Abstract
Objective
To determine the familiality of primary ovarian insufficiency (POI) at population level through examination of multigenerational genealogical information linked to electronic medical records.Design
Case-control study.Setting
Not applicable.Patient(s)
Women with POI were identified using International Classification of Disease 9 and 10 codes in electronic medical records (1995-2021) from 2 major health care systems in Utah and reviewed for accuracy. Cases were linked to genealogy information in the Utah Population Database (UPDB). All included POI cases (n = 396) were required to have genealogy information available for at least 3 generations of ancestors. The risk of POI in relatives was compared with population rates for POI matched by age, sex, and birthplace.Intervention(s)
Not applicable.Main outcome measure(s)
Relative risk of POI in first-, second-, and third-degree relatives.Result(s)
We identified 396 validated cases of POI with an associated 2,132 first-degree relatives, 5,245 second-degree relatives, and 10,853 third-degree relatives. We found an increased risk of POI among the extended relatives of cases. Specifically, first-degree relatives demonstrated an 18-fold increased risk of POI compared with controls relative risk ([RR],18.52 95% confidence interval [CI], 10.12-31.07), second-degree relatives demonstrated a 4-fold increase (RR, 4.21; CI, 1.15-10.79), and third-degree relatives demonstrated a 2.7-fold increase (RR, 2.65; CI, 1.14-5.21]).Conclusion(s)
This is the first population-based study to assess the familial clustering of POI. The data demonstrate excess familiality, familial clustering of POI in excess compared with matched population rates of disease, among first-, second-, and third-degree relatives. These findings support a genetic contribution to POI.Free full text

Primary Ovarian Insufficiency Has Strong Familiality: Results of a Multigenerational Genealogical Study
Abstract
Objective:
To determine the familiality of primary ovarian insufficiency (POI) on a population level through examination of multigenerational genealogical information linked to electronic medical records.
Design:
Case control study
Subjects:
Women with POI were identified using ICD9 and 10 codes in electronic medical records (1995–2021) from two major healthcare systems in Utah and reviewed for accuracy. Cases were linked to genealogy information in the Utah Population Database (UPDB). All included POI cases (n=396) were required to have genealogy information available for at least three generations of ancestors. The risk of POI in relatives was compared to population rates for POI matched by age, sex, and birthplace.
Main outcome measured:
Relative risk of POI in first-, second- and third-degree relatives.
Results:
We identified 396 validated cases of POI with an associated 2,132 first-degree relatives, 5,245 second-degree relatives and 10,853 third-degree relatives. We found an increased risk of POI among the extended relatives of cases. Specifically, first-degree relatives demonstrated an 18-fold increased risk of POI compared to controls (RR=18.52 [95% confidence interval 10.12–31.07]), second-degree relatives demonstrated a 4-fold increase (RR=4.21 [1.15–10.79]), and third-degree relatives demonstrated a 2.7-fold increase (RR=2.65 [1.14–5.21]).
Conclusion:
This is the first population-based study to assess the familial clustering of POI. The data demonstrate excess familiality, familial clustering of POI in excess compared to matched population rates of disease, among first-, second-, and third-degree relatives. These findings support a genetic contribution to POI.
Capsule:
In a multigenerational study, we found an increased risk of primary ovarian insufficiency among first-, second- and third-degree relatives of POI cases, suggesting that POI has strong familiality.
Introduction:
Although early menopause and primary ovarian insufficiency (POI) may be detrimental to fertility, these traits are heritable. There is a strong relationship between age at menopause in mothers and daughters, with an odds ratio (OR) of 6 (95% confidence intervals 3.4–10.7) for early menopause in daughters whose mothers had early menopause (1). One small clinical study demonstrated a 31% prevalence of familial POI in patients based on recall (2). A recent population study in Finland estimated an OR of 4.6 (3.3–6.5) for POI in mothers, daughters and sisters of women with POI (3). However, population studies have not been performed to determine the true prevalence of familial POI beyond assessment of first-degree relatives.
The etiology of POI is thought to be largely genetic and it is clear that some genetic causes are familial (4, 5). The most common familial genetic causes include FMR1 (FMRP translational regulator 1) premutations and autoimmune polyglandular syndromes (6,8). Rare recessive gene variants account for POI in consanguineous families, while rare heterozygous variants may be transmitted in a dominant fashion (4, 6–8). Polygenic risk scores created from genome-wide associated variants for age at natural menopause also demonstrate that common genetic variants partially contribute to POI presenting as early as 34 years (9). Therefore, examining POI in families may clarify patterns of inheritance and identify new risk genes when coupled with next generation sequencing studies.
