Abstract
Free full text

Mitochondrial Effects on the Physiological Characteristics of Lentinula edodes
Abstract
In the mating of filamentous basidiomycetes, dikaryotic mycelia are generated through the reciprocal movement of nuclei to a monokaryotic cytoplasm where a nucleus of compatible mating type resides, resulting in the establishment of two different dikaryotic strains having the same nuclei but different mitochondria. To better understand the role of mitochondria in mushrooms, we created four sets of dikaryotic strains of Lentinula edodes, including B2 ×
× E13 (B2 side) and B2
E13 (B2 side) and B2 ×
× E13 (E13 side), B5
E13 (E13 side), B5 ×
× E13 (B5 side) and B5
E13 (B5 side) and B5 ×
× E13 (E13 side), E8
E13 (E13 side), E8 ×
× H3 (E8 side) and E8
H3 (E8 side) and E8 ×
× H3 (H3 side), and K3
H3 (H3 side), and K3 ×
× H3 (K3 side) and K3
H3 (K3 side) and K3 ×
× H3 (H3 side). The karyotypes and mitochondrial types of the dikaryotic strains were successfully identified by the A mating type markers and the mitochondrial variable length tandem repeat markers, respectively. Comparative analyses of the dikaryotic strains on the mycelial growth, substrate browning, fruiting characteristics, and mitochondrial gene expression revealed that certain mitochondria are more effective in the mycelial growth and the production of fruiting body, possibly through the activated energy metabolism. Our findings indicate that mitochondria affect the physiology of dikaryotic strains having the same nuclear information and therefore a selection strategy aimed at mitochondrial function is needed in the development of new mushroom strain.
H3 (H3 side). The karyotypes and mitochondrial types of the dikaryotic strains were successfully identified by the A mating type markers and the mitochondrial variable length tandem repeat markers, respectively. Comparative analyses of the dikaryotic strains on the mycelial growth, substrate browning, fruiting characteristics, and mitochondrial gene expression revealed that certain mitochondria are more effective in the mycelial growth and the production of fruiting body, possibly through the activated energy metabolism. Our findings indicate that mitochondria affect the physiology of dikaryotic strains having the same nuclear information and therefore a selection strategy aimed at mitochondrial function is needed in the development of new mushroom strain.
1. Introduction
Mitochondria are important eukaryotic organelles that play central roles in cellular energy metabolism. They contribute to cell growth and differentiation by generating cellular signals and supplying chemical precursors of essential biological molecules [1–3]. They are also involved in programmed cell death through the intrinsic apoptotic pathways [2,3]. Given the importance of mitochondrial functions, inheritance of mitochondria has been one of the interesting subjects in eukaryotic genetics.
The mitochondrial DNA (mtDNA) is inherited mostly from the maternal origin in higher eukaryotes, independently of nuclear DNA because of its implicative bacterial origin [4]. However, the mtDNA inheritance in fungal systems is largely different from plants and animals. In an ascomycete yeast Saccharomyces cerevisiae, mitochondria in the daughter cell can inherit from one or both parents depending on the budding site [5,6], whereas basidiomycete yeasts, such as Cryptococcus neoformans and Ustilago maydis, show uniparental inheritance [7,8]. Mating type-specific transcription factors, Sxi1a/Sxi2a and Mat2 are shown to involve in the mitochondrial inheritance in C. neoformans [9,10]. The mitochondrial inheritance in the filamentous basidiomycetes is even more unique. During the mating between two monokaryotic cells through hyphal fusion, the nuclei in both monokaryotic cells undergo mitotic division and one of the divided nuclei enters into the partner monokaryotic cytoplasm reciprocally. The newly acquired nucleus undergoes further nuclear division and is transferred successively to the next connected monokaryotic cells through clamp connections to establish dikaryotic cells throughout the mycelia [11]. This process eventually produces two different types of dikaryotic cells which contain the same nuclei but differ in mitochondria originating from each mating monokaryotic cell.
Lentinula edodes is a filamentous basidiomycete and is one of the representative commercial mushrooms. Breeding of new cultivars through traditional mating with cultivated or wild strains of different genetic backgrounds has been of interest. We previously reported the diversity of mating types in the cultivated and wild strains of L. edodes together with the development of mating type markers which can represent the nuclear type of each nucleus in the dikaryotic cells [12,13]. We also found that mitochondria in the different strains of L. edodes carry different variations in the variable number tandem repeats (VNTR) through which the identity of a certain mitochondrial type can be verified [14]. Analyses of nuclear mobility and mitochondrial inheritance using the mating type marker and the mitochondrial VNTR marker in the mating between dikaryotic and monokaryotic strains (Di-Mon mating) have revealed that a specific nucleus inside the dikaryotic cell preferentially enters into the monokaryotic cytoplasm, in which mitochondria from monokaryotic strain reside, for the generation of a new dikaryotic strain [14].
