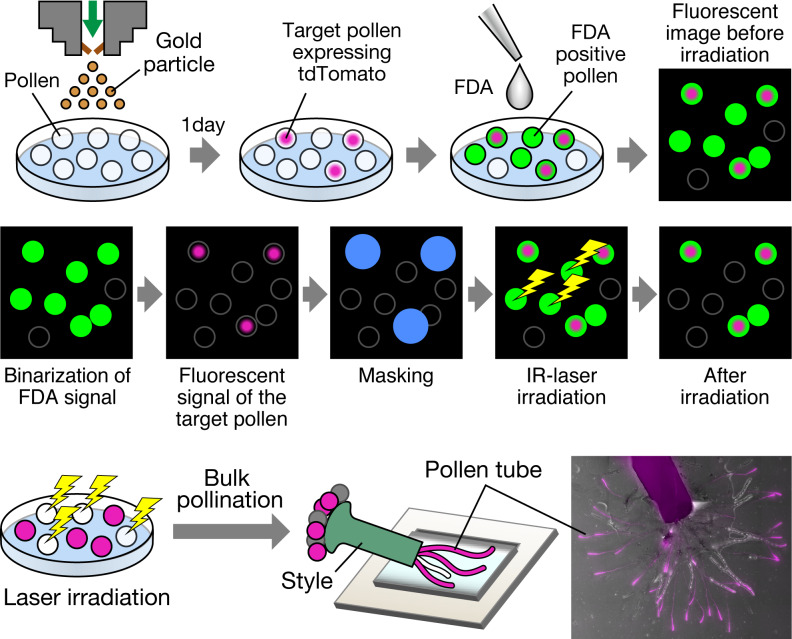
Abstract
Free full text

Target pollen isolation using automated infrared laser-mediated cell disruption
Peer Review Summary
Abstract
Single-cell analysis is important to understand how individual cells work and respond at the cell population level. Experimental single-cell isolation techniques, including dilution, fluorescence-activated cell sorting, microfluidics, and micromanipulation, have been developed in recent decades. However, such applications typically require large cell populations and skilled professionals. Additionally, these methods are unsuitable for sequential analysis before and after cell isolation. In this study, we propose a method for target cell isolation using automated infrared laser-mediated disruption of pollen grains in pollen populations. Germination of the target pollen was observed at the same location as that before laser irradiation, and germinated pollen grains were enriched in the cell population. Pollination of laser-irradiated bulk pollen populations also showed that the target pollen preferentially germinated on the stigma. This method is expected to facilitate physiological analyses of target cells at the single-cell level and effectively produce seeds derived from target pollen.
1. Introduction
A cell is a fundamental unit in living organisms. In cell biology, analysing the selection of specific target cells apart from other cells is necessary to gain a better understanding of their biological characteristics such as gene function, cell structure, and development. Methods used to separate target cells from other cells promote the study of biology, physiology, biochemistry, and taxonomy at the single-cell level (Gross et al., 2015). Target cell isolation techniques can be divided into two major categories: (1) selection of targeted cells and (2) elimination of non-target cells. Single-cell isolation and sorting techniques have been refined in recent decades (Hu et al., 2016). Fluorescence-activated cell sorting and microfluidic droplet-based platforms have been widely used for high-throughput isolation and selection of targeted cells; however, these require a large number of cells, which interferes with cell location and phenotypic profile. Micromanipulation, which involves manual cell picking with micropipettes using a microscope, is a simple approach for single-cell isolation; however, its throughput is limited and requires skilled professionals. In the latter category of isolation techniques, the most commonly known method of eliminating non-target cells is the selection of Escherichia coli using antibiotics. Traditionally, antibiotics and antibiotic-resistance genes have been used to select E. coli strains harbouring recombinant plasmids. However, some cell species are insensitive or resistant to antibiotics (Alanis, 2005). Therefore, a means of isolating target cells without relying on conventional methods is required.