Taken together, the heterogeneous genetics of POI and the potentially high prevalence of familial cases suggest that further examination of inheritance patterns may shed light on familial POI risk. Based on the detrimental effect of POI on fertility, the extent that POI is encountered in extended relatives needs investigation. We hypothesized that POI would exhibit excess familial clustering from first- through third-degree relatives when studied on a population level and would indicate patterns of inheritance. We addressed the hypothesis through examination of multigenerational genealogical information linked to individuals diagnosed with POI identified in electronic medical records.
MATERIALS AND METHODS
Cases
Women ≤40 years with POI were identified using the electronic medical records (EMR) from 1995–2021 at the University of Utah Health Science Center and Intermountain Healthcare. Together, these two health care systems serve approximately 85% of all residents in the state of Utah (10). Cases of POI were initially identified using International Classification of Disease (ICD)- 9 (256.3, 256.31, 256.39) and ICD-10 codes (E28.3, E28.31, E28.39, E28.310 and E28.319), EMR text indicating POI diagnoses, and/or consistent lab values (elevated FSH > 20 IU/L or AMH <0.08 ng/mL in a woman under the age of 40 years at the time of the laboratory draw).
We excluded patients who had medication, ICD or current procedural terminology (CPT) codes indicating hysterectomy, oophorectomy, endometriosis with pelvic surgery, pelvic radiation, or chemotherapy before the diagnosis of POI and those with rheumatologic disorders treated with cyclophosphamide. We also excluded ICD codes indicating Turner syndrome (ICD-9 758.6 and ICD-10 Q96). Following initial list generation, the charts of all probable cases were individually reviewed by a medical or reproductive endocrinologist (C.K.W. or L.E.V.) for appropriate inclusion into the subject group. Additional subjects were removed based on the exclusion criteria missed by automated exclusion. FSH and AMH levels were used to corroborate the diagnosis, removing diagnoses that were not POI, such as hypogonadotropic hypogonadism. Next, the specific encounter associated with the ICD code was reviewed paying close attention to the type of physician making the diagnosis and signs and symptoms of POI including vasomotor and genitourinary symptoms, irregular menses, osteoporosis and infertility.
Pedigree Creation
The Utah Population Database (UPDB) is a unique database that links genealogy information dating back to the 1800s to medical record information and other demographic data sources (10). In total, over 11 million individuals are represented in the UPBD and approximately 2.2M of those individuals have at least three generations of genealogical data available (11). This database allows for powerful linkage of multigenerational cohorts and identification of similar disease states among families. To use the UPDB, medical record numbers were converted to UPDB identification numbers by an independent oversight group. The UPDB IDs were then linked to genealogy information contained within the UPDB. For this multigenerational study, all subjects included in the final analysis were required to have at least three generations of genealogy information available (proband, both parents, and all four grandparents). Three generations of data make it more likely that complete family data is available for any woman.
Ethics
The University of Utah and Intermountain Healthcare Institutional Review Boards and the Resource for Genetic and Epidemiologic Research (RGE), overseers of UPDB data, approved this study.
Calculation of relative risk:
Familiality is the familial clustering of disease in significant excess compared to matched population rates of disease. To assess familial clustering, we estimated the relative risk of POI in first, second and third-degree relatives. Relative risk (RR) was estimated as the ratio of the observed number of cases of POI for a specific relative type (e.g., first-degree relatives) compared to the expected number of POI cases for the specific relative type. The expected number of affected female relatives was calculated based on population rates of POI. We calculated the population rate of POI for each 5-year birth cohort represented by the POI cases and birth place (Utah or outside of Utah) within the University of Utah Health Sciences and Intermountain Healthcare. The rate was defined by the total number of POI cases within a cohort divided by the total cohort size. The number of expected cases was calculated as the sum of each cohort -specific POI risk for each individual in a set of relatives of a specific type (e.g., first-degree relatives). The population rate will be conservative if POI cases in the population were not captured by our initial ICD 9 and 10 code screens. Approximate 95% confidence intervals and exact hypothesis tests of the null hypothesis (RR=1.0) were constructed assuming that the number of POI cases found among the relatives follows a Poisson distribution. Relative risk was calculated in first-, second- and third-degree relatives of cases. Of note, first-degree relatives are defined as mothers, sisters and daughters of cases. Second-degree relatives include grandmothers, aunts, nieces, half-sisters, and granddaughters. Third-degree relatives include great grandmothers, great granddaughters, and first cousins. Because medical record data only extends back to 1995, included third-degree relatives were mostly first cousins.