Knowing that the nuclear and mitochondrial markers can identify the origins of the nucleus and mitochondria, the conventional mating was performed through the hyphal fusion of monokaryons and then the nuclear and the mitochondrial types in the newly formed dikaryon were analyzed using the nuclear and mtDNA markers. We found that the nuclei in the two monokaryon reciprocally transferred to the cytoplasm of the partner monokaryon where the mitochondria were preserved. This process inevitably generated two dikaryons with the same nuclei but different mitochondria. In the present study, we provide experimental evidence on this unique process and also provide the effect of the mitochondria on the phenotypic characteristics of mushroom mycelia and fruiting bodies.
2. Materials and methods
2.1. Strains and culture conditions
Monokaryotic strains were generated from basidiospores of L. edodes as previously described [12]. The monokaryotic B2 and B5 strains were from Chamaram cultivar, whereas E8 and E13 were from the SJ701 cultivar. The H3 and K3 strains were from the SJ707 and KFRI619 cultivars, respectively. Mating types of the monokaryotic strains were A5B12 for B2 strain, A5B11 for B5 strain, A1B4 for E8 and E13 strains, A5B12 for H3 strain, and A3B4 for K3 strain. The strains were grown on potato dextrose agar (PDA; Oxoid, Hampshire, UK) or in potato dextrose broth (PDB; BD Difco, Sparks, MD) at 25 °C.
°C.
2.2. Mating between monokaryotic strains
Mating between monokaryotic strains was performed on PDA plate by placing two mycelia of compatible mating types at a distance of 0.5 cm. The mating plate was incubated for 10 days at 25
cm. The mating plate was incubated for 10 days at 25 °C. The generation of dikaryotic strain through the mating was confirmed by the occurrence of clamp connections throughout the mycelia. The established dikaryotic strains were isolated at the farthest growing edges of opposite sides from the place where hyphal fusion occurred (Figure 1(A)).
°C. The generation of dikaryotic strain through the mating was confirmed by the occurrence of clamp connections throughout the mycelia. The established dikaryotic strains were isolated at the farthest growing edges of opposite sides from the place where hyphal fusion occurred (Figure 1(A)).
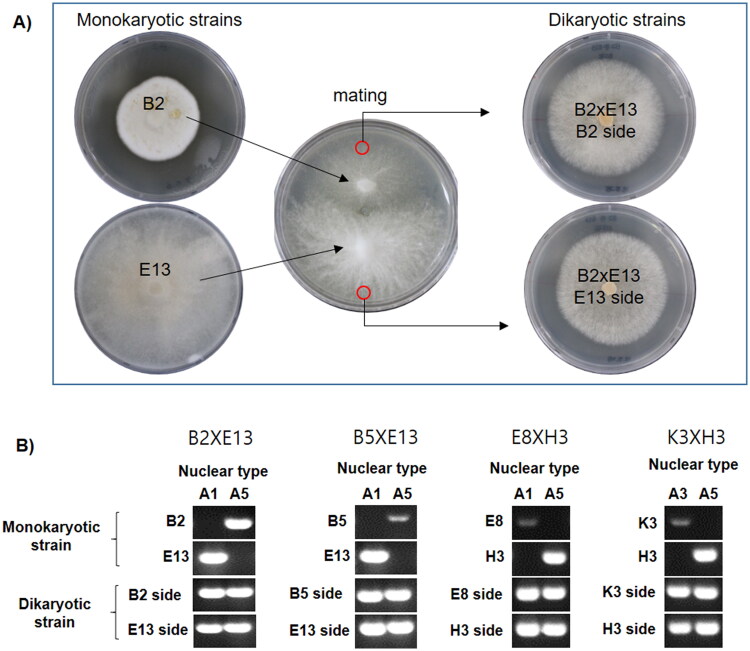
Generation of dikaryotic strains of Lentinula edodes. (A) Isolation of two dikaryotic strains having the same nuclei but different cytoplasm. The isolation sites of mated dikaryons are depicted as red circles. (B) Investigation of karyotypes using A mating types as nuclear markers. The nuclear types were confirmed by more than three independent PCR analysis.
2.3. Analysis of nuclear and mitochondrial types
The nuclear types in the monokaryotic and dikaryotic cells were identified by PCR using the primer sets (Supplementary Table S1) targeting the A mating type variable sequence region as previously reported [12]. The mitochondrial type inside the mycelial cells was identified using VNTR markers [14]. PCR analyses for the nuclear or mitochondrial typing were conducted using primer sets described in Supplementary Table S1. For the PCR analyses, total DNA was isolated using a genomic DNA extraction kit (AccuPrep Plant Genomic DNA Extraction Kit; Bioneer, Daejeon, Korea) from the mycelial powder, which was prepared by grinding frozen mycelia with a pestle and mortar. PCR was performed using the isolated DNA and the primer sets under the following conditions: initial incubation at 95 °C for 5
°C for 5 min, followed by 25 cycles of denaturation at 95
min, followed by 25 cycles of denaturation at 95 °C for 30
°C for 30 s, annealing at 55
s, annealing at 55 °C for 30
°C for 30 s, and polymerization at 72
s, and polymerization at 72 °C for 30
°C for 30 s, and final extension at 72
s, and final extension at 72 °C for 5
°C for 5 min. Analysis of the PCR product was conducted by agarose gel (2%) electrophoresis.
min. Analysis of the PCR product was conducted by agarose gel (2%) electrophoresis.
