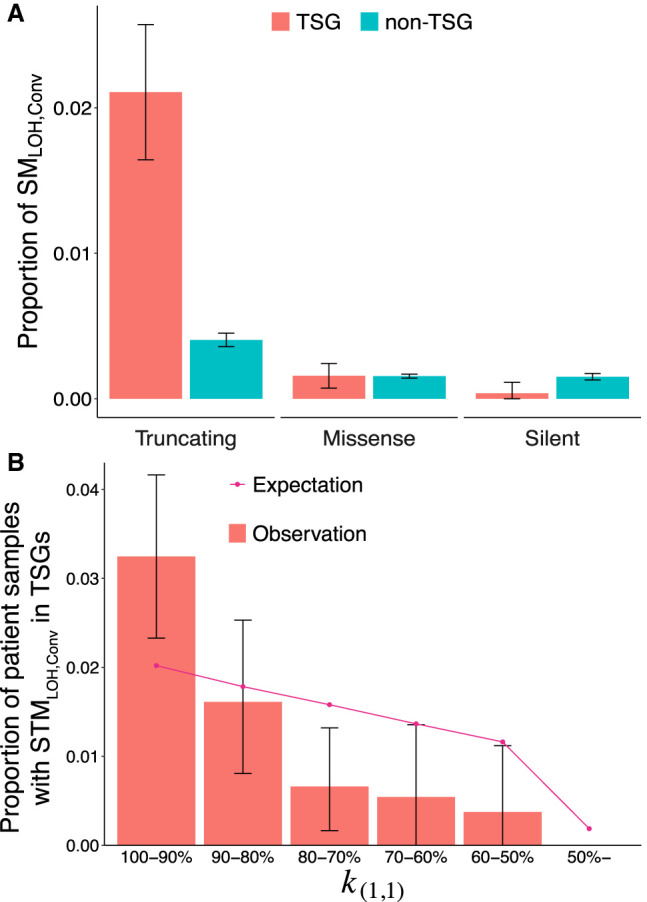
Rate of gene conversion in TSG. (A) Proportions of SMLOH,Convs in TSGs and non-TSGs. The somatic mutations in regions of (α1, α2) = (1, 1) were classified into the TSG (red) and non-TSG (blue) categories, and the proportion of SMLOH,Conv in each category was calculated. (**) P < 0.01, (NS) nonsignificant. (B) The proportion of patient samples with at least one STMLOH,Conv in TSG is plotted for each category of k(1,1), representing the degree of genome instability. The error bars represent the 95% confidence intervals. The pattern in A holds when the data were separated into k(1,1)-high and k(1,1)-low regions (k(1,1) > 90% and the rest) (see Supplemental Fig. S5).
By looking at individual genes, it was found that four cancer-driver genes (listed in COSMIC) (Futreal et al. 2004, Sondka et al. 2018) had significantly high gene conversion rates (FDR < 0.05, one-tailed exact test) (Table 2; for a list of all significant genes, see Supplemental Table S7). We also tested whether gene conversion more likely transfers damaging mutations (truncating and missense mutations). We found that three of the four genes in Table 2 (RNF43, ACVR2A, JAK1) constitute the top three highest proportions of SMLOH,Convs in damaging mutations (Table 2; Supplemental Table S7). RNF43 had the highest gene conversion rate, which plays a major role in colorectal cancer development (Sondka et al. 2018; Tsukiyama et al. 2020). It would be interesting to point out that, compared with cancer types, colon adenocarcinoma (COAD) is a cancer type with a very high gene conversion rate (Supplemental Fig. S3B; Supplemental Table S8), suggesting that gene conversion plays a particularly important role in this cancer type.
Table 2.
Cancer-driver genes with significantly high SMLOH,Conv

From this result, it was proposed that gene conversion should be a prominent mechanism for LOH for patients with no drastic karyotype changes. To test this hypothesis, 6285 patient samples were first categorized according to the degree of genome instability measured by k(1,1), the proportion of the (1,1) region in the genome (Supplemental Fig. S4). A high k(1,1) means that most of the genome consists of the (1,1) region; that is, the chromosomal instability is low. Then, we computed the proportion of patient samples with at least one STMLOH,Conv in TSGs, and the data are binned in Figure 5B. It is found that the proportion is highest in the category of patient samples with k(1,1) > 90% and that it decreases as k(1,1) decreases. Note that this decline could be partly because patient samples with a higher k(1,1) have more chances to have STMLOH,Convs in TSGs. To exclude this effect, we compared the observation with the expectation. The expectation was obtained by assuming that the number of STMLOH,Convs is proportional to k(1,1). It was found that the observed decline is much stronger than the expectation (P = 0.0009, one-tailed χ2 test) (Fig. 5B), supporting our hypothesis that STMLOH,Convs in TSGs play a significant role in tumorigenesis, particularly in genomes with low chromosomal instability.