Pollen grains are male gametophytes in flowering plants. Pollen grains can be easily collected from anthers as isolated cells and can be conveniently used for experiments as a uniform isolated cell population. Biolistic delivery, also referred to as particle bombardment, is widely used to transiently introduce and express exogenous genes in cells (Sanford, 2000). Recently, we reported a method for the biolistic delivery of genes into the pollen of Nicotiana benthamiana, which is easy to handle and has abundant pollen grains (Nagahara et al., 2021). However, the efficiency of gene introduction into pollen by bombardment is low, and pollen isolation using antibiotic-resistance genes cannot be used (Hoffmann et al., 1988). Therefore, a method for isolating gene-introduced pollen for efficient observation and pollination is required.
Herein, we propose a method for target cell isolation by the selective disruption of cells in a specific medium using infrared (IR) laser irradiation of N. benthamiana pollen. Target pollen grains were transiently labelled by the expression of fluorescent proteins that were mechanically extracted from the pollen population. Automatic enrichment of labelled cells is possible with this method. In addition, this method facilitates the enrichment of target cells while maintaining cell location and phenotypic profile, such as pollen germination in the culture medium. It also allows pollination of the extracted pollen grains, which suggests that seeds may be produced efficiently.
2. Methods
2.1. Plant materials and growth conditions
In this study, wild-type and transgenic N. benthamiana, AtUBQ10p::H2B-mClover, 35Sp::H2B-tdTomato, AtUBQ10p::tdTomato, and wild-type Nicotiana tabacum ‘Petit Havana SR1’ described in a previous study were used (Nagahara et al., 2021). Seeds were sterilised for 10 min using a solution containing 1% (v/v) sodium hypochlorite (Fujifilm Wako Pure Chemical, Osaka, Japan) and 0.02% (v/v) Triton X-100 (Nacalai Tesque, Kyoto, Japan). After washing with sterilised water, the seeds were sown on a medium containing 1× Murashige and Skoog Basal Medium (#M0404; MERCK, Darmstadt, Germany), 3% (w/v) sucrose (Fujifilm Wako Pure Chemical), and 0.8% (w/v) Bacto Agar (Thermo Fisher Scientific, Waltham, MA, USA) adjusted to pH 5.8 with KOH. Plants were germinated and grown in a growth chamber at 25–30°C after cold treatment at 4°C for 2–3 d. Two-week-old seedlings were transferred to the soil and grown in a greenhouse at 25–30°C under long-day conditions (16 hr light/8 hr dark).
2.2. Plasmid construction
The plasmid vector AtUBQ10p::H2B-tdTomato (MUv2458) used for particle bombardment was provided by Dr. Minako Ueda (Tohoku University). The fluorescent protein tdTomato was fused with Arabidopsis thaliana H2B (HISTONE 2 B; At1g07790), which was driven by the A. thaliana UBQ10 (UBIQUITIN 10; At4g05320) promoter (634 bp). Each fragment was cloned into the pGreen0029 vector (Hellens et al., 2000).
2.3. Pollen number counting
A single anther from wild-type N. benthamiana was suspended in 100 μL pollen culture medium. The pollen culture medium had the following composition: 0.5 M sucrose, 3 mM glutamine, 10 g/L lactalbumin hydrolysate, 10 mM KNO3, 1 mM Ca (NO3)2·4H2O, 1 mM MgSO4·7 H2O, 0.16 mM H3BO3, 1 mM uridine, 0.5 mM cytidine, and 1 mM phosphate buffer (pH 7.0) (Tupý et al., 1991). Of these, 10 μL of the pollen suspension was used for pollen counting using a cell counter (TC20; Bio-Rad Laboratories, Hercules, CA, USA) to calculate pollen density. Two anthers each from five flowers were analysed to estimate the average pollen number in a single anther.