Calculation of the Genealogical Indexing of Familiality (GIF):
We tested for excess relatedness of POI using the Genealogical Index of Familiality (GIF) (12). The GIF statistic is a measure of the average pairwise relatedness of all possible pairs of POI cases compared to the average relatedness of 1000 sets of matched controls (13, 14). Control subjects were matched on sex, 5-year birth cohort, and birthplace (Utah or not). The GIF was specifically developed to evaluate relatedness among subjects in genealogical research and has been widely used in other large familiality studies (12, 15). The pairwise relatedness is measured using the Malecot coefficient of kinship, which measures the probability that a pair of individuals share a homologous piece of a chromosome identical-by-descent (IBD) from a common ancestor (11, 14) and multiple possible paths of relatedness are considered in the calculation (13). The 1000 control sets provide an empirical distribution to which the case GIF is compared to determine statistical significance. The GIF test determines whether the relatedness among cases is significantly greater than the relatedness of matched controls. As shared environmental factors can influence disease risk among more closely related individuals, we also calculated a distant GIF, which is calculated similarly to the GIF except first and second-degree relatives are eliminated from the analysis.
Identification of high-risk pedigrees:
We identified high-risk POI pedigrees as those pedigrees with a significant excess number of POI cases among descendants. We identified any shared ancestors of the POI cases and all related POI descendants of those ancestor founders. For pedigrees that completely overlapped, only the largest pedigree was retained. We compared the observed number of POI cases to the expected number of POI cases for all descendants based on population rates of POI matched for 5-year birth cohort, sex, and birth place (Utah or not). The expected number of POI cases for each pedigree was calculated as the cohort specific POI rate for each member of the pedigree, summed across all pedigree members. Pedigrees with a statistical excess (p<0.05) number of observed POI cases compared to the expected number of POI cases were called high-risk. The charts of these subjects were reviewed again to determine whether any cause of POI had been excluded or identified. For example, some subjects had data available for FMR1 premutation testing, karyotype and/or adrenal antibodies as part of the clinical assessment. The electronic records of the high-risk pedigrees were also further reviewed for possible affected relatives that were not identified in the ICD9/10 code-based searches. These additional individuals were not included in the familiality analyses but served to increase the number of affected individuals within the high-risk pedigrees.
RESULTS
The initial search at the University of Utah yielded 2,239 potential POI cases, with 1,440 (positive predictive value 0.64) confirmed POI upon chart review and 117 (5%) without sufficient data to confirm. The search at Intermountain Healthcare identified 2,682 POI cases, with 1,444 (PPV 0.54) confirmed to both have POI and be distinct from the University of Utah data set. The confirmed POI cases, age 31.3±8.0 years, with 98% born in Utah, were carried forward in the analysis (Supplementary Table 1).
After combining the University of Utah and Intermountain POI cases and linking them to genealogy information in the UPDB, there were 396 unique women with POI who also had three generations of genealogical data available in the UPDB. These 396 individuals diagnosed with POI had 2,132 first-degree relatives, 5,245 second-degree relatives and 10,853 third-degree relatives.
We observed an excess number of POI cases among relatives of our POI cohort compared with age, sex and birthplace matched controls (Table 1). Among first-degree relatives, we observed 14 cases of POI and expected to observe 0.76 cases. Specifically, we observed 2 mothers but expected 0.12, 11 sisters but expected 0.58 and 3 daughters but expected 0.06 (Supplementary Table 2). Among second-degree relatives, we observed four cases of POI and expected 0.95 cases. Among third-degree relatives, we observed 8 cases of POI and expected to observe 3.02 cases. In total, we found that 6.3% of our 396 POI cases had an affected relative defined as the proband having an affected first-, second-, or third-degree relative.
Table 1.