Discussion
Inactivation of TSGs is crucial to carcinogenesis, which is commonly achieved through LOH (Michor et al. 2004). Deletion and UPD have been well documented as the mechanisms of LOH, partly because they are relatively easy to detect; a large deletion may be detected as an abnormal karyotype, and UPD could be recognized as a chromosome-wide absence of heterozygous sites (Rajagopalan et al. 2003; Sieber et al. 2003; Tuna et al. 2009). In contrast, although having a similar effect to UPD, gene conversion is difficult to detect because it affects only a short region. In this work, as a mechanism of LOH, we hypothesized that somatic gene conversion could be as important as deletion and UPD. To test this hypothesis, a simple algorithm to detect somatic gene conversion from short-read data was developed. By applying it to 6285 patient samples, 4978 somatic mutations were found in which LOH is most likely achieved through gene conversion (i.e., SMLOH,Conv). This number is large enough to account for 14.8% of the total SMLOHs, which could be a conservative number because our method can detect gene conversion that occurred only after somatic mutation arose, and gene conversions that erased somatic mutations are also undetectable. Our results further show an important role of somatic gene conversion in cancer cell development by showing the enrichment of LOH somatic mutations through gene conversion in TSGs compared with non-TSG genes. It is suggested that gene conversion could play a significant role in carcinogenesis and that it is important to pay more attention to the variant allele frequencies of somatic mutations in (1,1) regions to fully understand the genome evolution leading to cancer.
It is considered that chromosomal instability plays a crucial role in carcinogenesis (Pino and Chung 2010). Chromosomal instability causes an imbalance in chromosome number (aneuploidy) and an increased LOH rate, thereby accelerating the TSG inactivation rate. Indeed, many patients have drastic karyotype changes in their genomes. Alternatively, there are some cancer patients without drastic karyotype changes (Li et al. 2020). One might think that, in such a patient, a TSG must acquire two independent mutations to lose its function as Knudson's two-hit theory (Knudson 1971) describes (Fig. 1B). However, we showed that somatic gene conversion could provide a major route for LOH of somatic mutation in TSGs in normal diploid regions (i.e., regions of (1, 1)), potentially accounting for a significant proportion of chromosomal instability–negative tumors. We emphasize the role of somatic gene conversion in carcinogenesis, especially for chromosomal instability–negative patients, and propose that somatic gene conversion may be a major mechanism of LOH.
It should be noted that our method is designed to be quite conservative so as not to catch too many false positives. One of the major reasons is that our method requires a large number of reads to detect statistically reliable gene conversion. As a consequence, our estimate can detect gene conversion that occurred in the early stages of cancer development, and it is difficult to identify young gene conversions that are present in low frequencies. Therefore, our estimate of the somatic gene conversion rate should be underestimated.
LOH of somatic mutations in copy number–neutral (1,1) regions, which we defined as SMLOH,Conv, is also caused by biallelic parallel mutations, that is, multiple mutations that occur to the same position. Demeulemeester et al. (2022) pointed out that biallelic mutation (i.e., “two alleles independently mutate to the same alternate bases” according to the investigators’ definition) should play a role to create LOH, so that the square of the mutation rate would predict the number of SMLOHs caused by biallelic mutations. To take this into account, we computed the expected number of biallelic SNVs based on the mutation rate considering mutation rate variation owing to the trinucleotide-based mutational spectrum. Figure 6 shows the expected number of biallelic mutations and the number of SMLOHs observed in most mutated 300 patient samples (see also Supplemental Table S9). The number of SMLOH,Convs we detected per patient sample is much larger than the expectation owing to biallelic mutations, which is overall <10% of SMLOH,Conv (Supplemental Table S9). This result suggests a significant role of somatic gene conversion, although a part of the SMLOH,Convs may be explained by biallelic mutations.
