Abstract
Objective
To distinguish among predictors of seroconversion, progression to multiple autoantibodies and from multiple autoantibodies to type 1 diabetes in young children.Research design and methods
Genetically high-risk newborns (n = 8,502) were followed for a median of 11.2 years (interquartile range 9.3-12.6); 835 (9.8%) developed islet autoantibodies and 283 (3.3%) were diagnosed with type 1 diabetes. Predictors were examined using Cox proportional hazards models.Results
Predictors of seroconversion and progression differed, depending on the type of first appearing autoantibody. Male sex, Finnish residence, having a sibling with type 1 diabetes, the HLA DR4 allele, probiotic use before age 28 days, and single nucleotide polymorphism (SNP) rs689_A (INS) predicted seroconversion to IAA-first (having islet autoantibody to insulin as the first appearing autoantibody). Increased weight at 12 months and SNPs rs12708716_G (CLEC16A) and rs2292239_T (ERBB3) predicted GADA-first (autoantibody to GAD as the first appearing). For those having a father with type 1 diabetes, the SNPs rs2476601_A (PTPN22) and rs3184504_T (SH2B3) predicted both. Younger age at seroconversion predicted progression from single to multiple autoantibodies as well as progression to diabetes, except for those presenting with GADA-first. Family history of type 1 diabetes and the HLA DR4 allele predicted progression to multiple autoantibodies but not diabetes. Sex did not predict progression to multiple autoantibodies, but males progressed more slowly than females from multiple autoantibodies to diabetes. SKAP2 and MIR3681HG SNPs are newly reported to be significantly associated with progression from multiple autoantibodies to type 1 diabetes.Conclusions
Predictors of IAA-first versus GADA-first autoimmunity differ from each other and from the predictors of progression to diabetes.Free full text

Predictors of the Initiation of Islet Autoimmunity and Progression to Multiple Autoantibodies and Clinical Diabetes: The TEDDY Study
Abstract
OBJECTIVE
To distinguish among predictors of seroconversion, progression to multiple autoantibodies and from multiple autoantibodies to type 1 diabetes in young children.
RESEARCH DESIGN AND METHODS
Genetically high-risk newborns (n = 8,502) were followed for a median of 11.2 years (interquartile range 9.3–12.6); 835 (9.8%) developed islet autoantibodies and 283 (3.3%) were diagnosed with type 1 diabetes. Predictors were examined using Cox proportional hazards models.
RESULTS
Predictors of seroconversion and progression differed, depending on the type of first appearing autoantibody. Male sex, Finnish residence, having a sibling with type 1 diabetes, the HLA DR4 allele, probiotic use before age 28 days, and single nucleotide polymorphism (SNP) rs689_A (INS) predicted seroconversion to IAA-first (having islet autoantibody to insulin as the first appearing autoantibody). Increased weight at 12 months and SNPs rs12708716_G (CLEC16A) and rs2292239_T (ERBB3) predicted GADA-first (autoantibody to GAD as the first appearing). For those having a father with type 1 diabetes, the SNPs rs2476601_A (PTPN22) and rs3184504_T (SH2B3) predicted both. Younger age at seroconversion predicted progression from single to multiple autoantibodies as well as progression to diabetes, except for those presenting with GADA-first. Family history of type 1 diabetes and the HLA DR4 allele predicted progression to multiple autoantibodies but not diabetes. Sex did not predict progression to multiple autoantibodies, but males progressed more slowly than females from multiple autoantibodies to diabetes. SKAP2 and MIR3681HG SNPs are newly reported to be significantly associated with progression from multiple autoantibodies to type 1 diabetes.
CONCLUSIONS
Predictors of IAA-first versus GADA-first autoimmunity differ from each other and from the predictors of progression to diabetes.
Introduction
The detection of multiple islet autoantibodies, recognized as the initial stage of type 1 diabetes (1), is usually preceded by presence of a single autoantibody. Some individuals are diagnosed with diabetes with a single autoantibody; others may progress to multiple autoantibodies, but not to clinical diabetes, over an extended period of time. Progression depends on the age at seroconversion and the number of autoantibodies present (2–11). A family history of type 1 diabetes and genetic factors are less predictive (3,9,11) of progression than they are of seroconversion.
Multiple reports describe different patterns of islet autoantibody initial presentation, subsequent spreading, and their relationship to diabetes onset (12,13). There seems to be a number of factors that contribute to the initiation of islet autoimmunity and affect the risk of progression through to type 1 diabetes. One of the factors is which islet autoantibody appears first (4,13). This article focuses on the identification of predictors for detection of a first autoantibody (seroconversion from autoantibody negative to autoantibody positive) and the progression from a single autoantibody to multiple autoantibodies and from multiple autoantibodies to type 1 diabetes in The Environmental Determinants of Diabetes in the Young (TEDDY) study. The hypothesis is that there are different predictors at each step, that class II HLA genes are more related to initiation of seroconversion than progression to diabetes once islet autoantibodies are manifest. Differences in the significance of other predictors may shed light on environmental exposures or gene-environment interactions that are not uniform across disease stages.
Research Design and Methods
Participants
TEDDY is a prospective cohort study funded by the National Institutes of Health with the primary goal of identifying environmental causes of type 1 diabetes. It includes six clinical research centers: three in the U.S., Colorado, Georgia/Florida, and Washington state, and three in Europe, Finland, Germany, and Sweden. Detailed study design and methods have previously been published (14–16). Written informed consent was obtained for all study participants from a parent or primary caretaker, separately, for genetic screening and participation in the prospective follow-up.
The high-risk HLA genotypes for participants screened from the general population were as follows: DRB1*04-DQA1*03-DQB1*03:02/DRB1*03-DQA1*05-DQB1*02:01 (DR3/4), DRB1*04-DQA1*03-DQB1*03:02/DRB1*04-DQA1*03-DQB1*03:02 (DR4/4), DRB1*04-DQA1*03-DQB1*03:02/DRB1*08-DQA1*04-DQB1*04:02 (DR4/8), and DRB1*03-DQA1*05-DQB1*02:01/DRB1*03-DQA1*05-DQB1*02:01 (DR3/3). Additional genotypes were included for first-degree relatives (FDRs) of a subject with type 1 diabetes: DRB1*04-DQA1*03-DQB1*03:02/DRB1*04-DQA1*03-DQB1*02:02 (DR4/4b), DRB1*04-DQA1*03-DQB1*03:02/DRB1*01- DQA1*01-DQB1*05:01 (DR4/1), DRB1*04-DQA1*03-DQB1*03:02/DRB1*13-DQA1*01-DQB1*06:04 (DR4/13), DRB1*04-DQA1*03-DQB1*03:02/DRB1*09- DQA1*03-DQB1*03:03 (DR4/9), and DRB1*03-DQA1*05-DQB1*02:01/DRB1*09-DQA1*03-DQB1*03:03 (DR3/9). Genotyping was confirmed by reverse blot hybridization at the central HLA Reference Laboratory at Roche Molecular Systems, Oakland, CA (16), along with the INS-23Hph1 (rs689), CTLA4-T17A (rs231775), and PTPN22-R620W (rs2476601) single nucleotide polymorphism (SNP) primer pairs. The study was approved by local institutional review or ethics boards and is monitored by an External Evaluation Committee formed by the National Institutes of Health.
