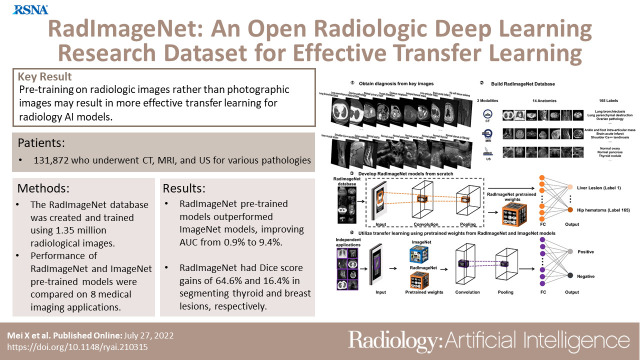
Abstract
Purpose
To demonstrate the value of pretraining with millions of radiologic images compared with ImageNet photographic images on downstream medical applications when using transfer learning.Materials and methods
This retrospective study included patients who underwent a radiologic study between 2005 and 2020 at an outpatient imaging facility. Key images and associated labels from the studies were retrospectively extracted from the original study interpretation. These images were used for RadImageNet model training with random weight initiation. The RadImageNet models were compared with ImageNet models using the area under the receiver operating characteristic curve (AUC) for eight classification tasks and using Dice scores for two segmentation problems.Results
The RadImageNet database consists of 1.35 million annotated medical images in 131 872 patients who underwent CT, MRI, and US for musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, abdominal, and pulmonary pathologic conditions. For transfer learning tasks on small datasets-thyroid nodules (US), breast masses (US), anterior cruciate ligament injuries (MRI), and meniscal tears (MRI)-the RadImageNet models demonstrated a significant advantage (P < .001) to ImageNet models (9.4%, 4.0%, 4.8%, and 4.5% AUC improvements, respectively). For larger datasets-pneumonia (chest radiography), COVID-19 (CT), SARS-CoV-2 (CT), and intracranial hemorrhage (CT)-the RadImageNet models also illustrated improved AUC (P < .001) by 1.9%, 6.1%, 1.7%, and 0.9%, respectively. Additionally, lesion localizations of the RadImageNet models were improved by 64.6% and 16.4% on thyroid and breast US datasets, respectively.Conclusion
RadImageNet pretrained models demonstrated better interpretability compared with ImageNet models, especially for smaller radiologic datasets.Keywords: CT, MR Imaging, US, Head/Neck, Thorax, Brain/Brain Stem, Evidence-based Medicine, Computer Applications-General (Informatics) Supplemental material is available for this article. Published under a CC BY 4.0 license.See also the commentary by Cadrin-Chênevert in this issue.Free full text

RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning
Abstract
Purpose
To demonstrate the value of pretraining with millions of radiologic images compared with ImageNet photographic images on downstream medical applications when using transfer learning.
Materials and Methods
This retrospective study included patients who underwent a radiologic study between 2005 and 2020 at an outpatient imaging facility. Key images and associated labels from the studies were retrospectively extracted from the original study interpretation. These images were used for RadImageNet model training with random weight initiation. The RadImageNet models were compared with ImageNet models using the area under the receiver operating characteristic curve (AUC) for eight classification tasks and using Dice scores for two segmentation problems.
Results
The RadImageNet database consists of 1.35 million annotated medical images in 131 872 patients who underwent CT, MRI, and US for musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, abdominal, and pulmonary pathologic conditions. For transfer learning tasks on small datasets—thyroid nodules (US), breast masses (US), anterior cruciate ligament injuries (MRI), and meniscal tears (MRI)—the RadImageNet models demonstrated a significant advantage (P < .001) to ImageNet models (9.4%, 4.0%, 4.8%, and 4.5% AUC improvements, respectively). For larger datasets—pneumonia (chest radiography), COVID-19 (CT), SARS-CoV-2 (CT), and intracranial hemorrhage (CT)—the RadImageNet models also illustrated improved AUC (P < .001) by 1.9%, 6.1%, 1.7%, and 0.9%, respectively. Additionally, lesion localizations of the RadImageNet models were improved by 64.6% and 16.4% on thyroid and breast US datasets, respectively.
872 patients who underwent CT, MRI, and US for musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, abdominal, and pulmonary pathologic conditions. For transfer learning tasks on small datasets—thyroid nodules (US), breast masses (US), anterior cruciate ligament injuries (MRI), and meniscal tears (MRI)—the RadImageNet models demonstrated a significant advantage (P < .001) to ImageNet models (9.4%, 4.0%, 4.8%, and 4.5% AUC improvements, respectively). For larger datasets—pneumonia (chest radiography), COVID-19 (CT), SARS-CoV-2 (CT), and intracranial hemorrhage (CT)—the RadImageNet models also illustrated improved AUC (P < .001) by 1.9%, 6.1%, 1.7%, and 0.9%, respectively. Additionally, lesion localizations of the RadImageNet models were improved by 64.6% and 16.4% on thyroid and breast US datasets, respectively.