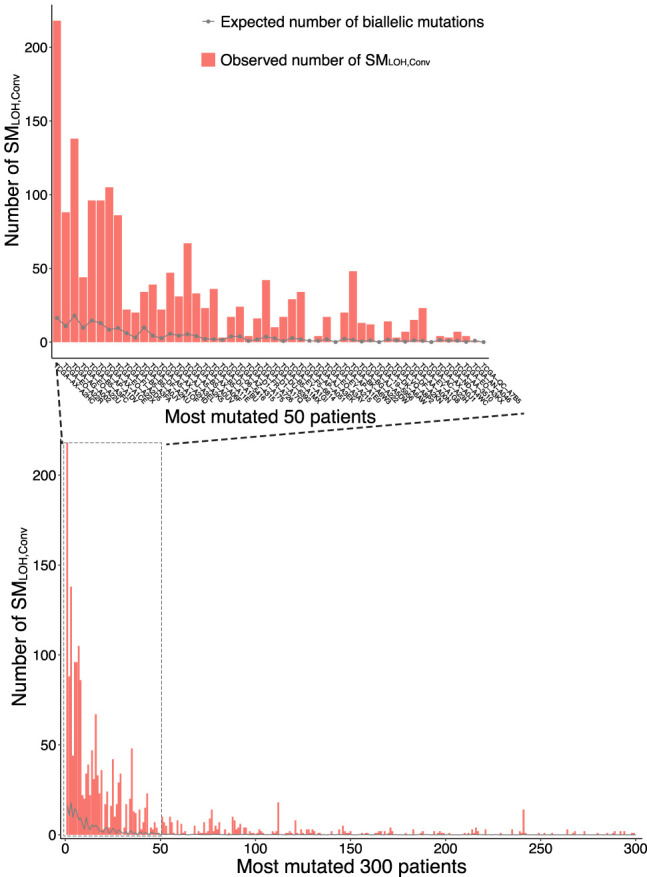
Potential contribution of biallelic parallel mutation to SMLOHs for patients in the top 300 most SMLOHs. The inner panel is a close-up for the top 50 patients.
According to Demeulemeester et al. (2022), the observed SMLOH was well explained by the square of the mutation rate, at least in some patients. But this result does not rule out the contribution of gene conversion completely, especially when LOH of germline variants owing to gene conversion was repeatedly reported (Zhang et al. 2006; Auclair et al. 2007), which was also detected by our method (see Supplemental Fig. S6). A potential reason for the inconsistency between our result and that of Demeulemeester et al. (2022) could be in the statistical power owing to the read coverage, which is roughly twice higher in the WXS we analyzed than that of the whole-genome sequence data (The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium 2020) used by Demeulemeester et al. (2022). The quantitative evaluation of the relative contribution of gene conversion and parallel mutation would be subject to further investigation.
It is interesting to point out that homologous recombination is a DSB repair mechanism and that homologous recombination deficiency promotes carcinogenesis (Moynahan and Jasin 2010; Ceccaldi et al. 2016; Polak et al. 2017; Nguyen et al. 2020). In contrast, as homologous recombination causes gene conversion, the presence of homologous recombination could also contribute to carcinogenesis through gene conversion. Thus, we suggest that homologous recombination should have two roles that counteract in terms of cancer development.
Our documentation of somatic gene conversion might contribute to our understanding of DSBs. It has been very difficult to detect DSBs: It is feasible only when DSBs cause structural changes (Hu et al. 2016; Wei et al. 2016, Li et al. 2020). In this study, we comprehensively identified somatic gene conversions in short-read data. As gene conversion results from DSB repairs in somatic cell division, our results could be considered footprint of DSBs. If so, we identified, on average, 306,000 footprints of DSBs per patient, which is several times larger than the number of DSBs detectable from structural changes. We suggest that detecting gene conversion should be an efficient method to understand how often and where DSBs occur in the genome.
Methods
Screening for somatic mutations
Thirty-two cancer types in the 33 types (excluding LAML) defined in TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) were investigated. LAML was excluded because purity could not be estimated for this type of blood cancer. Mutation annotation format (MAF) files for the 32 cancer types were downloaded, consisting of already extracted and annotated somatic mutations from WXSs per patient (downloaded from https://portal.gdc.cancer.gov/). The MAF files include somatic mutations identified using four different software programs, MuTect2 (Cibulskis et al. 2013), VarScan 2 (Koboldt et al. 2012), MuSE (Fan et al. 2016), and SomaticSniper (Larson et al. 2012), and we used somatic mutations whose quality was guaranteed (“PASS” in the “filter” column) by all four software programs. If a patient has more than one sample, we used the one with the largest number of mutations resulting in 3,349,768 somatic mutations in 9482 patient samples. These data were subjected to the following screening processes (all patient samples are listed in Supplemental Table S1).