2.4. Analysis of mitochondrial gene expression
The mushroom mycelia, which were cultivated in PDB (50 mL) at 25
mL) at 25 °C for 7 days, were harvested and ground by a mortar and pestle in liquid nitrogen. Total RNA was extracted from the ground mycelia using an RNA extraction kit (AccuPrep Universal RNA Extraction Kit; Bioneer). cDNA for the mitochondrial genes were synthesized using TOPscript cDNA Synthesis Kit (Enzynomics, Daejeon, Korea) with a mixture of primers specific to mitochondrial genes (Supplementary Table S1). Droplet digital PCR (ddPCR) was conducted by QX200 Droplet Digital PCR system (Bio-rad, Hercules, CA). Each test was performed in 20
°C for 7 days, were harvested and ground by a mortar and pestle in liquid nitrogen. Total RNA was extracted from the ground mycelia using an RNA extraction kit (AccuPrep Universal RNA Extraction Kit; Bioneer). cDNA for the mitochondrial genes were synthesized using TOPscript cDNA Synthesis Kit (Enzynomics, Daejeon, Korea) with a mixture of primers specific to mitochondrial genes (Supplementary Table S1). Droplet digital PCR (ddPCR) was conducted by QX200 Droplet Digital PCR system (Bio-rad, Hercules, CA). Each test was performed in 20 µl of the reaction mixture, which consisted of 10
µl of the reaction mixture, which consisted of 10 µl of QX200 ddPCR EvaGreen Supermix (Bio-Rad) and 10
µl of QX200 ddPCR EvaGreen Supermix (Bio-Rad) and 10 µl of primers (100
µl of primers (100 nM) and cDNA template (1
nM) and cDNA template (1 ng) mixture. ddPCR was performed under the following conditions: initial denaturation at 95
ng) mixture. ddPCR was performed under the following conditions: initial denaturation at 95 °C for 5
°C for 5 min, 40 cycles of amplification reaction at 95
min, 40 cycles of amplification reaction at 95 °C for 30
°C for 30 s, and 60
s, and 60 °C for 60
°C for 60 s. The fluorescence signal was monitored using QX200 Droplet Reader (Bio-Rad). Each reaction with negative control (without template) was performed triplicate. The statistical analysis of the data sets was performed using one-way ANOVA test.
s. The fluorescence signal was monitored using QX200 Droplet Reader (Bio-Rad). Each reaction with negative control (without template) was performed triplicate. The statistical analysis of the data sets was performed using one-way ANOVA test.
2.5. Investigation of fruiting characteristics
For the fruiting of the dikaryotic L. edodes strains with different mitochondrial types, a sawdust substrate (1 kg) containing oak tree sawdust (380
kg) containing oak tree sawdust (380 g), rice bran (20
g), rice bran (20 g), and water (600
g), and water (600 ml) in a polyethylene (PE) bag was prepared after sterilization for 30
ml) in a polyethylene (PE) bag was prepared after sterilization for 30 min at 121
min at 121 °C. The substrate bag was incubated for 30
°C. The substrate bag was incubated for 30 days at 25
days at 25 °C in the dark after inoculation with the mushroom mycelia cultured in PDB (20
°C in the dark after inoculation with the mushroom mycelia cultured in PDB (20 ml). When the mycelia were fully propagated to the substrate, browning was induced by additional 60
ml). When the mycelia were fully propagated to the substrate, browning was induced by additional 60 days of incubation at 20
days of incubation at 20 °C under white light. After the browning, the PE bag was removed and the mycelial substrate mass was soaked in water for 12
°C under white light. After the browning, the PE bag was removed and the mycelial substrate mass was soaked in water for 12 h at 19
h at 19 °C to induce fruiting. The soaked substrate was then moved to a culture room where temperature and relative humidity were maintained at 23
°C to induce fruiting. The soaked substrate was then moved to a culture room where temperature and relative humidity were maintained at 23 °C and 80%, respectively. Fruiting bodies were harvested 15
°C and 80%, respectively. Fruiting bodies were harvested 15 days after the induction. The degree of browning was measured in triplicate using NIH ImageJ program (https://imagej.nih.gov/ij/) averaging the total surface color density of the substrate surface after browning. The color density of the pileus (cap) of fruiting bodies was also obtained in triplicate using the NIH ImageJ by measuring the color density ranging from pale brown to dark brown. For the calculation of the relative color density, the palest part of the cap among the test strains was assigned as “0” and the darkest part was as “1”.
days after the induction. The degree of browning was measured in triplicate using NIH ImageJ program (https://imagej.nih.gov/ij/) averaging the total surface color density of the substrate surface after browning. The color density of the pileus (cap) of fruiting bodies was also obtained in triplicate using the NIH ImageJ by measuring the color density ranging from pale brown to dark brown. For the calculation of the relative color density, the palest part of the cap among the test strains was assigned as “0” and the darkest part was as “1”.