Conclusion
RadImageNet pretrained models demonstrated better interpretability compared with ImageNet models, especially for smaller radiologic datasets.
Keywords: CT, MR Imaging, US, Head/Neck, Thorax, Brain/Brain Stem, Evidence-based Medicine, Computer Applications–General (Informatics)
Supplemental material is available for this article.
Published under a CC BY 4.0 license.
See also the commentary by Cadrin-Chênevert in this issue.
Introduction
ImageNet (1,2) is a dataset composed of millions of images of the natural world. ImageNet, as an open source dataset, has been a central resource for deriving sophisticated models in computer vision. Unlike ImageNet, publicly available medical imaging databases for research purposes are in scarcity because of the difficulty of curation, anonymization, or annotations of clinical data (3). Only a few well-curated annotated medical imaging datasets with high-quality ground truth pathologic labels are publicly available. The U.K. Biobank (4,5) contains multimodal images in more than 100 000 participants. However, most U.K. Biobank participants are healthy. The Cancer Imaging Archive (6) contains a large database of national lung screening trials, which consists of low-dose helical CT images in 53
000 participants. However, most U.K. Biobank participants are healthy. The Cancer Imaging Archive (6) contains a large database of national lung screening trials, which consists of low-dose helical CT images in 53 454 participants (7). The National Institutes of Health ChestX-ray8 contains 108
454 participants (7). The National Institutes of Health ChestX-ray8 contains 108 948 chest radiographs in 32
948 chest radiographs in 32 717 patients, with eight disease labels (8). Most current publicly available medical imaging datasets are limited in sample size, diversity of disease labels, or modality variety for artificial intelligence (AI) practice or lack pathologic findings.
717 patients, with eight disease labels (8). Most current publicly available medical imaging datasets are limited in sample size, diversity of disease labels, or modality variety for artificial intelligence (AI) practice or lack pathologic findings.
Limited sample size may create barriers to developing successful AI models. In cases of limited sample size, transfer learning (9) is a commonly used deep learning approach whereby a model designed for one problem can be reused to initiate a different but related task in deep learning. Due to the lack of annotated images and limited resources of computing power to train new models from scratch, transfer learning has become a popular method in deep learning, which can thus speed up the training process with fewer input data and improve the performance and generalizability of a deep learning model (10). Transfer learning with models trained using ImageNet has been extensively explored in medical imaging AI applications. The architectures of ResNet (11), Inception networks (12,13), and DenseNet (14) pretrained with ImageNet have been widely adopted and used in medical imaging applications for COVID-19 diagnosis at chest CT (15), classification of fibrotic lung disease (16), classification of skin cancer (17), and detection of acute intracranial hemorrhage (18).
Despite the high performance of many medical imaging models pretrained with ImageNet, previous works (19–21) have shown that pretrained models developed from medical source databases could achieve better performance than pretrained models from ImageNet. Successful transfer learning requires a reasonably large sample size, diversity of images, and similarity between the training and the target application images (22). In this study, we aim to create and evaluate a large-scale, diverse medical imaging dataset, RadImageNet, to generate pretrained convolutional neural networks (CNNs) trained solely from medical imaging to be used as the basis of transfer learning for medical imaging applications.
Materials and Methods
Study Patients
The institutional review boards waived the requirement for written informed consent for this retrospective, Health Insurance Portability and Accountability Act–compliant study, which evaluated de-identified data and involved no potential risk to patients. To avert any potential breach of confidentiality, no link between the patients, data provider, and data receiver was made available. A third party issued a certification of de-identified data transfer from the data provider to the data receiver. The RadImageNet dataset was collected between January 2005 and January 2020 from 131 872 patients at an outpatient radiology facility.
872 patients at an outpatient radiology facility.
Collection of RadImageNet Key Images
Each study was interpreted by a reading radiologist during daily clinical practice. A total of 20 board-certified, fellowship-trained radiologists participated in the original clinical interpretation. The radiologists had between 1 and 40 years of postfellowship experience at the time of clinical interpretation. As part of the interpretation of each study, the reading radiologist chose individual images representative of the pathologic finding or findings shown in each examination. The pathologic finding label was assigned to each of these “key images,” and a region of interest was created to localize the imaging findings. These original clinical interpretations were retrospectively extracted from the key images and provided the basis for the RadImageNet classes. The key images could be axial, sagittal, and/or coronal views and could include any sequence. The only requirement was that the image clearly represented substantial pathologic findings in the study.