Heritability of primary ovarian insufficiency (POI) among relatives of women with POI (n=396).
| Relationship | Number | Relative Risk (95% confidence intervals) | p value |
|---|---|---|---|
| First Degree | 2,132 | 18.52 (10.12–31.07) | 1.13e–13 |
| Second Degree | 5,245 | 4.21 (1.15–10.79) | 0.016 |
| Third Degree | 10,853 | 2.65 (1.14–5.21) | 0.013 |
The GIF result that compares the overall relatedness of cases compared to the overall relatedness of 1000 matched control sets was significant (case GIF=5.24, control GIF=2.38; empirical p<0.0001). However, the distant GIF result that excludes first and second-degree relatives was not significant (empirical p=0.38) with the case GIF of 2.29 and control GIF of 2.24 (Figure 1). The largest contribution to the GIF statistic was from close relatives (1 meiosis is a mother-daughter relationship, 2 meioses are sisters or grandmother-granddaughter relationships, and 3 meioses is an aunt-niece relationship).
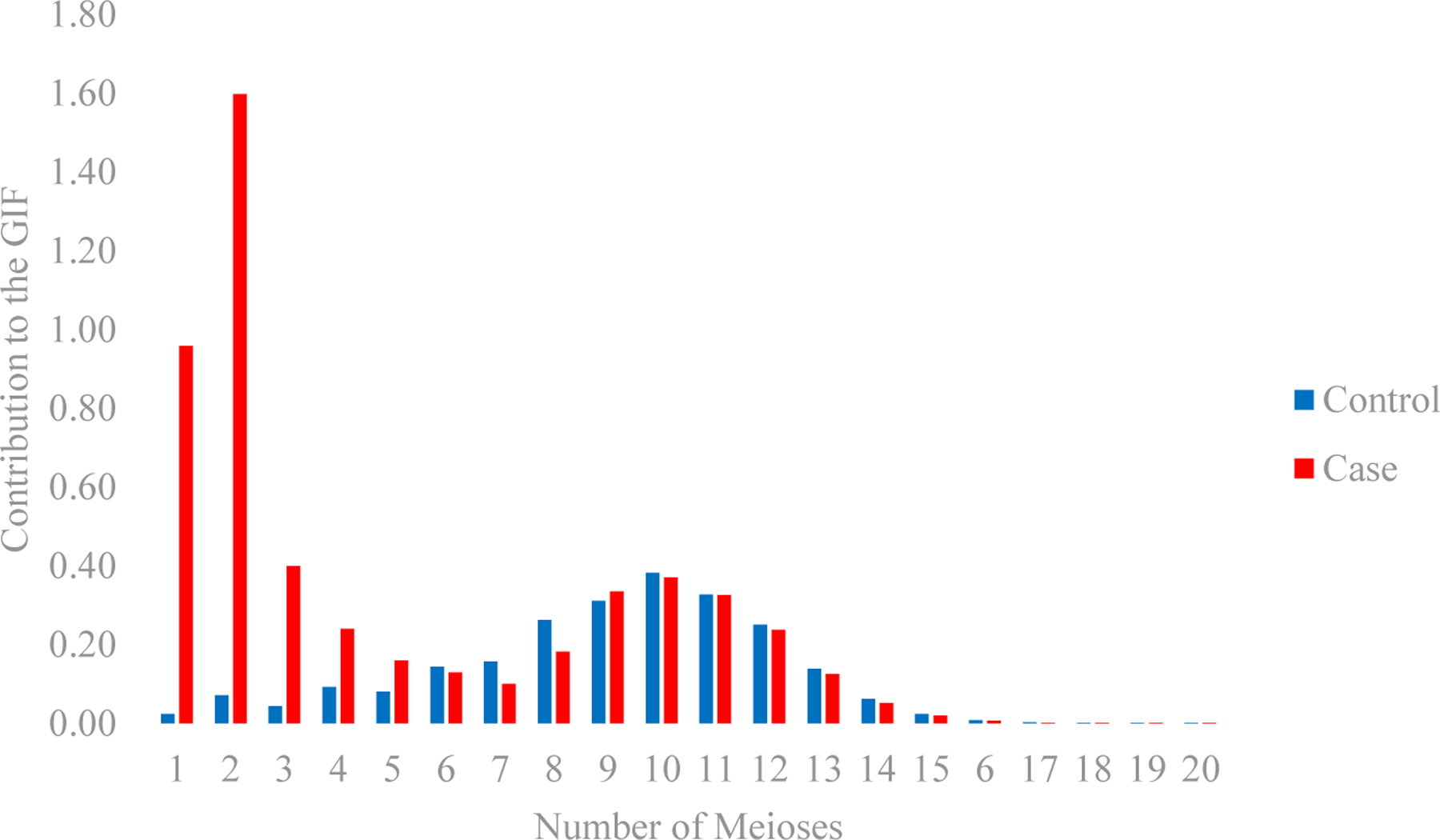
Contribution to the Genealogical Index of Familiality (GIF) Statistic for POI cases and 1000 sets of matched controls. The number of meiosis (x-axis) represents the pairwise genetic distance with reference to a particular proband: 1=parent or child, 2=sibling or grandparent, 3=aunt/uncle or niece/nephew, 4=first cousin or great aunt/uncle, 5=first cousin once removed, 6=second cousins.