3. Results
3.1. Generation of dikaryotic strains of the same nuclear composition but with different mitochondrial types
The dikaryotic strains generated by conventional mating were isolated at two different locations from the mating plate. For example, the monokaryotic strains B2 and E13 were cross-mated and the resulting dikaryotic strain B2 ×
× E13 was isolated at the B2 side and the E13 side, designated as B2
E13 was isolated at the B2 side and the E13 side, designated as B2 ×
× E13 (B2 side) and B2
E13 (B2 side) and B2 ×
× E13 (E13 side), respectively (Figure 1(A)). The establishment of dikaryon in the isolated mates was monitored through the detection of A mating type markers because each nucleus inside the dikaryotic cytoplasm carries its own A mating type thereby enabling the use of the A mating type as a marker of nuclear type. In the mating between the B2 and E13 strains, the A mating type analyses revealed that the B2 and E13 carried A5 and A1 mating type markers, respectively, whereas the dikaryotic strain B2
E13 (E13 side), respectively (Figure 1(A)). The establishment of dikaryon in the isolated mates was monitored through the detection of A mating type markers because each nucleus inside the dikaryotic cytoplasm carries its own A mating type thereby enabling the use of the A mating type as a marker of nuclear type. In the mating between the B2 and E13 strains, the A mating type analyses revealed that the B2 and E13 carried A5 and A1 mating type markers, respectively, whereas the dikaryotic strain B2 ×
× E13 carries both A5 and A1 mating type markers, regardless of the isolated positions (Figure 1(B)). The successful establishment of dikaryon was also confirmed in the B5
E13 carries both A5 and A1 mating type markers, regardless of the isolated positions (Figure 1(B)). The successful establishment of dikaryon was also confirmed in the B5 ×
× E13, E8
E13, E8 ×
× H3, and K3
H3, and K3 ×
× H3 (Figure 1(B)).
H3 (Figure 1(B)).
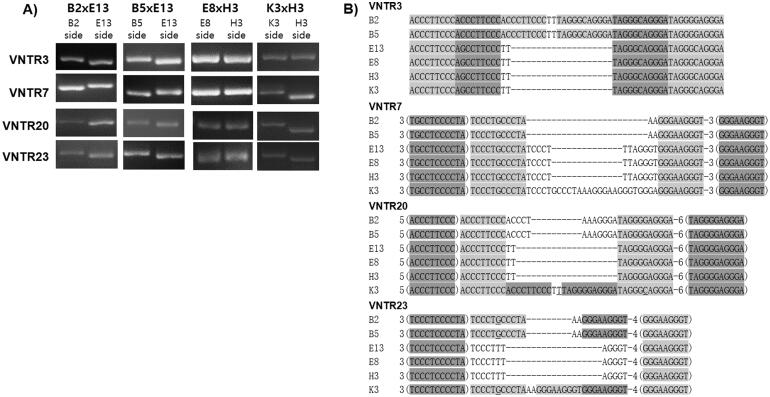
We next investigated the mitochondrial types of the monokaryotic and dikaryotic strains using the VNTR markers [14]. The marker analyses demonstrated that the mitochondrial type within the dikaryotic isolates from the same mating plate could be identified by the length difference (Figure 2(A)) as well as sequence difference (Figure 2(B)) in the VNTR markers, except for the E8 and H3 strains. The VNTR marker analyses revealed that the mitochondrial type within a mated dikaryotic strain was dependent on its location of the isolation. When the B2 ×
× E13 was isolated from the B2 side (Figure 1(A)), the dikaryotic cytoplasm contained the B2 type mitochondria, wherras the B2
E13 was isolated from the B2 side (Figure 1(A)), the dikaryotic cytoplasm contained the B2 type mitochondria, wherras the B2 ×
× E13 isolated from the E13 side carried the E13 type mitochondria (Figure 2). This also occurred in other dikaryotic strains (B5
E13 isolated from the E13 side carried the E13 type mitochondria (Figure 2). This also occurred in other dikaryotic strains (B5 ×
× E13 and K3
E13 and K3 ×
× H3), except for E8
H3), except for E8 ×
× H3 in which the VNTR marker failed to verify the mitochondrial types.
H3 in which the VNTR marker failed to verify the mitochondrial types.
3.2. Influence of mitochondria on the mycelial growth of dikaryotic strains
The mycelial growth of the dikaryotic strains isolated from the opposite sides of the mating plates was monitored for a week on PDA at 25 °C. The growth was largely different depending on the isolation site. The radial growth of the B2
°C. The growth was largely different depending on the isolation site. The radial growth of the B2 ×
× E13 (E13 side) was significantly faster than that of the B2
E13 (E13 side) was significantly faster than that of the B2 ×
× E13 (B2 side) (Figure 3). This was also observed in the B5
E13 (B2 side) (Figure 3). This was also observed in the B5 ×
× E13 (E13 side), E8
E13 (E13 side), E8 ×
× H3 (E8 side), and K3
H3 (E8 side), and K3 ×
× H3 (H3 side) over B5
H3 (H3 side) over B5 ×
× E13 (B5 side), E8
E13 (B5 side), E8 ×
× H3 (H3 side), and K3
H3 (H3 side), and K3 ×
× H3 (K3 side), respectively. The mycelial density of the B2
H3 (K3 side), respectively. The mycelial density of the B2 ×
× E13 (E13 side) was slightly higher than that of the B2
E13 (E13 side) was slightly higher than that of the B2 ×
× E13 (B2 side). The density difference was more evident in the B5
E13 (B2 side). The density difference was more evident in the B5 ×
× E13 strain. The B5 side isolate in the B5
E13 strain. The B5 side isolate in the B5 ×
× E13 showed slower growth but significantly higher density than the E13 side isolate. The E8
E13 showed slower growth but significantly higher density than the E13 side isolate. The E8 ×
× H3 (E8 side) showed fluffier growth than the E8
H3 (E8 side) showed fluffier growth than the E8 ×
× H3 (H3 side), whereas the K3
H3 (H3 side), whereas the K3 ×
× H3 showed similar growth morphology.