SNP genotyping was performed by the Center for Public Health Genomics at the University of Virginia, using the Illumina Immunochip, which is a custom array for genotyping of SNPs selected from regions of the human genome firmly associated with autoimmune diseases (17). The final selection of SNPs, including ~186,000 SNPs in 186 regions, for 12 autoimmune diseases, was decided by the Immunochip Consortium. In previous work in TEDDY, investigators examined whether any of 41 non-HLA SNPs previously shown to be associated with type 1 diabetes conferred risk for islet autoimmunity.
Islet Autoantibodies
Islet autoantibodies to insulin (IAA), GAD (GADA), or IA-2 (IA-2A) were measured in two laboratories with radio binding assays. In the U.S., all sera were assayed at the Barbara Davis Center for Childhood Diabetes at the University of Colorado Denver; in Europe, all sera were assayed at the University of Bristol, Bristol, U.K. Both laboratories demonstrated high sensitivity and specificity as well as concordance (19). All positive islet autoantibody samples and 5% of the negative samples were retested in the other reference laboratory and deemed confirmed if concordant. Persistent islet autoimmunity was defined as confirmed positivity for IAA, GADA, or IA-2A in at least two consecutive samples.
Statistical Methods
Time-to-event analyses using multivariable Cox proportional hazards models were applied to examine factors related to the risk of each disease stage leading to clinical diabetes: development of islet autoantibody (IA) positivity, progression from a single autoantibody to multiple autoantibodies, and progression from multiple autoantibodies to type 1 diabetes. Separate cause-specific proportional hazards models for competing risks were used to study the risk (cause-specific hazard ratios [HRs]) of first developing GADA positivity (GADA-first) and IAA-first, respectively through censoring IA events from other causes at the event time (20). The magnitude of the association is described with HRs with 95% CIs.
IA positivity was defined as confirmed positive autoantibodies to GADA, IAA, or IA-2A in at least two consecutive samples by both TEDDY laboratories. The time to the development of IA was the age at the initial of two or more consecutive positive tests. Children negative for IA were right censored at the date of the last negative sample drawn for autoantibodies.
Progression from a single autoantibody to multiple autoantibodies was defined as the development of a second IA in children positive for a single IA. The time-to-event variable was the duration calculated from the onset date of the first IA to the onset date of a second IA or right censored for children without multiple autoantibodies at the date of the last negative sample drawn for the other autoantibodies.
Similarly, progression from multiple autoantibodies to type 1 diabetes was defined as the development of type 1 diabetes in children positive for multiple IAs. The time-to-event variable was the duration calculated from the date at onset of multiple IAs to the diagnosis date of type 1 diabetes or right censored for those who did not develop type 1 diabetes by the date of the last TEDDY visit.
The factors under examination included those previously published for TEDDY (13,21). In addition, 19 SNPs were included in the multivariable Cox models. Population stratification (ancestral heterogeneity) was accounted for through inclusion of the top two principal components calculated from the Immunochip data as covariates in the Cox models (22). The 19 SNPs were identified from 45 SNPs in type 1 diabetes risk loci (rs689 in INS, 41 SNPs examined in TEDDY, and 3 SNPs recently available in TEDDY [rs11755527 in BACH2 and rs12444268, rs917997]) by forward selection procedures with Cox regression of them on each of the risks of IA, IAA-first, GADA-first, progression from a single autoantibody to multiple autoantibodies, and progression from multiple autoantibodies to type 1 diabetes, with inclusion criteria of P value <0.05 (11,18).
Age at seroconversion was normalized using a log-transformation and then included as a covariate in the Cox analyses of progression from a single autoantibody to multiple autoantibodies and from multiple autoantibodies to type 1 diabetes.
Constant risks were assumed for comparison of the average annual hazard rate of development of IA, IAA-first IA, GAD-first IA, progression from a single autoantibody to multiple autoantibodies, and progression from multiple autoantibodies to type 1 diabetes stratified by follow-up time period (≤2 years and >2 years) (23).
Data were analyzed with use of SAS software (version 9.4; SAS Institute, Cary, NC). Two-tailed P values <0.05 were considered to indicate statistical significance. No adjustment in type 1 error was made for multiple comparisons except in the context of the multivariable Cox regression model.
Data and Resource Availability
The data sets generated and analyzed during the current study will be made available in the NIDDK Central Repository at https://repository.niddk.nih.gov/studies/teddy.
Results
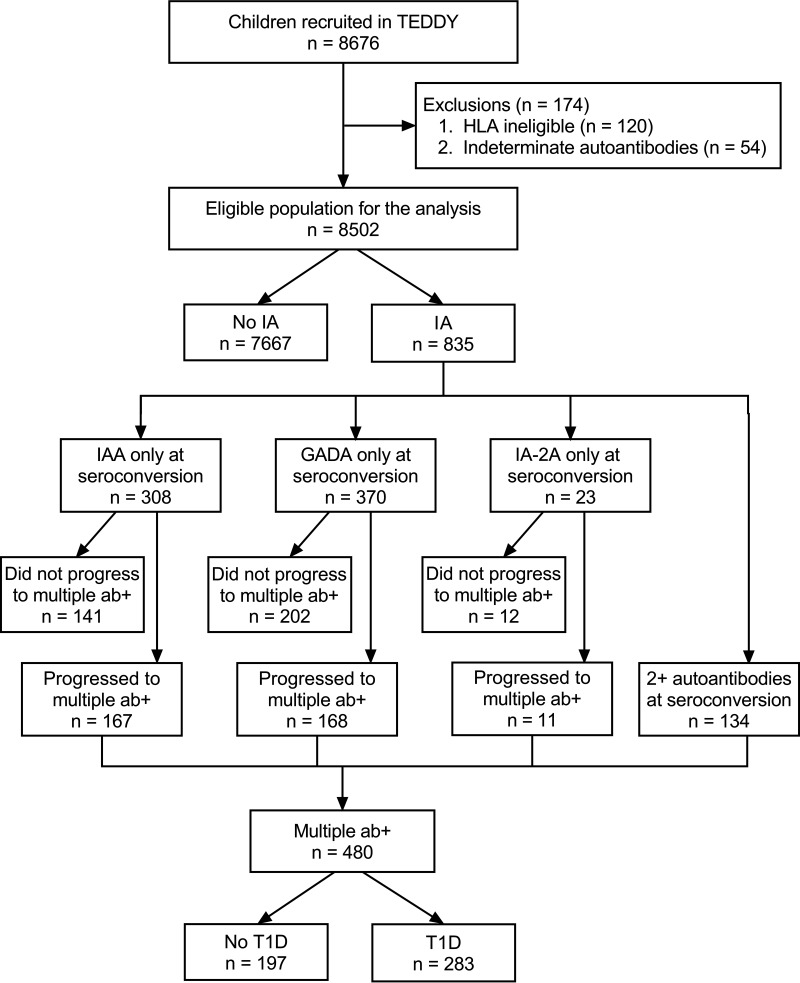
The TEDDY study has enrolled and followed, from 3 months of age, a cohort of 8,676 infants at elevated genetic risk for autoimmune type 1 diabetes; 174 children were excluded due to HLA ineligibility or indeterminate autoantibody status, leaving 8,502 in the analysis. Children were followed quarterly for a first-appearing islet autoantibody and progression to diagnosis of diabetes. Follow-up of children with one or more islet autoantibodies continued on this schedule, whereas children who were autoantibody negative were followed semiannually after 4 years of age. The median age at the last follow-up was 11.2 years (interquartile range 9.3–12.6 years, range 2 months–15.3 years).