2.4. Biolistic delivery of plasmid DNA into pollen
Pollen grains were used for biolistic delivery of plasmid vectors into cells. Pollen grains were obtained from buds 2 to 4 d before anthesis to analyse laser conditions and from just flowering flowers to analyse pollen germination and pollination. Pollen at 2–4 d before anthesis is suitable for analysing laser conditions for cell isolation because it cannot germinate pollen tubes. One-sixth to one-half of anthers were used in each experiment. Fresh anthers were squashed onto a wet filter with liquid pollen culture medium to disperse and absorb pollen. Biolistic delivery was performed as previously described (PDS-1000/He; Bio-Rad Laboratories; Nagahara et al., 2021). After bombardment, 1,000 μL of liquid pollen culture medium was added to the filter to suspend the pollen. The pollen suspension was then transferred to a 35-mm glass base dish (3910-035; Iwaki, AGC Techno Glass, Shizuoka, Japan) and cultured overnight in a chamber at 25°C. Gene introduction was confirmed based on the expression of fluorescent proteins (Figure 1a). Red fluorescent protein expression signals derived from the introduced plasmid DNA were observed using an inverted fluorescence microscope (emission filter: Cy3 570–610, Eclipse Ti2-E; Nikon, Tokyo, Japan) 18 hr after bombardment.
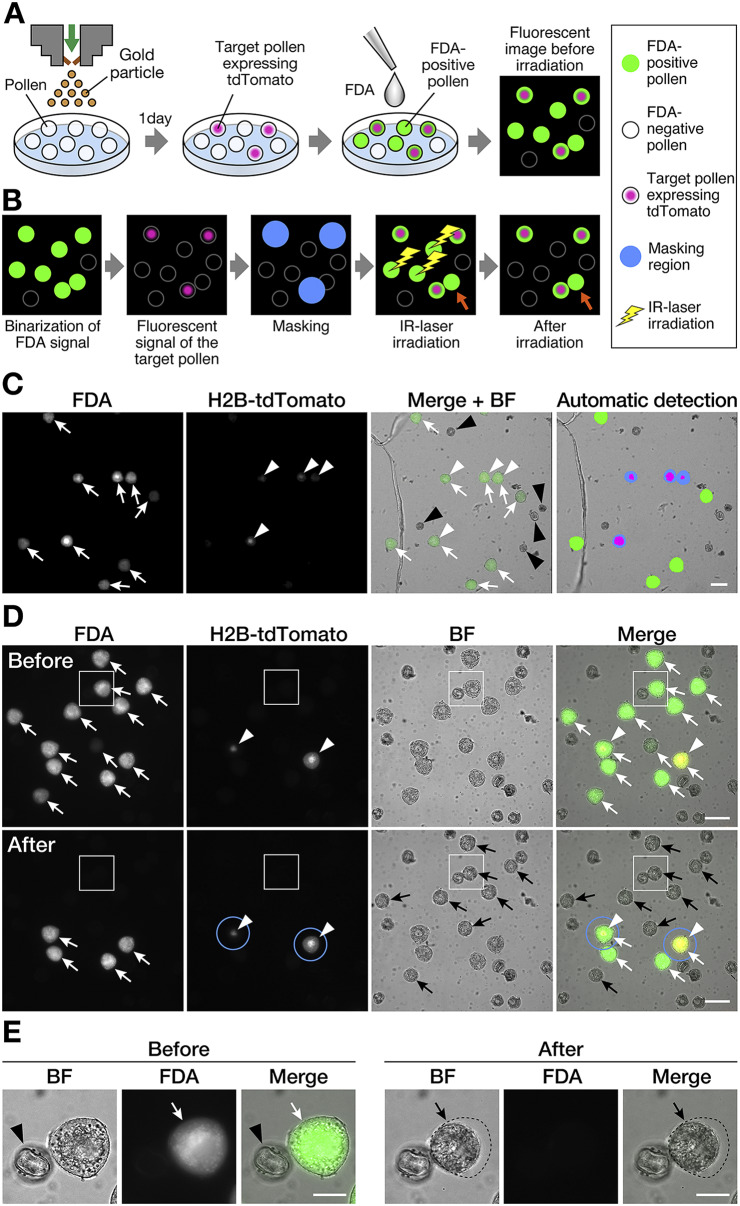
Visualisation of target cells and laser irradiation of non-target cells. (a) Visualisation of target cells and non-target cells. Biolistic delivery of gold particles with plasmid DNA into the pollen was performed to visualise the target pollen. The nuclei of the plasmid DNA-introduced pollen were labelled using red fluorescent proteins (H2B-tdTomato). Fluorescein diacetate (FDA) solution was added to the medium immediately before laser irradiation. Non-fluorescent FDA absorbed by pollen is converted to the green fluorescent metabolite fluorescein, which is used as an indicator of viable pollen grains. (b) Automated cell disruption for target cell isolation by infrared (IR) laser irradiation. First, green and red fluorescence signals were detected through image capture before irradiation. FDA-positive pollen grains were defined and binarised by the captured image to detect a living cell. The target pollen expressing red fluorescent proteins were also detected by the captured image. To protect these target grains from laser irradiation, a diameter of 60 μm from the centre of the target cell was masked (blue-filled circle). Based on