Normal Studies
To better investigate the characteristics of abnormal key images for model development, 8528 normal studies with 263 039 images were included. Normal studies were identified on the basis of a SQL query of the picture archiving and communication system database, whereby studies with a report containing “normal” or “unremarkable” in the impression were flagged. The findings and impressions of these studies were reviewed by a board-certified radiologist to confirm that the reports did indicate a completely normal study. Studies with abnormal but not clinically significant findings were excluded. All diagnostic sequences and images of these studies were included.
039 images were included. Normal studies were identified on the basis of a SQL query of the picture archiving and communication system database, whereby studies with a report containing “normal” or “unremarkable” in the impression were flagged. The findings and impressions of these studies were reviewed by a board-certified radiologist to confirm that the reports did indicate a completely normal study. Studies with abnormal but not clinically significant findings were excluded. All diagnostic sequences and images of these studies were included.
Study Design
This study was designed in four phases (Fig 1). First, key images and associated diagnoses were annotated by radiologists. Second, the images and diagnoses were further grouped by modalities, anatomic regions, and labels according to their imaging patterns to construct the medical imaging–only database RadImageNet. Third, four neural networks as pretrained models were trained from scratch based on RadImageNet. Finally, the pretrained models from RadImageNet and ImageNet were used and compared on eight medical imaging applications using area under the receiver operating characteristic curve (AUC) values and Dice scores if ground truth segmentation masks were available.
RadImageNet Model Training
The same architectures of Inception-ResNet-v2 (13), ResNet50 (11), DenseNet121 (14), and InceptionV3 (12) networks were employed to train RadImageNet models from scratch by using randomly initiated weights as the starting point. The RadImageNet dataset was split into 75% training set, 10% validation set, and 15% test set. Images in the same patient were always included in the same set.
Rather than importing the weights from existing models, we randomly initiated the weights to develop the individual models. All images were resized to 224 × 224 pixels and used as the inputs of the neural networks. A global average pooling layer, a dropout layer at a rate of 0.5, and the output layer activated by the softmax function were added after the CNNs. The models returned a list of probabilities that the image corresponded to one of the 165 labels. The RadImageNet pretrained models and codes can be accessed at https://github.com/BMEII-AI/RadImageNet.
Comparison of RadImageNet and ImageNet Pretrained Models
We applied RadImageNet and ImageNet pretrained models for transfer learning on eight external downstream applications. The eight tasks included the following: classification between malignant and benign thyroid nodules at US with 288 malignant images and 61 benign images (23); classification between malignant and benign breast lesions at US with 210 malignant images and 570 benign images (24); anterior cruciate ligament (ACL) and meniscus tear detection at MRI with 570 ACL tear images, 452 non-ACL tear images, 506 meniscal tear images, and 3695 nonmeniscal tear images (25); pneumonia detection on chest radiographs including 6012 pneumonia chest radiographs and 20 672 nonpneumonia images (26); differentiation of patients with COVID-19 from community-acquired pneumonia with 21
672 nonpneumonia images (26); differentiation of patients with COVID-19 from community-acquired pneumonia with 21 872 COVID-19 images and 36
872 COVID-19 images and 36 894 community-acquired pneumonia images (27); classification between patients with and without COVID-19 at chest CT with 4190 COVID-19–positive and 4860 COVID-19–negative images (15); and hemorrhage detection at head CT with 107
894 community-acquired pneumonia images (27); classification between patients with and without COVID-19 at chest CT with 4190 COVID-19–positive and 4860 COVID-19–negative images (15); and hemorrhage detection at head CT with 107 933 hemorrhage images and 465
933 hemorrhage images and 465 671 nonhemorrhage images (28). Detailed class distributions of the downstream datasets are reported in Tables E3 and E4 (supplement). The RadImageNet and ImageNet models were evaluated using receiver operating curve analysis. A total of 24 scenarios were simulated to fine-tune the models for each application. The four CNNs were trained with varied learning rates and different numbers of freezing layers. Unfreezing of all layers was conducted with learning rates of 0.001 and 0.0001, while the freezing of all layers and unfreezing of the top 10 layers were conducted with learning rates of 0.01 and 0.001. The average AUC and SD of these 24 settings were compared between RadImageNet and ImageNet pretrained models.
671 nonhemorrhage images (28). Detailed class distributions of the downstream datasets are reported in Tables E3 and E4 (supplement). The RadImageNet and ImageNet models were evaluated using receiver operating curve analysis. A total of 24 scenarios were simulated to fine-tune the models for each application. The four CNNs were trained with varied learning rates and different numbers of freezing layers. Unfreezing of all layers was conducted with learning rates of 0.001 and 0.0001, while the freezing of all layers and unfreezing of the top 10 layers were conducted with learning rates of 0.01 and 0.001. The average AUC and SD of these 24 settings were compared between RadImageNet and ImageNet pretrained models.