H3 showed similar growth morphology.
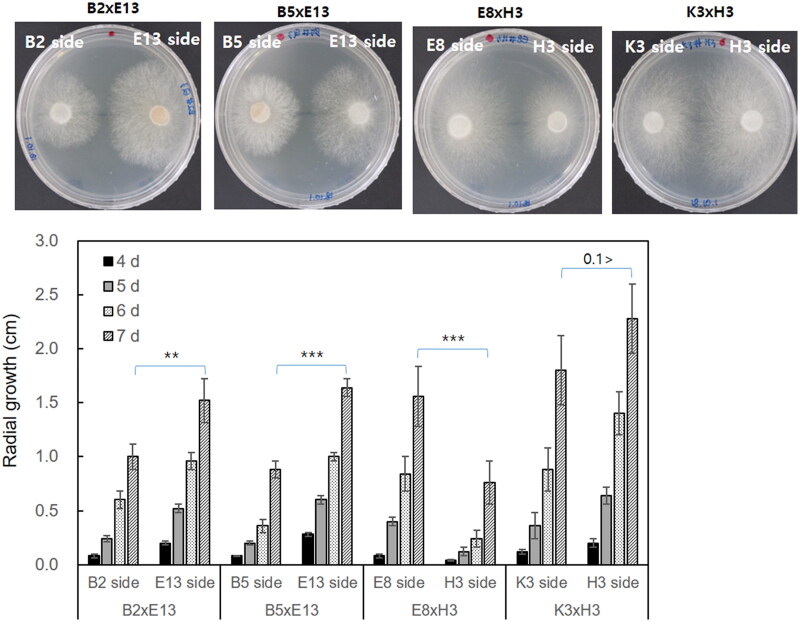
Comparison of mycelial growth of the dikaryotic strains. The dikaryotic strains having the same nuclei but different mitochondria were placed on the same PDA plates. The plates were incubated at 25 °C for seven
°C for seven days (upper panel, the mycelial growth after seven days of incubation) while monitoring the mycelial diameter in centimeter (bottom panel). The radial growth was measured after four days, five days, six days, and seven days of incubation. Error bars indicate the standard deviations of the means of triplicated data. The statistical significance was tested by one-way ANOVA and shown on the bar using asterisks (* for p
days (upper panel, the mycelial growth after seven days of incubation) while monitoring the mycelial diameter in centimeter (bottom panel). The radial growth was measured after four days, five days, six days, and seven days of incubation. Error bars indicate the standard deviations of the means of triplicated data. The statistical significance was tested by one-way ANOVA and shown on the bar using asterisks (* for p ≤
≤ 0.05, ** for p
0.05, ** for p ≤
≤ 0.01, *** for p
0.01, *** for p ≤
≤ 0.001, and **** for p
0.001, and **** for p ≤
≤ 0.0001).
0.0001).
3.3. Effect of mitochondria on the fruiting body formation
The mitochondrial effect on the fruiting characteristics of L. edodes was investigated by cultivation in the sawdust-based complex substrate with a commercial formulation using the eight dikaryotic strains described above. The mycelia were fully propagated after incubation for 30 d at 25 °C in the dark, regardless of the strains. The mycelial bags were further incubated for 60 d under light for browning, which is an essential procedure practiced in L. edodes cultivation for efficient substrate degradation and the fruiting body production through the expression of genes such as glucanase, chitinase, laccase, and hydrophobins [15]. As a result, we found that the browning was differently affected by mitochondrial types. The mitochondrial effect was the most eminent in the E8
°C in the dark, regardless of the strains. The mycelial bags were further incubated for 60 d under light for browning, which is an essential procedure practiced in L. edodes cultivation for efficient substrate degradation and the fruiting body production through the expression of genes such as glucanase, chitinase, laccase, and hydrophobins [15]. As a result, we found that the browning was differently affected by mitochondrial types. The mitochondrial effect was the most eminent in the E8 ×
× H3 strains in which the E8
H3 strains in which the E8 ×
× H3 (E8 side) strain showed a thorough browning (color density: 0.65
H3 (E8 side) strain showed a thorough browning (color density: 0.65 ±
± 0.05) on the surface of the substrate, whereas practically no browning was observed in E8
0.05) on the surface of the substrate, whereas practically no browning was observed in E8 ×
× H3 (H3 side) (color density: 0.08
H3 (H3 side) (color density: 0.08 ±
± 0.02) (Table 1, Supplementary Figure S1). The B5
0.02) (Table 1, Supplementary Figure S1). The B5 ×
× E13 strains also showed significant differences in degree of browning by exhibiting color densities of 0.59
E13 strains also showed significant differences in degree of browning by exhibiting color densities of 0.59 ±
± 0.08 and 0.43
0.08 and 0.43 ±
± 0.10, respectively, for B5
0.10, respectively, for B5 ×
× E13 (B5 side) and B5
E13 (B5 side) and B5 ×
× E13 (E13 side). The mitochondrial effect was less distinct in other strains.