the x, y location of these defined pollen grains, IR laser irradiation was performed targeting the centre of each non-target pollen. After irradiation, the target cells were observed to maintain both FDA- and protein-derived fluorescence signals. Orange arrows indicate the non-target pollen attached to the target pollen, which was omitted from laser irradiation by masking. (c) Automated cell detection for target pollen isolation using programmable software. Fluorescence and brightfield images were captured for the detection of fluorescent signals and pollen shape. The detection program was applied to the images to automatically discriminate between non-target pollen with laser irradiation and tdTomato-expressing target pollen without laser irradiation. The detected area was automatically coloured with the following colours: green, FDA-positive pollen; magenta, tdTomato-expression; blue, masking area. (d and e) Brightfield and fluorescence images of pollen before and after IR laser irradiation. FDA-negative pollen grains were considered non-viable and were omitted from laser irradiation. The pollen with nuclei labelled with H2B-tdTomato were defined as target pollen for cell isolation. Blue solid line circles denote the masking diameter of 60 μm from the centre of the target cell. After laser irradiation, non-target pollen showed no FDA signal and shrinkage. The pollen close to the tdTomato-expressing target pollen in the masking circle was not irradiated, and thus, the FDA signal was retained. (e) Magnified images of the area are demarcated by the white-bordered square in (d). The shape of pollen before laser irradiation is shown as a black dotted-line circle. White arrow, FDA-positive pollen; white arrowhead, tdTomato-expression; black arrow, FDA-negative pollen; black arrowheads, FDA-negative shrunken pollen after laser disruption. Scale bars, (c and d) 50 μm and (e) 20 μm.
2.5. Visualisation of viable pollen using fluorescein diacetate (FDA) solution
After confirming fluorescent protein expression, a fluorochromatic test, also known as FDA staining, was used to assess cell viability (Heslop-Harrison et al., 1984). A 1% (w/v) solution of FDA (Merck) in acetone was added to the pollen suspension in the culture medium (Figure 1a) at a final concentration 0.019 μM at 18 hr after bombardment. After more than 10 min, cell viability was assessed based on green fluorescence signals using an inverted fluorescence microscope (emission filter: FITC 510–560, Eclipse Ti2-E; Nikon).