Each downstream application dataset was split into 75% training set, 10% validation set, and 15% test set. Images in one patient were always in the same set. Binary cross-entropy was selected as the loss function. The input images were downscaled to 256 × 256 pixels for the trade-off between accuracy and efficiency. A global average pooling layer, a dropout layer, and an output layer activated by the softmax function were introduced after the last layer of the pretrained models. Models were trained for 30 epochs. The models with the lowest validation loss in such epochs were saved for further evaluation and comparison on the test set. Experiments on thyroid, breast, ACL, and meniscus applications were conducted using fivefold cross-validation because of the small size of the dataset. The distribution and data split for each application are shown in Tables E3 and E4 (supplement).
Gradient-weighted Class Activation Mapping
To understand the model interpretability, we used gradient-weighted class activation mapping (Grad-CAM) to visualize where the models make predictions in an image. Grad-CAM highlights the important regions in an image by using the gradients of the target layer that flows into the final convolutional layer to generate a localization map (29). For both RadImageNet and ImageNet models, the output layer was the target layer, whereas conv_7b_ac, conv5_block3_out, relu, and mixed10 were each selected as the final convolutional layer to generate the Grad-CAM for the Inception-ResNet-v2, ResNet50, DenseNet121, and InceptionV3 networks, respectively. Due to the varied recognition rates of CNN models for thyroid and breast classifications, a threshold of 180 was used to calculate Dice scores. This means that pixel values of a Grad-CAM that were greater than 180 for thyroid and breast US images were considered as predicted positives for further calculation.
Statistical Analysis
The paired t test (30) was used to calculate the two-sided P value comparing Dice scores between the RadImageNet and ImageNet models. Each image in the thyroid and breast datasets is independent. The normality of the distribution of Dice scores generated by RadImageNet and ImageNet models was confirmed by the Shapiro test (31,32). The DeLong method (33) was used to evaluate the 95% CI of the AUC and to calculate the two-sided P value for the comparison of RadImageNet and ImageNet models. Statistical significance was defined as a P value less than .05. The statistics of AUC comparisons were computed in the pROC package (1.18.0) in R (version 4.1.3; R Foundation for Statistical Computing) (34). The Shapiro test and paired t test were performed in the statsmodels package (0.13.2) in Python 3.8.5.
Results
The RadImageNet Database
The RadImageNet dataset includes 1.35 million annotated CT, MRI, and US images of musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, and pulmonary pathologic findings. For direct comparison with ImageNet (the initial size for the ImageNet challenge was 1.3 million images), we collected the most frequent modalities and anatomic regions on the same scale. The RadImageNet database used for comparison to ImageNet consists of three radiologic modalities, eleven anatomic regions, and 165 pathologic labels (Fig 2 and Table E1 [supplement]). The performance of the models on the test set is reported in Figure E1 and Table E2 (supplement).

Representative images and data structure of the RadImageNet database. (A) Overview of RadImageNet modalities and anatomic regions. RadImageNet was constructed with CT, MRI, and US images, including CT of the chest, abdomen, and pelvis, MRI of the ankle, foot, knee, hip, shoulder, brain, spine, abdomen, and pelvis, and US of the abdomen, pelvis, and thyroid. These images represent the diversity and fundamental structure of the RadImageNet database. (B–D) The components of the RadImageNet database subdivided by modalities, anatomic regions, classes, and number of associated images within each anatomic region for (B) CT studies, (C) US studies, and (D) MRI studies.
Performance on Small Datasets
The thyroid dataset contains 349 US images with radiologist-generated annotations collected from an open access thyroid image dataset (23). The breast dataset includes 780 breast US images (24) acquired for the detection of breast cancer. The knee MRI dataset consists of 1021 ACL tear and 4201 meniscal tear images (25). RadImageNet models demonstrated average AUCs of 0.85 ± 0.09 (SD) (P < .001), 0.94 ± 0.05 (P < .001), 0.97 ± 0.03 (P < .001), and 0.96 ± 0.02 (P < .001), compared with ImageNet models that showed values of 0.76 ± 0.14, 0.90 ± 0.10, 0.91 ± 0.08, and 0.92 ± 0.06 on the thyroid, breast, ACL tear, and meniscal tear datasets, respectively (Fig 3). The difference in AUCs between RadImageNet and ImageNet and the associated 95% CIs for the thyroid, breast, ACL tear, and meniscal tear datasets were 9.4% (2.6%, 16.2%), 4.0% (−0.6%, 8.7%), 4.8% (1.7%, 8.8%), and 4.5% (1.8%, 7.1%), respectively.