As of 29 February 2020, 835 children (9.8%) have developed islet autoantibodies: 701 with a single autoantibody (308 IAA, 370 GADA, 23 IA-2) and 134 with multiple autoantibodies. Of the 701 who developed a single autoantibody first, 346 (49.4%) progressed to multiple autoantibodies and 189 (54.6%) of these progressed to diagnosis of diabetes. Of those with multiple autoantibodies at seroconversion, 94 (70.1%) progressed to diabetes (Fig. 1). Omitted are 40 children who were diagnosed with diabetes but had no autoantibodies detected; most had limited follow-up prior to dropping out of the study. An additional 42 children were excluded with a single autoantibody (24 IAA, 13 GADA, and 5 IA-2) who were diagnosed with diabetes without having multiple autoantibodies observed.
The proportion of individuals seroconverting to IAA-first (31.2%) as compared with GADA-first (18.6%) was significantly higher among those from Finland than at other sites (P = 0.0001), among siblings of children with type 1 diabetes (7.5% vs. 2.7%, P = 0.004), and among those with DR4/8 (20.5% vs. 12.2%, P = 0.003) or FDR-specific genotypes (4.5% vs. 1.4%, P = 0.012) as compared with other groups. Only children with a DR3/3 genotype (22.2% vs. 9.7%, P = 0.00001) and those from Sweden had a significantly higher proportion of children seroconverting to GADA-first (38.1%) as compared with IAA-first (30.5%, P = 0.039) (Table 1).
Table 1
Characteristics of TEDDY children
| No IA (n = 7,667) | IA overall (n = 835) | IAA-first (n = 308) | GADA-first (n = 370) | Multiple ab+ (n = 480) | Type 1 diabetes (n = 283) | |
|---|---|---|---|---|---|---|
| Age at onset (years), mean (SD) | NA | 4.45 (3.39) | 3.38 (3.20) | 5.40 (3.39) | 4.35 (3.13) | 6.52 (3.46) |
| Age at onset (years), median (IQR) | NA | 3.32 (1.57, 7.15) | 2.03 (1.03, 4.57) | 4.79 (2.35, 8.29) | 3.31 (1.82, 6.47) | 6.43 (3.30, 9.26) |
| Country | ||||||
 U.S. U.S. | 3,339 (43.6) | 287 (34.4) | 98 (31.8) | 141 (38.1) | 162 (33.8) | 94 (33.2) |
 Finland Finland | 1,603 (20.9) | 201 (24.1) | 96 (31.2) | 69 (18.6) | 128 (26.7) | 82 (29.0) |
 Germany Germany | 514 (6.7) | 60 (7.2) | 20 (6.5) | 19 (5.1) | 39 (8.1) | 27 (9.5) |
 Sweden Sweden | 2,211 (28.8) | 287 (34.4) | 94 (30.5) | 141 (38.1) | 151 (31.5) | 80 (28.3) |
| Family history | ||||||
 FDR: mother FDR: mother | 298 (3.9) | 41 (4.9) | 12 (3.9) | 17 (4.6) | 27 (5.6) | 18 (6.4) |
 FDR: father FDR: father | 368 (4.8) | 86 (10.3) | 32 (10.4) | 40 (10.8) | 60 (12.5) | 35 (12.4) |
 FDR: sibling FDR: sibling | 111 (1.4) | 40 (4.8) | 23 (7.5) | 10 (2.7) | 26 (5.4) | 19 (6.7) |
 General population General population | 6,890 (89.9) | 668 (80.0) | 241 (78.2) | 303 (81.9) | 367 (76.5) | 211 (74.6) |
| Sex | ||||||
 Female Female | 3,813 (49.7) | 378 (45.3) | 137 (44.5) | 172 (46.5) | 214 (44.6) | 136 (48.1) |
 Male Male | 3,854 (50.3) | 457 (54.7) | 171 (55.5) | 198 (53.5) | 266 (55.4) | 147 (51.9) |
| HLA genotype | ||||||
 DR3/4 DR3/4 | 2,915 (38.0) | 403 (48.3) | 144 (46.8) | 181 (48.9) | 261 (54.4) | 162 (57.2) |
 DR4/4 DR4/4 | 1,506 (19.6) | 155 (18.6) | 57 (18.5) | 57 (15.4) | 98 (20.4) | 52 (18.4) |
 DR4/8 DR4/8 | 1,340 (17.5) | 128 (15.3) | 63 (20.5) | 45 (12.2) | 64 (13.3) | 35 (12.4) |
 DR3/3 DR3/3 | 1,662 (21.7) | 120 (14.4) | 30 (9.7) | 82 (22.2) | 37 (7.7) | 20 (7.1) |
 FDR specific* FDR specific* | 244 (3.2) | 29 (3.5) | 14 (4.5) | 5 (1.4) | 20 (4.2) | 14 (4.9) |
Data are presented as n (%) unless otherwise indicated. Multiple ab+, positive for multiple autoantibodies; NA, not applicable.
First Appearance of IA
Predictors for the appearance of IA differed depending on the type of first appearing autoantibody. Males, as compared with females, were at higher risk for IA (HR 1.30; 95% CI 1.13, 1.49; P < 0.001), and this was among those presenting with IAA-first (HR 1.35; 95% CI 1.07, 1.70; P = 0.013). The IAA-first–related association was also found with respect to increased IA risk where there was a sibling affected by diabetes (HR 5.06; 95% CI 3.22, 7.96; P < 0.001), for DR4 allele as compared with DR3/3 (HRs ranging from 2.10 to 2.68, all P ≤ 0.001), for individuals from Finland as compared with the U.S. (HR 2.19; 95% CI 1.47, 3.28; P < 0.001), for the protective effects of the introduction of probiotics within the first 28 days of life (HR 0.48; 95% CI 0.28, 0.84; P = 0.010), and for the SNPs rs689_A (INS) (HR 0.58; 95% CI 0.45, 0.73; P < 0.001) and rs2327832_G (TNFAIP3) (HR 0.76; 95% CI 0.61, 0.95; P = 0.015). The SNP rs7202877_C (CTRB2) (HR 1.28; 95% CI 1.01, 1.61; P = 0.039) was associated with increased IAA risk.
Increased weight z score at 12 months was a predictor (HR 1.17; 95% CI 1.05, 1.30; P = 0.003), as were the SNPs rs12708716_G (CLEC16A) (HR 0.81; 95% CI 0.69, 0.95; P = 0.010) and rs11258747_A (PRKCQ) (HR 1.19; 95% CI 1.00, 1.41; P = 0.045), for GADA-first. The SNPs rs2292239_T (ERBB3) (HR 1.17; 95% CI 1.00, 1.36; P = 0.054), rs11755527_C (BACH2) (HR 1.15; 95% CI 1.00, 1.33; P = 0.056), and rs7804356_G (SKAP2) (HR 0.83; 95% CI 0.69, 1.00; P = 0.054) were only marginally significant for GADA-first. Those with the FDR-specific genotypes included in the cohort were at significantly lower risk as compared with those with the DR3/3 genotype (HR 0.016; 95% CI 0.06, 0.46; P = 0.001).