First, samples with low purity were removed to secure the quality of analysis (detail in Supplemental Note). To estimate the purity of each patient sample, we used the consensus measurement of purity estimations (CPE) from Aran et al. (2015) if available; otherwise, the value of “percentage_tumor_nuclei” from TCGA was used (see https://docs.gdc.cancer.gov/API/Users_Guide/Search_and_Retrieval/). Then, samples with estimated purity < 0.7 were excluded, resulting in 6861 patient samples.
We obtained (α1, α2) for all somatic mutations using ASCAT (Van Loo et al. 2010). We first screened out patient samples for which the estimates of α1 and α2 for the entire genome might be unreliable. To do so, we obtained another estimate of copy number, β, from the sequencing depth of the WXS data (see below for details). Then, β/α were computed at each site and the density distributions of β/α were observed, where the average should be around one if the two estimates (α and β) are consistent with each other. It was found that, in most patient samples, β/α showed a normal-like distribution with mean = 1 (Supplemental Fig. S7A,B), whereas the distributions for several patient samples were much wider with multiple peaks (Supplemental Fig. S7C,D), suggesting that there were extensive local copy number changes and estimates of ASCAT (i.e., α1 and α2) may not be very reliable. Therefore, we excluded such patient samples with large variances of β/α (standard deviation of β/α > 0.5) (see Supplemental Fig. S7E), and 6296 patient samples remained (Supplemental Table S2).
This screening based on β/α can also be applied to individual somatic mutations to remove local regions where α and β are inconsistent with each other because the estimates by ASCAT based on a genotyping array is insensitive to relatively short duplications/deletions (e.g., on the order of 1 kb). We again looked at the density distribution of β/α for each of the 6296 patient samples and excluded somatic variants in regions with β/α in the top and bottom 10 percentiles. After these screening processes, 1,875,968 somatic mutations remained. They were then classified into three categories, (α1, α2) = (1, 0), (2, 0), and (1, 1), and the others with α1 + α2 > 2 were excluded in the following analyses. We obtained 1.33 million somatic mutations (1,134,589 SNVs and 191,738 indels) in the regions of (1, 1) and 34,479 and 66,612 somatic mutations for (1, 0) and (2, 0), respectively (Supplemental Table S2).
Computing β, a local estimate of copy number
Our analysis relies on β, allowing us to evaluate local copy number variation. β is an estimate of the copy number for a local genomic region, which should be two in a normal diploid region. β can be estimated for every single site by counting the number of reads in tumor (Stumor) and normal (Snormal) WXS data using MAF files for Mutect2 (Cibulskis et al. 2013). Let q be the value that ASCAT produces as “ploidy,” which is considered the genome-wide average of copy number, and k denotes the ratio of the genome-wide coverage of the tumor WXS to that of the normal WXS. Then, 2k/q explains the genome-wide effect on the Stumor/Snormal ratio, except for a local copy number variation at the focal site. Therefore, β can be estimated as
Estimating VAF′, VAF considering purity
VAF was adjusted by considering purity (VAF′). VAF′ can be calculated as
where p is purity, aDP is read count of alternative allele at the site, and DP is count of all reads at the site.
Linkage analysis for evidence of gene conversion
We explored further evidence for gene conversion by examining the linkage between the focal somatic mutation and an adjacent germline-heterozygote variant, as illustrated in Figure 3C. For each site with SMLOH,Conv, germline variants in the surrounding region of the focal site were searched using VarScan 2 (Koboldt et al. 2012) with the default parameters, and 625 such germline variants were extracted, confirmed with more than five reads. Of these, at 462 sites, SMLOH,Convs were confirmed to link to both alleles at the germline-heterozygote site, as illustrated in Figure 3C.