Performance of the RadImageNet pretrained models and ImageNet pretrained models on small datasets. The first column demonstrates swamp plots of 24 simulated experiments for RadImageNet and ImageNet models. The error bars represent the means and SDs on behalf of all area under the receiver operating characteristic curve (AUC) values. The second and third columns are receiver operating characteristic curves for the average of 24 simulations of RadImageNet models and ImageNet models, respectively. The shades indicate SDs obtained by averaging false-positive rates and true-positive rates from each experiment. The gains were calculated on the basis of the changes between RadImageNet and ImageNet models from ImageNet models. Two-sided P values were calculated by comparing the paired models between RadImageNet and ImageNet. AUC comparisons were evaluated by the DeLong test. (A) Thyroid US showed a 9.4% gain. (B) Breast US showed a 4.0% gain. (C) Anterior cruciate ligament (ACL) MRI showed a 4.8% gain. (D) Meniscus MRI showed a 4.5% gain.
Performance on Larger Datasets
The COVID-19 dataset consists of 9050 chest CT images in patients with and without COVID-19 pneumonia (15). The pneumonia dataset consists of 26 685 chest radiographs (26). The SARS-CoV-2 dataset consists of 58
685 chest radiographs (26). The SARS-CoV-2 dataset consists of 58 766 chest CT images with and without SARS-CoV-2 pneumonia (27). The hemorrhage dataset consists of 573
766 chest CT images with and without SARS-CoV-2 pneumonia (27). The hemorrhage dataset consists of 573 614 head CT images with and without intracranial hemorrhage (28). On these four applications, the RadImageNet models demonstrated average AUCs of 0.82 ± 0.03 (P < .001), 0.85 ± 0.02 (P < .001), 0.97 ± 0.03 (P < .001), and 0.91 ± 0.04 (P < .001), outperforming ImageNet models, which showed mean AUCs of 0.76 ± 0.09, 0.83 ± 0.05, 0.95 ± 0.07, and 0.91 ± 0.05, respectively (Fig 4). The differences in AUCs between RadImageNet and ImageNet and the 95% CIs were indicated as 6.1% (2.2%, 10.0%), 1.9% (−0.2%, 4.0%), 1.7% (−1.4%, 4.7%), and 0.9% (−1.7%, 3.4%) for detection of COVID-19, pneumonia, SARS-CoV-2, and hemorrhage, respectively.
614 head CT images with and without intracranial hemorrhage (28). On these four applications, the RadImageNet models demonstrated average AUCs of 0.82 ± 0.03 (P < .001), 0.85 ± 0.02 (P < .001), 0.97 ± 0.03 (P < .001), and 0.91 ± 0.04 (P < .001), outperforming ImageNet models, which showed mean AUCs of 0.76 ± 0.09, 0.83 ± 0.05, 0.95 ± 0.07, and 0.91 ± 0.05, respectively (Fig 4). The differences in AUCs between RadImageNet and ImageNet and the 95% CIs were indicated as 6.1% (2.2%, 10.0%), 1.9% (−0.2%, 4.0%), 1.7% (−1.4%, 4.7%), and 0.9% (−1.7%, 3.4%) for detection of COVID-19, pneumonia, SARS-CoV-2, and hemorrhage, respectively.