Having a father with type 1 diabetes (as compared with the general population) was a predictor (HR 2.41; 95% CI 1.87, 3.11; P < 0.001), as were the SNPs rs2476601_A (PTPN22) (HR 1.44 [95% CI 1.15, 1.80], P = 0.002, and HR 1.42 [95% CI 1.15, 1.76], P = 0.001) and rs3184504_T (SH2B3) (HR 1.27 [95% CI 1.08, 1.50], P = 0.005, and HR 1.34 [95% CI 1.15, 1.56], P < 0.001), for IA for both IAA-first and GADA-first, respectfully. Four SNPs, rs763361_A (CD226) (HR 1.13; 95% CI 1.02, 1.25; P = 0.016), rs1990760_G (IFIH1) (HR 0.88; 95% CI 0.79, 0.97; P = 0.014), rs11203203_A (UBASH3A) (HR 1.12; 95% CI 1.01, 1.24; P = 0.039), and rs4948088_A (COBL) (HR 0.74; 95% CI 0.57, 0.97; P = 0.031), were significant for IA but not for IAA-first or GADA-first (Table 2).
Table 2
Association between factors and risk of developing islet autoantibodies, IAA-first, and GADA-first
| Factor | Comparison | IA (n = 701) | IAA-first (n = 308) | GADA-first (n = 370) | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| Sex | Male vs. female | 1.30 (1.13, 1.49) | <0.001 | 1.35 (1.07, 1.70) | 0.013 | 1.22 (0.99, 1.50) | 0.066 |
| Family history | FDR father vs. GP | 2.41 (1.87, 3.11) | <0.001 | 2.23 (1.45, 3.42) | <0.001 | 2.95 (2.06, 4.22) | <0.001 |
| FDR sibling vs. GP | 2.96 (2.11, 4.14) | <0.001 | 5.06 (3.22, 7.96) | <0.001 | 1.60 (0.82, 3.12) | 0.172 | |
| FDR mother vs. GP | 1.30 (0.92, 1.84) | 0.141 | 1.01 (0.53, 1.90) | 0.985 | 1.40 (0.82, 2.39) | 0.218 | |
| HLA genotype | DR3/4 vs. DR3/3 | 1.86 (1.51, 2.29) | <0.001 | 2.61 (1.73, 3.93) | <0.001 | 1.26 (0.96, 1.64) | 0.091 |
| DR4/4 vs. DR3/3 | 1.40 (1.10, 1.79) | 0.007 | 2.10 (1.33, 3.32) | 0.001 | 0.77 (0.54, 1.09) | 0.134 | |
| DR4/8 vs. DR3/3 | 1.41 (1.09, 1.83) | 0.010 | 2.68 (1.69, 4.24) | <0.001 | 0.79 (0.54, 1.16) | 0.225 | |
| FDR specific vs. DR3/3 | 0.73 (0.46, 1.15) | 0.178 | 1.62 (0.80, 3.27) | 0.178 | 0.16 (0.06, 0.46) | 0.001 | |
| Country | Finland vs. U.S. | 1.53 (1.20, 1.96) | 0.001 | 2.19 (1.47, 3.28) | <0.001 | 1.04 (0.72, 1.51) | 0.829 |
| Germany vs. U.S. | 1.01 (0.74, 1.40) | 0.935 | 1.07 (0.63, 1.81) | 0.813 | 0.63 (0.36, 1.11) | 0.110 | |
| Sweden vs. U.S. | 1.19 (0.99, 1.43) | 0.060 | 1.21 (0.88, 1.65) | 0.239 | 1.16 (0.90, 1.50) | 0.257 | |
| Probiotics introduction age | <28 days vs. ≥28 days | 0.66 (0.48, 0.91) | 0.010 | 0.48 (0.28, 0.84) | 0.010 | 0.73 (0.45, 1.20) | 0.215 |
| Weight z score at 12 months | 1.14 (1.06, 1.23) | <0.001 | 1.12 (0.99, 1.25) | 0.062 | 1.17 (1.05, 1.30) | 0.003 | |
| rs689_A (INS) | 0.73 (0.64, 0.84) | <0.001 | 0.58 (0.45, 0.73) | <0.001 | 1.00 (0.83, 1.21) | 0.997 | |
| rs2476601_A (PTPN22) | 1.42 (1.23, 1.63) | <0.001 | 1.44 (1.15, 1.80) | 0.002 | 1.42 (1.15, 1.76) | 0.001 | |
| rs3184504_T (SH2B3) | 1.26 (1.14, 1.40) | <0.001 | 1.27 (1.08, 1.50) | 0.005 | 1.34 (1.15, 1.56) | <0.001 | |
| rs12708716_G (CLEC16A) | 0.85 (0.76, 0.94) | 0.002 | 0.88 (0.74, 1.05) | 0.165 | 0.81 (0.69, 0.95) | 0.010 | |
| rs2292239_T (ERBB3) | 1.17 (1.05, 1.30) | 0.004 | 1.14 (0.96, 1.35) | 0.140 | 1.17 (1.00, 1.36) | 0.054 | |
| rs763361_A (CD226) | 1.13 (1.02, 1.25) | 0.016 | 1.10 (0.93, 1.29) | 0.264 | 1.09 (0.94, 1.26) | 0.262 | |
| rs1990760_G (IFIH1) | 0.88 (0.79, 0.97) | 0.014 | 0.90 (0.76, 1.07) | 0.236 | 0.86 (0.74, 1.01) | 0.063 | |
| rs11203203_A (UBASH3A) | 1.12 (1.01, 1.24) | 0.039 | 1.11 (0.93, 1.32) | 0.245 | 1.09 (0.93, 1.27) | 0.305 | |
| rs4948088_A (COBL) | 0.74 (0.57, 0.97) | 0.031 | 0.70 (0.44, 1.11) | 0.126 | 0.86 (0.59, 1.25) | 0.425 | |
| rs2327832_G (TNFAIP3) | 0.92 (0.81, 1.04) | 0.169 | 0.76 (0.61, 0.95) | 0.015 | 0.91 (0.76, 1.10) | 0.330 | |
| rs7202877_C (CTRB2) | 1.14 (0.98, 1.32) | 0.082 | 1.28 (1.01, 1.61) | 0.039 | 1.12 (0.89, 1.41) | 0.324 | |
| rs11258747_A (PRKCQ) | 1.09 (0.97, 1.23) | 0.127 | 1.16 (0.96, 1.39) | 0.127 | 1.19 (1.00, 1.41) | 0.045 | |
| rs12251307_A (RBM17, IL2RA) | 0.85 (0.73, 1.00) | 0.049 | 0.91 (0.71, 1.18) | 0.481 | 0.77 (0.60, 0.98) | 0.036 | |
| rs11755527_C (BACH2) | 1.03 (0.93, 1.14) | 0.551 | 0.88 (0.75, 1.04) | 0.146 | 1.15 (1.00, 1.33) | 0.056 | |
| rs7804356_G (SKAP2) | 0.95 (0.84, 1.07) | 0.402 | 0.99 (0.81, 1.20) | 0.903 | 0.83 (0.69, 1.00) | 0.054 | |
| rs12444268_A | 0.97 (0.87, 1.08) | 0.551 | 0.96 (0.80, 1.14) | 0.620 | 0.94 (0.80, 1.11) | 0.489 | |
| rs1534422_G (MIR3681HG) | 0.99 (0.90, 1.10) | 0.911 | 1.03 (0.87, 1.21) | 0.752 | 0.98 (0.85, 1.14) | 0.821 | |
| rs3825932_A (CTSH) | 0.93 (0.84, 1.03) | 0.179 | 0.90 (0.76, 1.07) | 0.218 | 1.03 (0.88, 1.21) | 0.688 | |
| rs1004446_A (INS) | 0.93 (0.83, 1.05) | 0.231 | 0.95 (0.79, 1.15) | 0.616 | 0.91 (0.77, 1.08) | 0.296 | |
| PC1* | 0.97 (0.87, 1.07) | 0.499 | 1.03 (0.86, 1.24) | 0.760 | 0.94 (0.82, 1.09) | 0.423 | |
| PC2* | 1.16 (1.05, 1.29) | 0.003 | 1.22 (1.03, 1.45) | 0.023 | 1.11 (0.97, 1.28) | 0.120 | |
Adjusted (cause-specific) HRs were estimated from multivariable proportional hazards models.