Note that the remaining 163 mutations do not necessarily show evidence for gene conversion because there is a possibility that the same gene conversion tract converted the examined germline variant as SMLOH,Conv (see Fig. 3D). In such a case, we attempted to move away to another germline variant, and a secondary germline variant was available for only 30 of the 163 SMLOH,Convs. For these 30, we examined whether the pattern of Figure 3C holds between SMLOH,Conv and the secondary germline variants. It turned out that eight of them showed evidence of gene conversion. Altogether, we conclude that a total of 462 + 8 = 470 (75.2%) SMLOH,Convs showed evidence for gene conversion with a linked germline variant.
Definition of TSG
TSGs were defined according to the COSMIC (Futreal et al. 2004; Sondka et al. 2018) definition. Our list of TSGs includes genes whose “role of Cancer” includes “TSG” and “mutation type” is not only “T” (translocation), which resulted in 242 TSGs.
Data sets
We used GRCh38 for the reference genome in this research. The WXS from the TCGA project was available through NCI Genomic Data Commons (GDC; https://portal.gdc.cancer.gov/). Corresponding SNP array data were downloaded from the GDC legacy archive (https://portal.gdc.cancer.gov/legacy-archive/). The definition of TSGs was according to the COSMIC (https://cancer.sanger.ac.uk/census). The meiotic recombination (crossing-over) rate in germline cells was according to HapMap project data (https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html).
Software availability
Codes used in this study are available at GitHub (https://github.com/Kazuki526/somatic_gene_conversion) and as Supplemental Code.
Acknowledgments
This work is partly supported by the Japan Agency for Medical Research and Development, Core Research for Evolutional Science and Technology (AMED-CREST), under grant number JP20gm1110010 to H.I. The supercomputing resource “SHIROKANE” was provided by the Human Genome Center at The University of Tokyo.
Author contributions: K.K.T. and H.I. designed research; K.K.T. performed research; K.K.T. analyzed data; and K.K.T. and H.I. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276617.122.
References
- Andersen CL, Wiuf C, Kruhøffer M, Korsgaard M, Laurberg S, Ørntoft TF. 2007. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis 28: 38–48. 10.1093/carcin/bgl086 [Abstract] [CrossRef] [Google Scholar]
- Aran D, Sirota M, Butte AJ. 2015. Systematic pan-cancer analysis of tumour purity. Nat Commun 6: 8971. 10.1038/ncomms9971 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Auclair J, Leroux D, Desseigne F, Lasset C, Saurin JC, Joly MO, Pinson S, Xu XL, Montmain G, Ruano E, et al. 2007. Novel biallelic mutations in MSH6 ANDPMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum Mutat 28: 1084–1090. 10.1002/humu.20569 [Abstract] [CrossRef] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26: 52–64. 10.1016/j.tcb.2015.07.009 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen J-M, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. 2007. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet 8: 762–775. 10.1038/nrg2193 [Abstract] [CrossRef] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. 10.1038/nbt.2514 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Demeulemeester J, Dentro SC, Gerstung M, Van Loo P. 2022. Biallelic mutations in cancer genomes reveal local mutational determinants. Nat Genet 54: 128–133. 10.1038/s41588-021-01005-8 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Durkin SG, Glover TW. 2007. Chromosome fragile sites. Annu Rev Genet 41: 169–192. 10.1146/annurev.genet.41.042007.165900 [Abstract] [CrossRef] [Google Scholar]
- Fan Y, Xi L, Hughes DS, Zhang J, Zhang J, Futreal PA, Wheeler DA, Wang W. 2016. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol 17: 178. 10.1186/s13059-016-1029-6 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat Rev Genet 4: 177–183. 10.1038/nrc1299 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. 2016. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification–mediated high-throughput genome-wide translocation sequencing. Nat Protoc 11: 853–871. 10.1038/nprot.2016.043 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hudson RR, Kaplan NL. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. 10.1093/genetics/111.1.147 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: a016618. 10.1101/cshperspect.a016618 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. 2020. Pan-cancer analysis of whole genomes. Nature 578: 82–93. 10.1038/s41586-020-1969-6 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- The International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million snps. Nature 449: 851–861. 10.1038/nature06258 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Knudson AG. 1971. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci 68: 820–823. 10.1073/pnas.68.4.820 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. 10.1101/gr.129684.111 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet 31: 241–247. 10.1038/ng917 [Abstract] [CrossRef] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. 2012. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28: 311–317. 10.1093/bioinformatics/btr665 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, et al. 2020. Patterns of somatic structural variation in human cancer genomes. Nature 578: 112–121. 10.1038/s41586-019-1913-9 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Michor F, Iwasa Y, Nowak MA. 2004. Dynamics of cancer progression. Nat Rev Cancer 4: 197–205. 10.1038/nrc1295 [Abstract] [CrossRef] [Google Scholar]