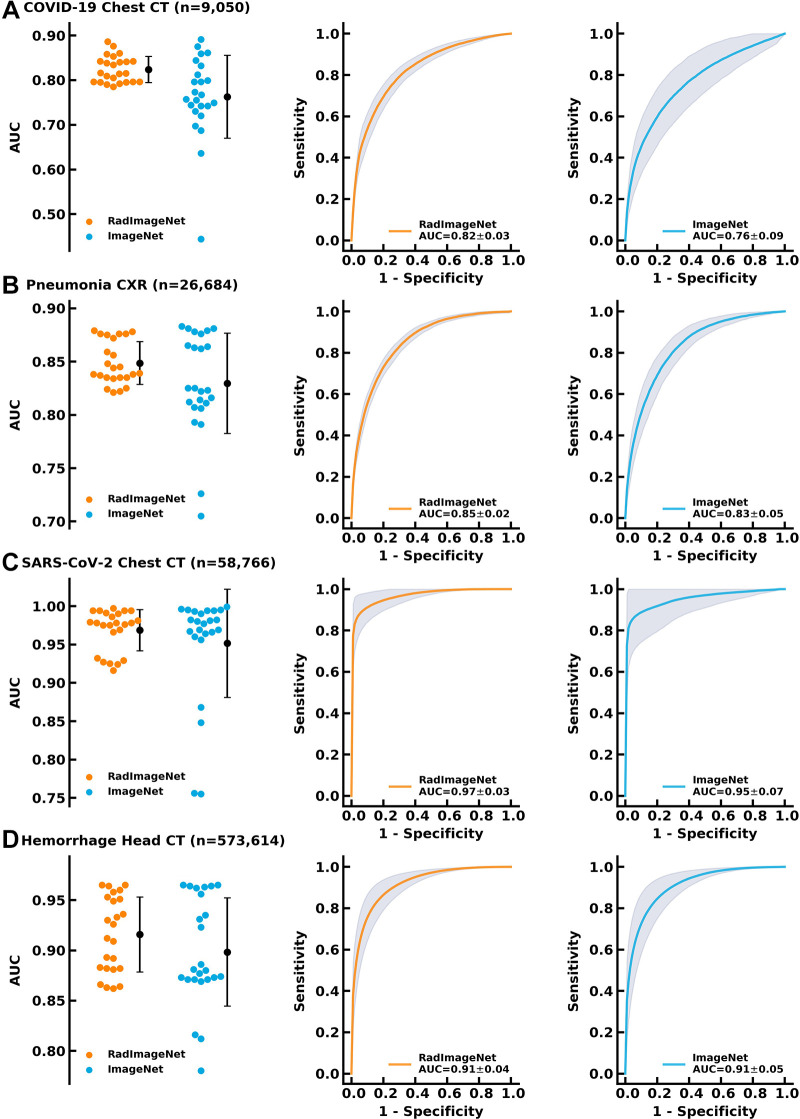
Performance of the RadImageNet pretrained models and ImageNet pretrained models on bigger datasets. The improvements from ImageNet are narrowed as compared with small datasets, but RadImageNet models are more consistent, showing smaller SDs among all simulations. (A) COVID-19 showed a 6.1% gain. (B) Pneumonia chest radiographs (CXR) showed a 1.9% gain. (C) SARS-CoV-2 CT showed a 1.7% gain. (D) Hemorrhage CT showed a 0.9% gain. AUC = area under the receiver operating characteristic curve.
Grad-CAM and Dice Scores
The thyroid and breast lesion US datasets contained graphical masks identifying the lesions. Dice score was used to evaluate the performance of the predicted CAM to the ground truth. Figure 5 shows CAM examples of RadImageNet models and ImageNet models. RadImageNet models achieved a Dice score of 0.29 for thyroid nodule detection, demonstrating a significant improvement of 64.6% from ImageNet models, which had a Dice score of 0.18 (P < .001). RadImageNet models illustrated a 0.16 Dice score for breast lesion detection, showing a significant gain of 16.4% from ImageNet models with a Dice score of 0.14 (P < .001).
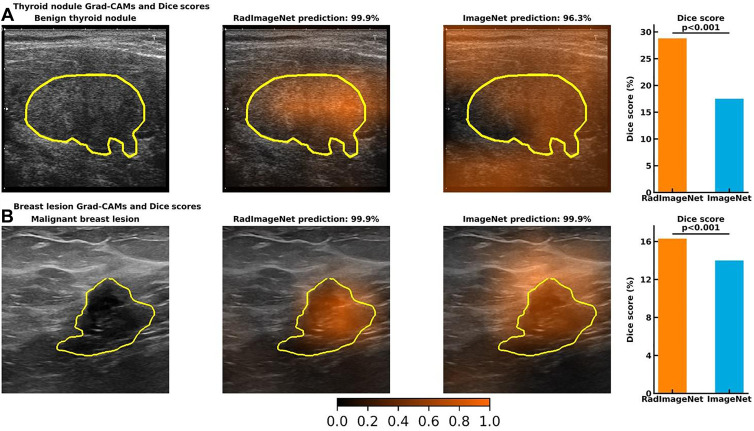
Visualizations of gradient-weighted class activation maps (Grad-CAMs) from accurately predicted images and Dice scores from the quantitative Grad-CAMs. The first column demonstrates the original image and the ground truth. The second and third columns show the Grad-CAMs from a RadImageNet model and an ImageNet model, respectively. The fourth column represents the average Dice score of 24 simulated RadImageNet and ImageNet models, with the error bar showing SDs. (A) A benign thyroid nodule. Both RadImageNet and ImageNet models accurately predicted the characteristics of this benign nodule, with the confidence of 99.9% and 96.3%, respectively. RadImageNet models achieved a Dice score of 28.8%, while ImageNet models had a Dice score of 17.5% (P < .001). (B) A malignant breast lesion. Both RadImageNet and ImageNet models accurately captured this malignant lesion, with confidence of 99.9% and 99.9%, respectively. RadImageNet models illustrated a 16.3% Dice score, outperforming ImageNet models, which showed a Dice score of 14.0% (P < .001). Two-sided P values for comparing the Dice scores were calculated by paired t test.