Progression From a Single Autoantibody to Multiple Autoantibodies and Multiple Autoantibodies to Type 1 Diabetes
Among those who seroconverted to a single autoantibody, there was no discernable difference in a multivariate proportional hazards model of the rate of progression to multiple autoantibodies between those who seroconverted to IAA-first vs. GADA-first (HR 1.00; 95% CI 0.77, 1.29; P = 0.99). Those who initially presented with multiple autoantibodies were at higher risk (HR 1.91; 95% CI 1.42, 2.56; P < 0.001) of progressing to diagnosis of diabetes than those with only a single autoantibody. Older age at either initial seroconversion or progression from a single autoantibody to multiple autoantibodies was significantly associated with a lower rate of progression to multiple autoantibodies (HR 0.65; 95% CI 0.57, 0.75; P < 0.001) or type 1 diabetes (HR 0.62; 95% CI 0.51, 0.75; P < 0.001), respectively. A father with type 1 diabetes (HR 1.42; 95% CI 1.00, 2.01; P = 0.049) or a sibling (HR 1.73; 95% CI 1.05, 2.85; P = 0.031) was associated with elevated risk (compared with risk for those without a first-degree relative with type 1 diabetes) for progressing from a single autoantibody to multiple autoantibodies but not diabetes. All of the DR4 allele genotypes were significant predictors compared with the DR3/3 genotype for progressing from a single autoantibody to multiple autoantibodies (HRs ranging from 1.80 to 2.63, 0.001 <P < 0.021), but none were significantly related to progression from multiple autoantibodies to type 1 diabetes. The HLA predictors for progression to multiple autoantibodies were significant only for those who first presented with IAA, not GADA, with the exception of DR3/4 and DR4/4 vs. DR3/3, which were significant for both and for those with a father with type 1 diabetes. Notably, male sex imparted a lower risk (HR 0.63; 95% CI 0.49, 0.83; P = 0.021) for progressing from multiple autoantibodies to type 1 diabetes, even though sex was not significantly associated with progression from a single autoantibody to multiple autoantibodies. Five SNPs, two with reduced risk and three with increased risk, were significant diabetes risk factors, although none were significant for the development of autoantibodies or progression from a single autoantibody to multiple autoantibodies: rs1004446_A (INS) (HR 0.81; 95% CI 0.65, 1.00; P = 0.053), rs1534422_G (MIR3681HG) (HR 1.32; 95% CI 1.09, 1.60; P = 0.005), rs2327832_G (TNFAIP3) (HR 1.40; 95% CI 1.12, 1.74; P = 0.003), rs3825932_A (CTSH) (HR 0.76; 95% CI 0.62, 0.93; P = 0.007), and rs7804356_G (SKAP2) (HR 1.26; 95% CI 1.01, 1.58; P = 0.043) (Table 3).
Table 3
Association between factors and risk of progression from single autoantibody to multiple autoantibodies and from multiple autoantibodies to type 1 diabetes
| Factor | Comparison | Single autoantibody to multiple autoantibodies | Multiple ab+ to type 1 diabetes, overall (n/N = 283/480) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall (n/N = 346/701) | Subgroup by autoantibody at seroconversion | ||||||||
| IAA-first (n/N = 167/308) | GADA-first (n/N = 168/370) | ||||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| Autoantibody at seroconversion | IA-2A vs. GADA | 1.12 (0.57, 2.22) | 0.736 | ||||||
| IAA vs. GADA | 1.00 (0.77, 1.29) | 0.992 | |||||||
| Initial ab | Multiple vs. single ab+ | 1.91 (1.42, 2.56) | <0.001 | ||||||
| Age (years) at seroconversion* | 0.65 (0.57, 0.75) | <0.001 | 0.63 (0.51, 0.77) | <0.001 | 0.64 (0.52, 0.80) | <0.001 | 0.62 (0.51, 0.75) | <0.001 | |
| Sex | Male vs. female | 1.04 (0.82, 1.31) | 0.752 | 1.04 (0.72, 1.51) | 0.823 | 0.89 (0.63, 1.26) | 0.505 | 0.63 (0.49, 0.83) | 0.001 |
| Family history | FDR father vs. GP | 1.42 (1.00, 2.01) | 0.049 | 1.52 (0.91, 2.54) | 0.111 | 1.35 (0.80, 2.27) | 0.260 | 0.99 (0.65, 1.51) | 0.970 |
| FDR sibling vs. GP | 1.73 (1.05, 2.85) | 0.031 | 1.97 (1.01, 3.86) | 0.047 | 0.97 (0.32, 2.97) | 0.958 | 1.36 (0.77, 2.39) | 0.290 | |
| FDR mother vs. GP | 1.50 (0.83, 2.71) | 0.182 | 1.81 (0.70, 4.65) | 0.219 | 1.37 (0.60, 3.12) | 0.460 | 1.55 (0.88, 2.74) | 0.127 | |
| HLA genotype | DR3/4 vs. DR3/3 | 2.63 (1.75, 3.94) | <0.001 | 5.42 (1.66, 17.67) | 0.005 | 2.44 (1.52, 3.92) | <0.001 | 1.12 (0.66, 1.89) | 0.680 |
| DR4/4 vs. DR3/3 | 2.28 (1.44, 3.60) | <0.001 | 5.82 (1.74, 19.48) | 0.004 | 1.84 (1.03, 3.30) | 0.041 | 1.00 (0.56, 1.79) | 0.987 | |
| DR4/8 vs. DR3/3 | 1.80 (1.09, 2.96) | 0.021 | 3.93 (1.17, 13.22) | 0.027 | 1.93 (0.95, 3.93) | 0.070 | 0.89 (0.48, 1.67) | 0.724 | |
| FDR specific vs. DR3/3 | 2.09 (0.98, 4.47) | 0.057 | 4.74 (1.16, 19.40) | 0.030 | 3.82 (0.80, 18.18) | 0.092 | 1.37 (0.59, 3.16) | 0.458 | |