E13 (E13 side). The mitochondrial effect was less distinct in other strains.
Table 1.
Fruiting characteristics of dikaryotic strains of Lentinula edodes.
| Dikayons | Relative degree of browning | Fruiting period from primordia | Weight (g) | Pileus | Stipe | ||||
|---|---|---|---|---|---|---|---|---|---|
| Relative color density | Diameter (cm) | Thickness (cm) | Length (cm) | Diameter (cm) | |||||
| B2xE13 | B2 side | 0.36 ± ± 0.15 0.15 | 5 d | 44.5 ± ± 9.4 9.4 | 0.29 ± ± 0.08 0.08 | 5.0 ± ± 0.6 0.6 | 1.6 ± ± 0.1 0.1 | 1.5 ± ± 0.1 0.1 | 3.1 ± ± 0.2 0.2 |
| E13 side | 0.39 ± ± 0.11 0.11 | 4 d | 61.0 ± ± 5.5 5.5 | 0.51 ± ± 0.07 0.07 | 4.8 ± ± 0.2 0.2 | 2.1 ± ± 0.1 0.1 | 1.2 ± ± 0.0 0.0 | 3.4 ± ± 0.1 0.1 | |
| B5xE13 | B5 side | 0.59 ± ± 0.08 0.08 | 4 d | 67.0 ± ± 15.0 15.0 | 0.45 ± ± 0.04 0.04 | 5.6 ± ± 0.4 0.4 | 1.9 ± ± 0.2 0.2 | 0.7 ± ± 0.4 0.4 | 3.2 ± ± 0.4 0.4 |
| E13 side | 0.43 ± ± 0.10 0.10 | 3 d | 98.0 ± ± 12.1 12.1 | 0.51 ± ± 0.12 0.12 | 5.6 ± ± 0.7 0.7 | 2.1 ± ± 0.1 0.1 | 1.1 ± ± 0.1 0.1 | 3.9 ± ± 0.6 0.6 | |
| E8xH3 | E8 side | 0.65 ± ± 0.05 0.05 | 4 d | 64.0 ± ± 10.5 10.5 | 0.70 ± ± 0.05 0.05 | 5.9 ± ± 1.0 1.0 | 2.0 ± ± 0.5 0.5 | 1.3 ± ± 0.1 0.1 | 2.3 ± ± 0.4 0.4 |
| H3 side | 0.08 ± ± 0.02 0.02 | 5 d | 44.0 ± ± 2.0 2.0 | 0.54 ± ± 0.08 0.08 | 5.5 ± ± 0.5 0.5 | 1.6 ± ± 0.2 0.2 | 1.4 ± ± 0.1 0.1 | 2.0 ± ± 0.2 0.2 | |
| K3xH3 | K3 side | 0.90 ± ± 0.03 0.03 | 4 d | 95.5 ± ± 11.5 11.5 | 0.75 ± ± 0.01 0.01 | 6.9 ± ± 0.3 0.3 | 3.0 ± ± 0.1 0.1 | 1.1 ± ± 0.0 0.0 | 5.3 ± ± 0.3 0.3 |
| H3 side | 0.83 ± ± 0.07 0.07 | 5 d | 64.0 ± ± 9.0 9.0 | 0.64 ± ± 0.03 0.03 | 5.4 ± ± 0.7 0.7 | 2.4 ± ± 0.3 0.3 | 1.0 ± ± 0.1 0.1 | 5.0 ± ± 0.5 0.5 | |
We next investigated the fruiting bodies of the dikaryotic strains through the induction of primordia after the browning. Eight to 10 primordia per dikaryotic strain were raised to mature fruiting bodies. The total cultivation period from the primordia to the mature fruiting bodies varied from three to five days (Table 1). Time-dependent observation of the fruiting process revealed more evident differences between the strains with different mitochondria. For example, the B5 ×
× E13 (B5 side) strain took more than four
E13 (B5 side) strain took more than four days of cultivation for full maturation, whereas B5
days of cultivation for full maturation, whereas B5 ×
× E13 (E13 side) needed only three
E13 (E13 side) needed only three days (Supplementary Figure S2).
days (Supplementary Figure S2).