Discussion
In our study, the RadImageNet database contained only grayscale medical images, while natural world images use three red-green-blue channels. Pretraining on grayscale images can allow the training of more generalizable low-level filters in the initial layers of the network. The RadImageNet models demonstrated higher performance in imaging recognition and consistency over 24 simulated tuning scenarios regardless of the sample size of the applications. Within the 24 scenarios, unfreezing all layers consistently achieved the best performance as compared with unfreezing partial layers and training only fully connected layers. A smaller learning rate at 0.0001 would be suggested when training all trainable parameters to potentially better capture global optimal performance. If computational power and turnaround time allowed, Inception-ResNet-v2, ResNet50, and DenseNet121 achieved higher performance than Inception V3. However, the superiority of RadImageNet models is more evident on small datasets. For the four small datasets—thyroid US, breast US, and ACL and meniscus MRI—the RadImageNet models demonstrated a significant advantage over ImageNet models as demonstrated by a 9.4% (P < .001), 4.0% (P < .001), 4.8% (P < .001), and 4.5% (P < .001) AUC improvement, respectively. In addition, RadImageNet models are more stable. RadImageNet contained both the modality (US and MRI) and similar classes (normal thyroid and thyroid nodules, ACL injury and meniscus injury) to the target data, indicating that source data similarity can contribute to extraordinary performance with a small dataset. For the four relatively larger datasets—pneumonia detection at chest radiography (26 684 images), COVID-19 CT (9050 images), SARS-CoV-2 CT (58
684 images), COVID-19 CT (9050 images), SARS-CoV-2 CT (58 766 images), and intracranial hemorrhage detection CT (573
766 images), and intracranial hemorrhage detection CT (573 614 images)—the RadImageNet models also illustrated improvements of AUC by 1.9% (P < .001), 6.1% (P < .001), 1.7% (P < .001), and 0.9% (P < .001), respectively. RadImageNet contained both the modality (CT and MRI) and similar classes (infections on chest CT images and brain injuries on brain MRI studies) but with no radiography data involved. This indicated that even though the similarity to the target data was not high, the diversity of sourcing data and the larger sample size of the target data could compensate for model performance.
614 images)—the RadImageNet models also illustrated improvements of AUC by 1.9% (P < .001), 6.1% (P < .001), 1.7% (P < .001), and 0.9% (P < .001), respectively. RadImageNet contained both the modality (CT and MRI) and similar classes (infections on chest CT images and brain injuries on brain MRI studies) but with no radiography data involved. This indicated that even though the similarity to the target data was not high, the diversity of sourcing data and the larger sample size of the target data could compensate for model performance.
Moreover, RadImageNet models showed better generalizability. For the three applications (breast US, pneumonia chest radiography, and hemorrhage CT) in which the classes, anatomic regions, or modalities were not all included in the RadImageNet database, RadImageNet models demonstrated comparable performance by showing improvements of 4.0%, 1.9%, and 0.9%, respectively. These improvements confirmed that RadImageNet models are applicable to medical datasets regardless of modalities, anatomic regions, and classes.
In addition to the superiority of RadImageNet models in classification tasks, the interpretation of the RadImageNet models results more closely matched ground truth as compared with the ImageNet models. The quantification of the class activation maps for thyroid and breast images showed that RadImageNet models are more explainable and more consistent than ImageNet models, demonstrating improvements in Dice scores relative to ImageNet models of 64.6% and 16.4%, respectively. These results further confirmed that the similarity of the features learned from the RadImageNet database contributed to a higher recognition rate with better interpretability of the models (Fig 5).
Our proposed RadImageNet models did have limitations. First, the evaluation of pathologic features on a single image by radiologists at a single radiology facility and RadImageNet models did not mimic clinical diagnostic workflow, likely resulting in the similar, but poor, diagnostic performance by both. Second, the images presented may have contained multiple pathologic features, but we only used one label. Moreover, the region of interest placed on the primary pathologic finding by the reading radiologist was not used in this study. Therefore, it is possible the radiologists or RadImageNet models correctly matched a pathologic finding shown on the image but not the one that corresponded to the primary label. Third, the RadImageNet models used reduced-resolution images in algorithm development because of processing limitations. These lower-resolution images may obscure small areas of pathologic findings. Fourth, the current 165 categories were grouped on the basis of the International Classification of Diseases, Tenth Revision, and imaging characteristics, thus the RadImageNet models could not be diagnostic models to assist human experts. Finally, the number of classes in the limited RadImageNet dataset used for comparison to ImageNet was less than the number in ImageNet.
In conclusion, RadImageNet pretrained models could serve as a better starting point for transfer learning approaches in medical imaging analysis. In future studies, higher-spatial-resolution images could result in higher performance for recognition of smaller foci of pathologic features. Other imaging modalities such as radiography and PET should be included. The number of classes of pathologic findings in RadImageNet can be further expanded. Moreover, performance could be improved by introducing the regions of interest, as defined by radiologists, to highlight pathologic appearance in the images, as well as by providing additional sequences and/or adjacent images for the development of diagnostic models. Finally, more fine-tuning the pretrained models and comparing them with the standard pretrained models will be further analyzed.