| Country | Finland vs. U.S. | 1.29 (0.84, 1.97) | 0.246 | 1.62 (0.89, 2.94) | 0.113 | 1.22 (0.61, 2.40) | 0.575 | 1.02 (0.63, 1.67) | 0.931 |
| Germany vs. U.S. | 0.69 (0.40, 1.18) | 0.177 | 0.75 (0.36, 1.56) | 0.447 | 0.71 (0.27, 1.90) | 0.496 | 1.24 (0.72, 2.14) | 0.432 | |
| Sweden vs. U.S. | 0.69 (0.52, 0.93) | 0.014 | 0.74 (0.46, 1.18) | 0.202 | 0.69 (0.46, 1.05) | 0.081 | 0.82 (0.59, 1.16) | 0.266 | |
| rs689_A (INS) | 0.70 (0.56, 0.88) | 0.002 | 0.83 (0.58, 1.18) | 0.292 | 0.62 (0.44, 0.86) | 0.005 | 1.05 (0.81, 1.36) | 0.704 | |
| rs2476601_A (PTPN22) | 1.31 (1.05, 1.63) | 0.017 | 1.08 (0.79, 1.49) | 0.631 | 1.79 (1.30, 2.47) | <0.001 | 1.15 (0.88, 1.51) | 0.307 | |
| rs3184504_T (SH2B3) | 0.98 (0.84, 1.15) | 0.809 | 0.91 (0.71, 1.17) | 0.467 | 1.08 (0.85, 1.38) | 0.531 | 1.02 (0.85, 1.22) | 0.834 | |
| rs12708716_G (CLEC16A) | 0.88 (0.73, 1.05) | 0.157 | 0.80 (0.59, 1.07) | 0.126 | 0.98 (0.74, 1.29) | 0.864 | 1.19 (0.97, 1.45) | 0.095 | |
| rs2292239_T (ERBB3) | 1.12 (0.94, 1.34) | 0.212 | 1.08 (0.80, 1.46) | 0.633 | 1.12 (0.87, 1.44) | 0.397 | 1.08 (0.89, 1.31) | 0.444 | |
| rs763361_A (CD226) | 0.86 (0.73, 1.01) | 0.070 | 0.84 (0.67, 1.07) | 0.158 | 1.00 (0.78, 1.29) | 1.000 | 1.12 (0.92, 1.35) | 0.265 | |
| rs1990760_G (IFIH1) | 0.99 (0.83, 1.18) | 0.928 | 1.22 (0.93, 1.59) | 0.145 | 0.87 (0.67, 1.12) | 0.282 | 1.12 (0.92, 1.37) | 0.255 | |
| rs11203203_A (UBASH3A) | 1.19 (1.01, 1.40) | 0.041 | 1.01 (0.77, 1.33) | 0.930 | 1.35 (1.06, 1.72) | 0.014 | 1.05 (0.87, 1.27) | 0.613 | |
| rs4948088_A (COBL) | 0.83 (0.52, 1.33) | 0.440 | 1.01 (0.47, 2.20) | 0.977 | 0.77 (0.41, 1.45) | 0.418 | 1.27 (0.77, 2.09) | 0.343 | |
| rs2327832_G (TNFAIP3) | 0.91 (0.74, 1.13) | 0.419 | 0.81 (0.58, 1.12) | 0.199 | 0.98 (0.70, 1.37) | 0.904 | 1.40 (1.12, 1.74) | 0.003 | |
| rs7202877_C (CTRB2) | 1.00 (0.79, 1.27) | 0.999 | 0.96 (0.68, 1.34) | 0.805 | 1.13 (0.78, 1.64) | 0.512 | 1.13 (0.86, 1.47) | 0.382 | |
| rs11258747_A (PRKCQ) | 1.10 (0.92, 1.33) | 0.298 | 1.14 (0.86, 1.51) | 0.365 | 1.04 (0.80, 1.36) | 0.749 | 0.96 (0.78, 1.18) | 0.713 | |
| rs12251307_A (RBM17, IL2RA) | 0.68 (0.51, 0.90) | 0.007 | 0.81 (0.54, 1.22) | 0.314 | 0.61 (0.39, 0.94) | 0.026 | 1.03 (0.77, 1.39) | 0.839 | |
| rs11755527_C (BACH2) | 1.11 (0.94, 1.30) | 0.224 | 1.44 (1.12, 1.84) | 0.004 | 0.92 (0.72, 1.19) | 0.546 | 0.85 (0.70, 1.03) | 0.101 | |
| rs7804356_G (SKAP2) | 0.87 (0.71, 1.07) | 0.189 | 0.92 (0.68, 1.23) | 0.574 | 0.72 (0.52, 0.98) | 0.038 | 1.26 (1.01, 1.58) | 0.043 | |
| rs12444268_A | 1.25 (1.05, 1.50) | 0.014 | 1.45 (1.09, 1.92) | 0.010 | 1.08 (0.83, 1.40) | 0.557 | 0.88 (0.71, 1.08) | 0.219 | |
| rs1534422_G (MIR3681HG) | 0.90 (0.76, 1.06) | 0.192 | 0.73 (0.56, 0.94) | 0.016 | 0.91 (0.71, 1.16) | 0.428 | 1.32 (1.09, 1.60) | 0.005 | |
| rs3825932_A (CTSH) | 0.92 (0.78, 1.08) | 0.306 | 0.83 (0.65, 1.06) | 0.139 | 1.01 (0.80, 1.29) | 0.907 | 0.76 (0.62, 0.93) | 0.007 | |
| rs1004446_A (INS) | 0.91 (0.75, 1.09) | 0.304 | 0.85 (0.64, 1.13) | 0.263 | 1.09 (0.82, 1.46) | 0.554 | 0.81 (0.65, 1.00) | 0.053 | |
| PC1† | 1.17 (0.95, 1.45) | 0.137 | 1.03 (0.78, 1.36) | 0.838 | 1.42 (0.99, 2.03) | 0.059 | 0.77 (0.60, 0.99) | 0.044 | |
| PC2† | 1.19 (0.98, 1.45) | 0.075 | 1.30 (1.04, 1.63) | 0.024 | 1.10 (0.78, 1.54) | 0.597 | 0.94 (0.75, 1.19) | 0.604 | |
Adjusted HRs were estimated from multivariable proportional hazards models. ab, autoantibody; Multiple ab+, positive for multiple autoantibodies; Multiple vs. single ab+, positive for multiple autoantibodies or a single autoantibody.
The Effect of Age
The effect of increasing age at seroconversion was highly statistically significantly associated with a decrease in the risk of seroconverting (and the type of first appearing autoantibody), progression from a single autoantibody to multiple autoantibodies, and progression from multiple autoantibodies to type 1 diabetes. Yet, progression from multiple autoantibodies to type 1 diabetes was not related to age of seroconversion with GADA-first (HR 0.99; 95% CI 0.62, 1.57; P = 0.97) but it was with IAA-first (HR 0.45; 95% CI 0.31, 0.67; P < 0.001) or when the initial seroconversion was to multiple autoantibodies (HR 0.44; 95% CI 0.28, 0.68; P < 0.001). Additionally, a closer inspection (Supplementary Figs. 1 and 2) reveals that among those children who did not progress within 2 years, the age effect was substantially reduced. That is, the hazard rate for IAA-first and that for progression from a single autoantibody to multiple autoantibodies was significantly higher in the first 2 years of follow-up than during the remainder follow-up period (Table 4), with the exception of the risk of developing GADA-first and the risk of progressing from multiple autoantibodies to type 1 diabetes.