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. 2005. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol 15: 43–49. 10.1016/j.semcancer.2004.09.007 [Abstract] [CrossRef] [Google Scholar]
- Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207. 10.1038/nrm2851 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nachman MW. 2002. Variation in recombination rate across the genome: evidence and implications. Curr Opin Genet Dev 12: 657–663. 10.1016/S0959-437X(02)00358-1 [Abstract] [CrossRef] [Google Scholar]
- Nguyen L, Martens JWM, Van Hoeck A, Cuppen E. 2020. Pan-cancer landscape of homologous recombination deficiency. Nat Commun 11: 5584. 10.1038/s41467-020-19406-4 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nowak MA, Michor F, Komarova NL, Iwasa Y. 2004. Evolutionary dynamics of tumor suppressor gene inactivation. Proc Natl Acad Sci 101: 10635–10638. 10.1073/pnas.0400747101 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pino MS, Chung DC. 2010. The chromosomal instability pathway in colon cancer. Gastroenterology 138: 2059–2072. 10.1053/j.gastro.2009.12.065 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kübler K, Mouw KW, et al. 2017. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 49: 1476–1486. 10.1038/ng.3934 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Raghavan M, et al. 2005. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res 65: 375–378. [Abstract] [Google Scholar]
- Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. 2003. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer 3: 695–701. 10.1038/nrc1165 [Abstract] [CrossRef] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sieber OM, Heinimann K, Tomlinson IP. 2003. Genomic instability: the engine of tumorigenesis? Nat Rev Cancer 3: 701–708. 10.1038/nrc1170 [Abstract] [CrossRef] [Google Scholar]
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. 2018. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 18: 696–705. 10.1038/s41568-018-0060-1 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Stark M, Hayward N. 2007. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res 67: 2632–2642. 10.1158/0008-5472.CAN-06-4152 [Abstract] [CrossRef] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. 10.1016/0092-8674(83)90331-8 [Abstract] [CrossRef] [Google Scholar]
- Tsukiyama T, Zou J, Kim J, Ogamino S, Shino Y, Masuda T, Merenda A, Matsumoto M, Fujioka Y, Hirose T, et al. 2020. A phospho-switch controls RNF43-mediated degradation of Wnt receptors to suppress tumorigenesis. Nat Commun 11: 4586. 10.1038/s41467-020-18257-3 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tubbs A, Nussenzweig A. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168: 644–656. 10.1016/j.cell.2017.01.002 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tuna M, Knuutila S, Mills GB. 2009. Uniparental disomy in cancer. Trends Mol Med 15: 120–128. 10.1016/j.molmed.2009.01.005 [Abstract] [CrossRef] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. 2010. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci 107: 16910–16915. 10.1073/pnas.1009843107 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. 2016. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164: 644–655. 10.1016/j.cell.2015.12.039 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. 10.1038/ng.2760 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zhang J, Lindroos A, Ollila S, Russell A, Marra G, Mueller H, Peltomaki P, Plasilova M, Heinimann K. 2006. Gene conversion is a frequent mechanism of inactivation of the wild-type allele in cancers from mlh1/msh2 deletion carriers. Cancer Res 66: 659–664. 10.1158/0008-5472.CAN-05-4043 [Abstract] [CrossRef] [Google Scholar]
Articles from Genome Research are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gr.276617.122
Read article for free, from open access legal sources, via Unpaywall:
https://genome.cshlp.org/content/32/6/1017.full.pdf
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/128918605
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ever since Knudson.
Trends Genet, 17(10):569-573, 01 Oct 2001
Cited by: 67 articles | PMID: 11585662
Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis.
Genes Chromosomes Cancer, 22(3):200-209, 01 Jul 1998
Cited by: 161 articles | PMID: 9624531
Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene.
Oncogene, 20(29):3814-3823, 01 Jun 2001
Cited by: 137 articles | PMID: 11439345
Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview.
Cell Physiol Biochem, 51(6):2647-2693, 12 Dec 2018
Cited by: 122 articles | PMID: 30562755
Review
Funding
Funders who supported this work.
AMED-CREST (1)
Grant ID: JP20gm1110010