Through chart review, we found additional affected first-degree (29 mothers, 14 sisters, 1 daughter) and second-degree relatives (3 maternal aunts, 2 maternal grandmothers, 2 maternal first cousins and 1 niece) that were not identified in our initial examination using ICD codes, EMR text and lab testing. These individuals were not included in the familiality analyses (Table 1) because of differences in ascertainment. However, they were added to the high-risk POI pedigree analysis to expand the identified high-risk pedigrees. Including these subjects, we identified 49 high risk pedigrees (Table 2).
Table 2.
Primary ovarian insufficiency (POI) hereditary patterns in high-risk families. Families with POI relatives up to 8 meioses were included.
| Family | Closest Relationships | Number | Age at Diagnosis (if available) | Mode of Inheritance | Family Diagnoses1 |
|---|---|---|---|---|---|
| First degree | 1–2 meiosis | 16 | 36.3±7.0 | 12 dominant or complex, 2 dominant or recessive, 2 unknown | Autoimmune (n=1)FMR1 premutation (n=1) Chr 9 inversion (n=1) |
| Second and Third degree | 3–8 meioses | 33 | 35.3±8.2 | Dominant (unclear if variable expressivity) or complex | FMR1 premutation (n=1) |
Twelve of the families had a possible dominant or complex mode of inheritance, with a mother and daughter affected (Table 2 and Supplementary Figure 1). Four families contained affected sister pairs, which could indicate dominant or recessive inheritance. Of the affected sister pairs, one sister had an inversion on chromosome 9 (46,XX,inv(9)(p12q13)) thought to be causal, although her sister was not tested based on likely causality. The closest relationship for the rest of the families was a third degree relative, which may represent a dominant inheritance pattern with/without incomplete expressivity or a complex inheritance pattern. There is evidence for female-only expressivity in some families with possible male carriers (Supplementary Figure 1). Of those distant relative families, the closest relationship for 4 families was 2nd cousins, for 5 families was 2nd cousins once removed, and for 21 families was 3rd cousins. One cousin in a 2nd cousin pair was diagnosed with an FMR1 premutation, although a more distantly related family member did not carry a premutation.
DISCUSSION
We present the first population-based study to evaluate the familial clustering of POI among multigenerational pedigrees. In our cohort, we observed an excess number of cases of POI among relatives of confirmed POI cases. The relationship appears to be strongest among first-degree relatives, although increased risk was also observed in second- and third-degree relatives. The data suggest that even when ovarian aging is aberrant, there is likely a familial component that may extend to distant relatives.
Previous POI heritability studies are limited by case identification, examination of first-degree relatives, only, or small numbers. In a Finnish population study, POI probands and affected first-degree relatives were identified by use of hormone therapy before age 40 years (3). While women with previous oophorectomy were excluded, women who had undergone treatment with chemotherapy or radiation therapy were not removed. Despite these differences, the prevalence of first-degree relatives with POI was very similar between the populations. In Utah, 3.5% of POI probands had an EMR documented POI case among first-degree relatives, whereas the rate in Finland was 2.6% (3). Further, the majority of the first-degree relationships were mother/daughter and the second most common was sharing among 2–3 sisters in both studies. Nevertheless, the RR of 18.52 (95% CI: 10.12–31.07) for POI in first-degree relatives in Utah was much higher than the OR of 4.6 (95% CI: 3.3–6.5) in Finland. Differences in risk may be attributable to the method of POI ascertainment. Finland had ten times the number of POI cases identified in Utah and may include iatrogenic POI, which would be less heritable. In the current study, we were able to expand family relationships to second- and third-degree relatives using a genealogic database, which has not previously been available on a population level. Limiting our analysis to those with three generations of relatives available ensured that we were able to accurately identify risks in third-degree relatives because it is more likely that complete data is available for the extended family in the genealogy database.