A comparison of the total weights of fruiting bodies revealed the mitochondrial effect on the mushroom yield (Table 1). For example, the dikaryotic strains isolated from the E13 side, which were B2 ×
× E13 (E13 side) and B5
E13 (E13 side) and B5 ×
× E13 (E13 side), showed better fruiting body weight than those from the other side, B2
E13 (E13 side), showed better fruiting body weight than those from the other side, B2 ×
× E13 (B2 side) and B5
E13 (B2 side) and B5 ×
× E13 (B5 side). Conversely, the dikaryotic strains from the H3 side, including E8
E13 (B5 side). Conversely, the dikaryotic strains from the H3 side, including E8 ×
× H3 (H3 side) and K3
H3 (H3 side) and K3 ×
× H3 (H3 side), showed lower fruiting body weight than the E8
H3 (H3 side), showed lower fruiting body weight than the E8 ×
× H3 (H8 side) and the K3
H3 (H8 side) and the K3 ×
× H3 (K3 side) strains (Table 1). Characteristics of pileus and stipe, including cap color, cap diameter, cap thickness, stipe length, and stipe diameter differed depending on the strains. The fruiting bodies of B2
H3 (K3 side) strains (Table 1). Characteristics of pileus and stipe, including cap color, cap diameter, cap thickness, stipe length, and stipe diameter differed depending on the strains. The fruiting bodies of B2 ×
× E13 (B2 side) had pale-brown caps with short stipes, while those of B2
E13 (B2 side) had pale-brown caps with short stipes, while those of B2 ×
× E13 (E13 side) were more dark and thick caps. The B5
E13 (E13 side) were more dark and thick caps. The B5 ×
× E13 (B5 side) strain had a grayish flat cap, whereas the B5
E13 (B5 side) strain had a grayish flat cap, whereas the B5 ×
× E13 (E13 side) showed a brownish convex cap with a longer stipes (Figure 4(A)). A more severe morphological difference was observed in E8
E13 (E13 side) showed a brownish convex cap with a longer stipes (Figure 4(A)). A more severe morphological difference was observed in E8 ×
× H3. Similar to the browning pattern, the E8 side strain (E8
H3. Similar to the browning pattern, the E8 side strain (E8 ×
× H3 (E8 side)) produced dark brown caps, whereas the H3 side did pale brown caps (Supplementary Figure S3). The fruiting bodies of both strains had short but thick stipes.
H3 (E8 side)) produced dark brown caps, whereas the H3 side did pale brown caps (Supplementary Figure S3). The fruiting bodies of both strains had short but thick stipes.
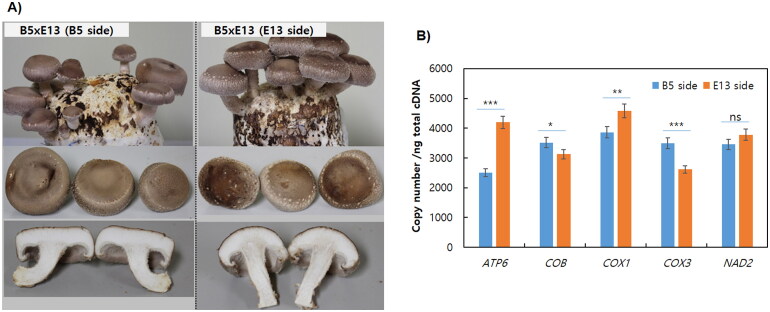
Fruiting body characteristics and mitochondrial gene expression of the B5XE13 strains. (A) Comparison of fruiting body characteristics. (B) Mitochondrial gene expression investigated by droplet digital PCR (ddPCR). The mushroom mycelia were cultivated for 7 d at 25 °C in PDB and total RNA was extracted from the ground mycelial powder. 1
°C in PDB and total RNA was extracted from the ground mycelial powder. 1 ng of cDNA generated by reverse transcription using a mixture of primers specific to the mitochondrial genes was subjected to ddPCR analysis. The data shown here are mean values of triplicated experiments. Error bars indicate the standard deviations of the means of triplicated data. The statistical significance was indicated by asterisks (* for p
ng of cDNA generated by reverse transcription using a mixture of primers specific to the mitochondrial genes was subjected to ddPCR analysis. The data shown here are mean values of triplicated experiments. Error bars indicate the standard deviations of the means of triplicated data. The statistical significance was indicated by asterisks (* for p ≤
≤ 0.05, ** for p
0.05, ** for p ≤
≤ 0.01, *** for p
0.01, *** for p ≤
≤ 0.001, and **** for p
0.001, and **** for p ≤
≤ 0.0001).
0.0001).
3.4. Mitochondrial gene expression
The expression of some representative mitochondrial genes in two dikaryotic strains having the same nuclei but different mitochondria, which were B5XE13 (B5 side) and B5XE13 (E13 side), was investigated by ddPCR analysis. The ddPCR showed that the mitochondrial genes COB, COX1, and NAD2 were expressed at similar levels in both B5XE13 (E13 side) and B5XE13 (B5 side), but the expression of COX3 was 1.34 folds higher in B5XE13 (B5 side) whereas the expression of ATP6 was 1.68 folds higher in B5XE13 (E13 side) (Figure 4(B)). This result implies that the function related to ATP generation is more active in the mitochondria of E13 in the dikaryotic B5XE13 strain, although it needs more elaborate analysis to draw a better conclusion.