Table 4
Annual hazard rate of development of IA, IAA-first, GADA-first, progression from single autoantibody to multiple autoantibodies, and progression from multiple autoantibodies to type 1 diabetes
| Annual hazard rate (95% CI) by follow-up time period | P | ||
|---|---|---|---|
| 0–2 years | >2 years | ||
| Development of IA | 0.018 (0.016, 0.020) | 0.011 (0.010, 0.012) | <0.001 |
| Development of IAA-first | 0.010 (0.008, 0.012) | 0.0030 (0.0026, 0.0036) | <0.001 |
| Development of GADA-first | 0.0046 (0.0036, 0.0058) | 0.0058 (0.0052, 0.0066) | 0.07 |
| Progression from single autoantibody to multiple autoantibodies | 0.30 (0.27, 0.34) | 0.05 (0.04, 0.07) | <0.001 |
| Progression from multiple autoantibodies to type 1 diabetes | 0.12 (0.10, 0.15) | 0.14 (0.12, 0.16) | 0.32 |
Data are presented assuming exponential survival distribution.
Conclusions
Type 1 diabetes is a chronic disease characterized by the loss of functional pancreatic islet β-cells. Appearance of islet autoantibodies preceded the clinical disease for a highly variable time from months to >15 years. In 2015, it was proposed that the disease is a continuum of identifiable stages prior to the clinical diagnosis of diabetes (1). Stage 1 was defined as the presence of two or more islet autoantibodies with normoglycemia, stage 2 as two or more islet autoantibodies with dysglycemia, and stage 3 as onset of clinical disease. Clinical intervention would be needed to prevent subsequent morbidity and mortality. It has also been noted that individuals at risk progress through these stages at different rates, determined in part by the age at which seroconversion to autoantibody positivity occurred and the number of autoantibodies present (3,24–27). Results of TEDDY (20) and TrialNet (10) have shown that some individuals who seroconvert to a single autoantibody will progress to having multiple autoantibodies and have subsequent increased type 1 diabetes risk. The type 1 diabetes risk in those who do not progress to multiple autoantibodies remains elevated as compared with those who have not seroconverted, as reported in TEDDY and elsewhere (28). Predictors for the initial occurrence of autoantibodies and for the progression to type 1 diabetes differ.
First, although family history of type 1 diabetes and HLA (at least among the high-risk genotypes included in TEDDY) are highly related to the initial seroconversion and progression to multiple autoantibodies, a major finding is that this association is lost with progression from multiple autoantibodies (stage 1 disease) to type 1 diabetes (stage 3 disease). It is notable that family history of type 1 diabetes as a predictor for progression from single to multiple autoantibodies is related to whether the affected family member is a sibling (for IAA-first) or a father (for either IAA-first or GADA-first). It has long been known that the risk for type 1 diabetes is eight times higher if a sibling has the disease compared with a fivefold increase when the father is affected. The present observation may suggest that this epidemiological observation may be explained by two different etiologies, one related to IAA-first and the other to GADA-first.
Second, age at seroconversion is a consistent predictor throughout, although for those with GADA-first, age is not significantly associated with progression to type 1 diabetes. An important finding is that risk of type 1 diabetes among those with multiple autoantibodies decreases with increasing age of initial seroconversion. These results confirm earlier findings in the German BABYDIAB study (2), and the Finnish Type 1 Diabetes Prediction and Prevention (DIPP) study (3,4). These studies were not limited to the TEDDY HLA-defined high-risk population, and the results combined therefore underline the importance of considering the type of initially appearing autoantibody and the age at development of multiple autoantibodies in evaluating predictors for progression to diagnosis of diabetes. One possible explanation may relate to the very infrequent appearance of IAA as the first appearing autoantibody after age 2 years and the relatively constant seroconversion to GADA after 5 years of age (11,13). It remains to be seen whether this trend remains for those developing multiple autoantibodies at older ages, since there are other reports (7) that describe decreasing type 1 diabetes risk with increasing age. With the caveats that TEDDY is a young population and follow-up is limited, it is clear, however, that the type 1 diabetes risk through the first two decades of life declines overall with increasing age at seroconversion and increasing time from seroconversion.
This study is not without its limitations. The relationship of predictors, age, and the progression through different type 1 diabetes stages might not be generalizable to other HLA-defined populations, even though we did not observe that HLA was associated with progression to diabetes diagnosis in children positive for multiple autoantibodies. Despite this study’s size of 8,502 children, the age range reported herein is still limited and there are still relatively few type 1 diabetes cases observed in the TEDDY cohort.
Yet, the TEDDY cohort represents, depending on country, ~40–50% of children expected to develop diabetes before 18 years of age (16). The age at screening for the presence of islet autoantibodies in both the general population and among FDR and its associated heterogeneity with respect to diabetes predictors (5,6) should be considered when subjects are to be enrolled in secondary prevention studies. It is of importance to note that ~10% of the TEDDY children had an FDR, a proportion consistent with the proportion with an FDR among population-based children and adolescents newly diagnosed with type 1 diabetes (29,30). The association between non-HLA genetic factors and the risk for a first appearing autoantibody, first observed after 6 (13,20) as well as 9 (11) years of follow-up, underscores the need for gene-environment interaction analyses, as INS polymorphism remained associated (protective) with IAA-first, but ERBB3_T does not, with an additional 5 years of follow-up. Conversely, CLEC16A was previously reported to be associated with IA and is now shown to be related to a protective effect for GADA-first. PTPN22 and SH2B3 remained associated with increased risk for either one. The INS gene polymorphism may be related to preproinsulin expression in the thymus and thereby affect central tolerance (31). CLEC16A regulates mitophagy and controls β-cell function (32). PTPN22 and SH2B3 polymorphisms are both associated with a number of autoimmune disorders including type 1 diabetes (33,34). A number of significant associations, not previously reported in TEDDY, have been found, for IA (CD226, IFIH1, UBASH3A), IAA-first (CTRB2), GADA-first (PRKCQ), and single autoantibody to multiple autoantibodies (SKAP2), and sometimes specific to whether there was IAA-first (MIR3681HG and rs12444268). SKAP2 and MIR3681HG were newly reported to be significantly associated with progression from multiple autoantibodies to type 1 diabetes. CTSH and TNFAIR3 had previously been reported to be significantly associated with progression to type 1 diabetes and remained significantly associated in this analysis.
Finally, caution should be exercised in interpreting statistically significant findings due to the number of comparisons that have been made (inflating the type 1 error). Additionally, the HRs reported herein with respect to IAA-first or GADA-first appearance are all derived separately from cause-specific models and assume independence of the event type. While some findings reported are novel, perhaps reflecting the increasing age of the cohort, others are confirmed in independently reported studies with a different population, lending strength to their credence. The continuous follow-up of the TEDDY children expanding the longitudinal observations with more observed cases of seroconversion and type 1 diabetes suggests confirmation of those relationships that have remained consistent after 6 (13,21), 9 (11), and now 12 years of follow-up. Adjusting the significance level for multiple comparisons in conducting epidemiological research, especially in the context of a multivariate analysis, has both supporters (35) and detractors (36,37). No matter what side of the argument the reader falls on, for proper interpretation the associations reported herein should be viewed in the larger context of the results of other studies and other populations.