A previous study using a smaller data set was able to examine POI in multi-generational families (2). The prevalence of familial POI was 31% in this study of 71 selected women (2). However, the subjects were ascertained based on presentation to a reproductive endocrinology physician, perhaps more likely in a family with multiple affected, and familial case counts were based purely on proband report and did not have any confirmatory laboratory data or office visits. These methods raise a concern for selection bias of both cases and family members. Our data suggest approximately 6.3% of cases are familial based on the proband having an affected first-, second-, or third-degree relative, although the prevalence may be higher. As the additional affected family members identified in our chart review were not included in familial risk estimates, our familial risk assessment numbers and the percentage that are familial are conservative estimates of extended familial POI risk in the population. The identification of additional affected relatives in the high-risk POI pedigrees we identified further strengthens these pedigrees as being high-risk.
The high-risk family analysis demonstrates several potential patterns of inheritance. Families with mothers and daughters affected point to a potential dominant or complex inheritance pattern. The affected sister pairs could represent recessive inheritance along with dominant or complex inheritance. There is obligate transmission through males, which may indicate female-specific expressivity (Supplemental Figure 1). The more distant relative relationships point to complex inheritance or dominant inheritance with/without incomplete penetrance. These subjects are being recruited for further clarification.
In addition, although not examined in depth, a unifying cause of POI was suggested for a small number of families. In families with affected first- degree relatives, polyglandular endocrinopathies and FMR1 premutations were potential etiologies. These diagnoses are part of recommended testing and, therefore, not surprising (16). The families with a dominant or complex mode of inheritance may harbor causal POI variants in known or novel genes. These families will be recruited for future genetic studies.
One key strength of our study is the observation of excess risk in second- and third-degree relatives, which helps control for environmental factors that may be present among first degree relatives living in the same conditions. In addition, our data is comprised of subjects living throughout a large state and throughout a 26-year time frame, decreasing the likelihood of specific environmental exposures affecting study subjects.
Another strength of our study is the rigor of our chart review. ICD 9/10 codes are used for billing and are not intended for diagnosis. In addition, they are often used when the goal is to rule-out a diagnosis. Our experience suggests that the use of ICD codes without thorough chart review will misclassify spontaneous POI 35–45% of the time, as we discovered in two independent medical systems. While any large dataset is subject to misclassification bias, we believe bias was significantly reduced in our case review methodology and suggest that validation will be critical for EMR-based POI studies.
Our study includes a very large dataset with concomitant genealogy data. The large number of individuals with genealogy data in the UPDB strengthened our ability to detect a difference in familial POI rates and allowed us to analyze both close and distant relatives. The inclusion of the GIF statistic further strengthens the relatedness results of our cases compared to the general population. Error could arise from genealogy errors in linking relatives, either over or under linking persons. These errors are lessened by continuous maintenance and updating of family relationships by the Pedigree and Population Resource of Huntsman Cancer Institute at the University of Utah.
Our study is not without limitations. There were several subjects with EMR documentation that a mother or other family member had POI or early menopause. However, the electronic records of those mothers, sisters and other family members did not contain an ICD 9/10 code indicating POI diagnosis. The ICD 9/10 codes only capture medical care, typically in the setting of infertility, vasomotor symptoms, irregular menses or amenorrhea. As we saw in our review of high-risk families (Table 2), some patients with POI did not present for care and thus could not be included in the familiality analysis. Therefore, using ICD 9/10 diagnostic codes will underrepresent the true POI prevalence. Further, it is unusual for records to indicate relatives with POI on the paternal side, which also limits the ability to determine POI familial clustering. We did not include the additional subjects in our heritability analysis because we did not review the charts of population controls. If we had been able to include subjects ascertained through self-reported family history, our results may have been even stronger.