4. Discussion
Mitochondria are intracellular organelles that play a key role in the cellular energy metabolism of eukaryotic organisms. In the mating of filamentous basidiomycetes, dikaryotic mycelia are generated by reciprocal movement of the nucleus to monokaryotic cytoplasm where a nucleus of compatible mating type resides. This process inevitably generates two different dikaryotic strains having the same nuclei but different mitochondria. For better understanding of the role of mitochondria in mushrooms, we made four sets of dikaryotic strains of L. edodes that have the same karyotype but different mitochondrial types and the physiological effect of mitochondria was investigated. Firstly, we were able to distinguish the nuclear and the mitochondrial types in the dikaryotic strains using the A mating type markers and the mitochondrial VNTR markers [14] (Figures 1 and and2).2). Investigation of the mycelial growth of the dikaryons on a solid medium reveals that mitochondria influence the mycelial growth rate as expected because of their function in energy generation. The dikaryons having mitochondria from E13 and E8, which are monokaryotic strains originating from the dikaryotic SJ701 strain, show better growth than that from B2 and B5 strains, which originated from the dikaryotic Chamaram strain [12] (Figure 3). Interestingly, B5XE13 (B5 side) produces more dense mycelia than B5XE13 (E13 side), suggesting that the mycelial morphology can be affected by nuclear-mitochondrial interaction. Indeed, mitochondrial functions are highly dependent on nuclear gene expression, including mitochondrial DNA replication, gene expression, stress-response, etc. [15]. In budding yeast, at least 12 nuclear genes, such as MIP1 (mtDNA polymerase), PIF1 (mtDNA helicase), and RIM1 (mitochondrial SSB protein), are involved in mtDNA maintenance [16]. Mitochondrial gene transcription and translation are also dependent on nuclear genes, including RPO41 (mtRNA polymerase), MTF1 (mitochondrial transcription factor), and tufM (mitochondrial elongation factor) [16]. Moreover, 38 nuclear gene products contribute to construct mitochondrial electron transport system (ETS) whereas only 7 mitochondrial proteins are incorporated in ETS [17]. Therefore, the mitochondrial effects demonstrated here are outcome of persistent interaction between nucleus and mitochondria.
In the mycelial propagation in a sawdust medium for fruiting, mitochondria affect the browning of the substrate, which is an important process in the fruiting body production [18,19]. The browning is related to the activities of nuclear DNA encoded β-tyrosinase [18,20], however, our study shows that mitochondria also affect the browning (Supplementary Figure S1). This can be related to the mitochondrial function which generates reactive oxygen species (ROS) as byproducts of energy metabolism. ROS are detrimental to mitochondria as well as cells and tissues causing oxidative breaks in DNA and RNA, and modifications in proteins [21]. ROS also plays a role in lignin oxidation which results in the production of polyphenolic oxidation products [22,23]. Therefore, mitochondria that produce more ROS can show a higher degree of substrate browning as shown in the B5XE13 (B5 side) and E8XH3 (E8 side) (Supplementary Figure S1). Investigation of the characteristics of the fruiting body, such as stipe length, cap morphology, and total yield, reveals that certain mitochondria are more effective in the production of the fruiting body. For example, the mitochondria from the E13 strains show better performance in the production of the fruiting body than the mitochondria from the B2 or B5 strains (Table 1). In addition, it was found that mitochondrial genes such as ATP6 and COX1 are better expressed in the strain that shows fast mycelial and fruiting body growth (Figure 4, B5XE13 (E13 side) vs. B5XE13 (B5 side), and Table 1). Taken together, our study shows that mitochondrion in mushrooms is an important factor governing physiological characteristics of mushrooms, such as mycelial growth rate, degree of substrate browning, and properties of the fruiting body. Our findings therefore imply that mitochondria should be given more consideration in the breeding of mushrooms.
References
Articles from Mycobiology are provided here courtesy of Korean Society of Mycology
Full text links
Read article at publisher's site: https://doi.org/10.1080/12298093.2022.2138226
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9645275
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/138126630
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1080/12298093.2022.2138226
Article citations
Potential marker genes for chronic obstructive pulmonary disease revealed based on single-cell sequencing and Mendelian randomization analysis.
Aging (Albany NY), 16(10):8922-8943, 23 May 2024
Cited by: 0 articles | PMID: 38787375
scFseCluster: a feature selection-enhanced clustering for single-cell RNA-seq data.
Life Sci Alliance, 6(12):e202302103, 03 Oct 2023
Cited by: 1 article | PMID: 37788907 | PMCID: PMC10547911
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nucleus-Selective Expression of Laccase Genes in the Dikaryotic Strain of Lentinula edodes.
Mycobiology, 45(4):379-384, 31 Dec 2017
Cited by: 4 articles | PMID: 29371806 | PMCID: PMC5780370
Isolation and Characterization of Monokaryotic Strains of Lentinula edodes Showing Higher Fruiting Rate and Better Fruiting Body Production.
Mycobiology, 43(1):24-30, 31 Mar 2015
Cited by: 7 articles | PMID: 25892911 | PMCID: PMC4397376
Establishment of uracil auxotrophic dikaryotic strains of Lentinula edodes by crossbreeding.
Breed Sci, 67(2):135-139, 17 Mar 2017
Cited by: 0 articles | PMID: 28588390 | PMCID: PMC5445960
Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes.
BMC Genomics, 20(1):121, 08 Feb 2019
Cited by: 30 articles | PMID: 30736734 | PMCID: PMC6368761
Funding
Funders who supported this work.
New Breeding Technologies Development Program (1)
Grant ID: PJ01697601

 a
a