Article Information
Acknowledgments. The authors thank Sarah Austin-Gonzalez with the Health Informatics Institute at the University of South Florida for assistance with preparing the figures. A special acknowledgment goes out to the TEDDY families for their continued participation in this wonderful study.
Funding. The TEDDY study is funded by grants U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, U01 DK128847, and HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, Centers for Disease Control and Prevention, and JDRF. This work was supported in part by the National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Awards to the University of Florida [UL1 TR000064] and the University of Colorado [UL1 TR002535]).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. All authors attested to meeting International Committee of Medical Journal Editors uniform requirements for authorship by making substantial contributions to conception and design of the manuscript; acquisition, analysis, and interpretation of data; drafting or revising the manuscript for intellectual content; and giving final approval of the published version. J.P.K. designed the study, proposed the analysis, interpreted the findings, and wrote the manuscript. X.L. performed the analysis and contributed to the manuscript. Å.L., W.A.H., M.J.R., J.-X.S., J.T., A.-G.Z., and B.A. designed the study and reviewed and edited the manuscript. J.P.K. and X.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Clinical trial reg. no. NCT00279318, clinicaltrials.gov
This article contains supplementary material online at https://doi.org/10.2337/figshare.20278056.
*A complete list of the TEDDY Study Group can be found in Supplementary Material.
Contributor Information
TEDDY Study Group
:
References
Articles from Diabetes Care are provided here courtesy of American Diabetes Association
Full text links
Read article at publisher's site: https://doi.org/10.2337/dc21-2612
Read article for free, from open access legal sources, via Unpaywall:
https://diabetesjournals.org/care/article-pdf/45/10/2271/688660/dc212612.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/136347630
Article citations
Evolving Concepts in Pathophysiology, Screening, and Prevention of Type 1 Diabetes: Report of Diabetes Mellitus Interagency Coordinating Committee Workshop.
Diabetes, 73(11):1780-1790, 01 Nov 2024
Cited by: 0 articles | PMID: 39167668
Review
Presentation and characteristics of children with screen-detected type 1 diabetes: learnings from the ELSA general population pediatric screening study.
BMJ Open Diabetes Res Care, 12(5):e004480, 26 Sep 2024
Cited by: 0 articles | PMID: 39327068 | PMCID: PMC11429353
Consensus guidance for monitoring individuals with islet autoantibody-positive pre-stage 3 type 1 diabetes.
Diabetologia, 67(9):1731-1759, 01 Sep 2024
Cited by: 1 article | PMID: 38910151 | PMCID: PMC11410955
Next-generation sequencing reveals additional HLA class I and class II alleles associated with type 1 diabetes and age at onset.
Front Immunol, 15:1427349, 09 Aug 2024
Cited by: 0 articles | PMID: 39185409 | PMCID: PMC11341356
Teplizumab's immunomodulatory effects on pancreatic β-cell function in type 1 diabetes mellitus.
Clin Diabetes Endocrinol, 10(1):23, 10 Aug 2024
Cited by: 0 articles | PMID: 39123252 | PMCID: PMC11316332
Review Free full text in Europe PMC
Go to all (23) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT00279318
SNPs (Showing 20 of 20)
- (5 citations) dbSNP - rs689
- (4 citations) dbSNP - rs3184504
- (4 citations) dbSNP - rs2292239
- (4 citations) dbSNP - rs2476601
- (4 citations) dbSNP - rs2327832
- (4 citations) dbSNP - rs11755527
- (4 citations) dbSNP - rs7804356
- (4 citations) dbSNP - rs12708716
- (3 citations) dbSNP - rs7202877
- (3 citations) dbSNP - rs763361
- (3 citations) dbSNP - rs4948088
- (3 citations) dbSNP - rs1990760
- (3 citations) dbSNP - rs11203203
- (3 citations) dbSNP - rs1004446
- (3 citations) dbSNP - rs1534422
- (3 citations) dbSNP - rs12444268
- (3 citations) dbSNP - rs3825932
- (3 citations) dbSNP - rs11258747
- (2 citations) dbSNP - rs12251307
- (1 citation) dbSNP - rs917997
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterisation of rapid progressors to type 1 diabetes among children with HLA-conferred disease susceptibility.
Diabetologia, 60(7):1284-1293, 31 Mar 2017
Cited by: 19 articles | PMID: 28364254
Genetic and Environmental Interactions Modify the Risk of Diabetes-Related Autoimmunity by 6 Years of Age: The TEDDY Study.
Diabetes Care, 40(9):1194-1202, 23 Jun 2017
Cited by: 111 articles | PMID: 28646072 | PMCID: PMC5566280
Associations between deduced first islet specific autoantibody with sex, age at diagnosis and genetic risk factors in young children with type 1 diabetes.
Pediatr Diabetes, 23(6):693-702, 18 Apr 2022
Cited by: 6 articles | PMID: 35403376 | PMCID: PMC9541564
Humoral beta-cell autoimmunity in relation to HLA-defined disease susceptibility in preclinical and clinical type 1 diabetes.
Am J Med Genet, 115(1):48-54, 01 May 2002
Cited by: 51 articles | PMID: 12116176
Review
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: UL1 TR000064
Grant ID: UL1 TR002535
NIDDK NIH HHS (20)
Grant ID: U01 DK063790
Grant ID: UC4 DK106955
Grant ID: UC4 DK063863
Grant ID: UC4 DK063865
Grant ID: UC4 DK117483
Grant ID: UC4 DK063829
Grant ID: UC4 DK063861
Grant ID: UC4 DK095300
Grant ID: U01 DK063836
Grant ID: U01 DK063863
Grant ID: U01 DK063865
Grant ID: U01 DK128847
Grant ID: UC4 DK063821
Grant ID: U01 DK063861
Grant ID: UC4 DK063836
Grant ID: UC4 DK100238
Grant ID: UC4 DK112243
Grant ID: U01 DK063821
Grant ID: U01 DK063829
Grant ID: U01 DK124166
NLM NIH HHS (1)
Grant ID: HHSN267200700014C
National Center for Advancing Translational Sciences (2)
Grant ID: UL1 TR000064
Grant ID: UL1 TR002535
National Institute of Diabetes and Digestive and Kidney Diseases (21)
Grant ID: U01 DK124166
Grant ID: U01 DK63863
Grant ID: UC4 DK100238
Grant ID: UC4 DK63829
Grant ID: UC4 DK63861
Grant ID: U01 DK63821
Grant ID: UC4 DK106955
Grant ID: U01 DK63790
Grant ID: U01 DK128847
Grant ID: UC4 DK63863
Grant ID: UC4 DK63865
Grant ID: U01 DK63865
Grant ID: UC4 DK117483
Grant ID: UC4 DK63821
Grant ID: U01 DK63829
Grant ID: U01 DK63836
Grant ID: U01 DK63861
Grant ID: UC4 DK112243
Grant ID: UC4 DK63836
Grant ID: HHSN267200700014C
Grant ID: UC4 DK95300