Of note, we used a broad definition of POI that included women with a final menses before the age of 45 years to ascertain additional affected relatives for high-risk pedigrees. Our approach is supported by previous studies demonstrating that POI and early menopause are found in the same pedigrees (17). POI has been defined as amenorrhea with a high FSH level before the age of 40 years, representing the earliest 1% of age at menopause (16). However, the distribution of age at menopause is continuous, making the statistical cutoff artificial. In addition, the architecture of POI includes recessive variants that cause primary amenorrhea, heterozygous rare variants accounting for early menopause across the spectrum of age and common variation partially explaining age at menopause starting at 34 years (4, 9). Taken together, an approach including ages that reflect the underlying genetic architecture rather than statistical cutoffs provides a physiologic basis for discovery, rather than using arbitrary cut points.
CONCLUSIONS
In summary, we identified strong familial clustering for primary ovarian insufficiency in a multigenerational, population-based cohort. This finding supports the concept that ovarian aging has a strong genetic component with heterogeneous inheritance patterns and emphasizes the importance of understanding the heritability of ovarian aging and risks for aberrant pathways.
Supplementary Material
Supplementary Information
Supplementary Figure
ACKNOWLEDGMENTS
We thank the Pedigree and Population Resource of Huntsman Cancer Institute, University of Utah (funded in part by the Huntsman Cancer Foundation) for its role in the ongoing collection, maintenance and support of the Utah Population Database (UPDB). We thank the University of Utah Center for Clinical and Translational Science (CCTS) (UL1TR002538), the Pedigree and Population Resource, University of Utah Information Technology Services and Biomedical Informatics Core for establishing the Master Subject Index between the Utah Population Database, the University of Utah Health Sciences Center and Intermountain Healthcare.
We also thank C. Matthew Peterson, MD, Megan Link, MD, and Joseph Letourneau, MD, for their assistance in patient recruitment. We thank M. Sean Esplin, MD, for research support at Intermountain Healthcare.
Funding Statement:
The work in this publication was supported by R56HD090159 and R01HD099487 (CKW). We also acknowledge partial support for the Utah Population Database through grant P30 CA2014 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure Statement: The authors have nothing to disclose.
Attestation Statement: Data regarding any of the subjects in the study has not been previously published unless specified. Data will be made available to the editors of the journal for review or query upon request.
REFERENCES
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/137592517
Article citations
Primary ovarian insufficiency: update on clinical and genetic findings.
Front Endocrinol (Lausanne), 15:1464803, 26 Sep 2024
Cited by: 0 articles | PMID: 39391877 | PMCID: PMC11466302
Review Free full text in Europe PMC
Female fertility preservation for family planning: a position statement of the Italian Society of Fertility and Sterility and Reproductive Medicine (SIFES-MR).
J Assist Reprod Genet, 41(9):2521-2535, 20 Jul 2024
Cited by: 0 articles | PMID: 39030346 | PMCID: PMC11405660
Family size for women with primary ovarian insufficiency and their relatives.
Hum Reprod, 38(10):1991-1997, 01 Oct 2023
Cited by: 1 article | PMID: 37632248 | PMCID: PMC10546072
DIS3 Variants are Associated With Primary Ovarian Insufficiency: Importance of Transcription/Translation in Oogenesis.
J Clin Endocrinol Metab, 108(9):2330-2335, 01 Aug 2023
Cited by: 0 articles | PMID: 36869713 | PMCID: PMC10686695
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Family size for women with primary ovarian insufficiency and their relatives.
Hum Reprod, 38(10):1991-1997, 01 Oct 2023
Cited by: 1 article | PMID: 37632248 | PMCID: PMC10546072
Population-based description of familial clustering of Chiari malformation Type I.
J Neurosurg, 128(2):460-465, 03 Feb 2017
Cited by: 17 articles | PMID: 28156254
Incidence and familial risk of premature ovarian insufficiency in the Finnish female population.
Hum Reprod, 37(5):1030-1036, 01 May 2022
Cited by: 8 articles | PMID: 35134918 | PMCID: PMC9071220
Significant evidence for a heritable contribution to cancer predisposition: a review of cancer familiality by site.
BMC Cancer, 12:138, 03 Apr 2012
Cited by: 40 articles | PMID: 22471249 | PMCID: PMC3350420
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR002538
NICHD NIH HHS (2)
Grant ID: R56 HD090159
Grant ID: R01 HD099487