Acknowledgments
We would like to thank the radiologists and IT staff at East River Medical Imaging. The RadImageNet database was provided by a third party. Requests for data should be directed to the provider at http://radimagenet.com. Public medical applications datasets are available as follows: (a) thyroid US: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/9287/92870W/An-open-access-thyroid-ultrasound-image-database/10.1117/12.2073532.full?SSO=1, (b) breast US: https://www.kaggle.com/aryashah2k/breast-ultrasound-images-dataset, (c) ACL and meniscus tear detection: https://stanfordmlgroup.github.io/competitions/mrnet/, (d) pneumonia detection: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge, (e) SARS-CoV-2 detection: http://ncov-ai.big.ac.cn/download?lang=en, and (f) intracranial hemorrhage detection: https://www.kaggle.com/c/rsna-intracranial-hemorrhage-detection.
Authors declared no funding for this work.
Disclosures of conflicts of interest: X.M. Member of Radiology: Artificial Intelligence trainee editorial board. Z.L. No relevant relationships. P.M.R. No relevant relationships. B.M. Consulting fees from Coridea. M.H. Portable knee MRI unit lent by Hyperfine to author’s institution, free of charge, for research purposes. A.D. No relevant relationships. A.J. No relevant relationships. C.C. No relevant relationships. K.E.L. Medical student research grant from Radiological Society of North America, payment made to NYU Langone; medical student scholarship from American College of Radiology, payment made to author. T.Y. No relevant relationships. Y.W. Grant support provided to author’s institution through NSF DMS-1752709 and NSF DMS-1720489 by the National Science Foundation. H.G. No relevant relationships. T.D. Radiologic data for the study was provided by RadImageNet and author is managing partner of RadImageNet. Z.A.F. Grants from Daiichi Sankyo, Amgen, Bristol Myers Squibb, and Siemens Healthineers; personal fees from Alexion, GlaxoSmithKline, and Trained Therapeutix Discovery; patents with Trained Therapeutix Discovery. Y.Y. Mount Sinai Seed Fund.
Abbreviations:
- ACL
- anterior cruciate ligament
- AI
- artificial intelligence
- AUC
- area under the receiver operating characteristic curve
- CNN
- convolutional neural network
- Grad-CAM
- gradient-weighted class activation mapping
References
Articles from Radiology: Artificial Intelligence are provided here courtesy of Radiological Society of North America
Full text links
Read article at publisher's site: https://doi.org/10.1148/ryai.210315
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9530758
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/133191815
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1148/ryai.210315
Article citations
Deep-learning model accurately classifies multi-label lung ultrasound findings, enhancing diagnostic accuracy and inter-reader agreement.
Sci Rep, 14(1):22228, 27 Sep 2024
Cited by: 0 articles | PMID: 39333570 | PMCID: PMC11437088
Overcoming data scarcity in biomedical imaging with a foundational multi-task model.
Nat Comput Sci, 4(7):495-509, 19 Jul 2024
Cited by: 1 article | PMID: 39030386 | PMCID: PMC11288886
Noise-induced modality-specific pretext learning for pediatric chest X-ray image classification.
Front Artif Intell, 7:1419638, 05 Sep 2024
Cited by: 0 articles | PMID: 39301479 | PMCID: PMC11410760
Construction and Validation of a General Medical Image Dataset for Pretraining.
J Imaging Inform Med, 15 Aug 2024
Cited by: 0 articles | PMID: 39147887
BrainSegFounder: Towards 3D foundation models for neuroimage segmentation.
Med Image Anal, 97:103301, 08 Aug 2024
Cited by: 1 article | PMID: 39146701
Go to all (45) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
RadImageNet and ImageNet as Datasets for Transfer Learning in the Assessment of Dental Radiographs: A Comparative Study.
J Imaging Inform Med, 24 Jul 2024
Cited by: 0 articles | PMID: 39048809
Exploring deep learning radiomics for classifying osteoporotic vertebral fractures in X-ray images.
Front Endocrinol (Lausanne), 15:1370838, 28 Mar 2024
Cited by: 0 articles | PMID: 38606087 | PMCID: PMC11007145
Construction and Validation of a General Medical Image Dataset for Pretraining.
J Imaging Inform Med, 15 Aug 2024
Cited by: 0 articles | PMID: 39147887
Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet.
PLoS Med, 15(11):e1002699, 27 Nov 2018
Cited by: 190 articles | PMID: 30481176 | PMCID: PMC6258509