2.6. Infrared laser-evoked gene operator (IR-LEGO) microscope system and image analysis
An optical IR-LEGO system was used for the cell disruption of non-target pollen. An Eclipse Ti2-E microscope (Nikon) with an IR-LEGO-490 mini/E system (Sigma-Koki, Saitama, Japan) was used. An objective lens (CFI PlanApo λ 20×, NA = 0.75, WD = 1.0; Nikon) was used for IR laser focusing, irradiation of the target cells with the IR laser, and imaging of the samples. When the power of the IR laser source was set to 300 mW, the laser power delivered to the samples in the focal plane was 145 mW, as measured using a power meter (Coherent, Santa Clara, CA, USA). The irradiation exposure time for cell disruption of each pollen was set from 1 to 10 ms. Images were captured using a scientific CMOS camera (ORCA-Fusion BT C15440-20UP; Hamamatsu Photonics, Shizuoka, Japan). Bandpass filters used for autofluorescence of pollen grains included fluorescein isothiocyanate (FITC) for the FDA signal, Cy3 for tdTomato fluorescence, and cyan fluorescent protein (CFP) (Nikon). ImageJ software version 1.53j (https://imagej.nih.gov/ij/index.html) was used to generate and analyse the images.
2.7. Automated cell disruption for target cell isolation using a programmable software
The protocols for automated image acquisition, target cell extraction, and laser irradiation routines for target cell isolation were produced using NIS-Elements JOBS Programmable Software (Nikon). NIS-Elements JOBS was used as a plug-in module for NIS-Elements AR (Nikon), which enabled the design of experiments with customised acquisition and analyses. In this study, a five-step program (including three automated detection, definition, and irradiation processes and two automated image capturing processes) was developed for the assessment of laser irradiation and consisted of the following steps: (a) image acquisition, (b) detection and definition of living pollen and its position on the microscope stage, (c) detection and masking of the target pollen, (d) IR laser irradiation of non-target pollen, and (e) image acquisition after irradiation (Figure 1a–c). In step a, the region of interest including the uniformly distributed pollen within an area of over 20 mm2, was defined as the irradiation area. Thereafter, automatic image capture was executed based on green fluorescence signals, such as the FDA signal, and red fluorescence signal with respect to tdTomato expression. In step b, the FDA-positive pollen was detected automatically. To detect a living cell, a fluorescence signal intensity >9,600 was defined as the threshold of FDA positivity. Thereafter, binarised images were obtained using a general analysis task. An FDA signal corresponding to a diameter of >12 μm was defined as a single pollen to exclude dust. The positional information of each FDA signal was recorded using a region list task. Similarly, a fluorescence signal intensity >65,535 was defined as the threshold of the target cells based on fluorescent protein expression. In step c, an area with a diameter of 60 μm from the centre of the target cell was masked and not irradiated to protect the target cells from laser irradiation. In step d, based on the x, y location of these defined cells, moving to the centre of each non-target pollen, shutter opening/closing for irradiation was performed. Finally, the irradiated area was captured using tiling images to assess the laser disruption of the non-target pollen in step e. Steps b–d were executed in a loop that was automatically performed for each tile. To define the FDA-positive/negative signals in steps a and e, the average signal intensity was set as the threshold by randomly selecting 10 pollen grains that produced almost no fluorescence. All capture and irradiation processes were performed using an inverted microscope (Ti2-E) at 24–28°C under dark conditions.
2.8. Examination of laser conditions for single pollen disruption
Laser conditions for single-pollen disruption were examined. After suspending N. benthamiana pollen in the culture medium, a 1% (w/v) solution of FDA in acetone was immediately added at a final concentration of 0.019 μM. Pollen was irradiated using an IR laser. The laser power delivered to the samples at the focal plane was changed from 24 to 194 mW, which was measured using a power meter (Coherent). The laser source range was 50–400 mW. The duration of irradiation varied between 1 and 20 ms. After irradiation, the time for which the signal intensity of the FDA fluorescence was less than the threshold was measured at 5 min. The fluorescence did not disappear after 5 min and was undetectable. The average timing was obtained for two to three pollen samples.
2.9. Pollination of the disrupted pollen population under the semi-in vivo assay
A 1:1 mixture of N. benthamiana wild-type and transgenic AtUBQ10p::tdTomato pollen was used for the laser irradiation. A half of the anthers were used per laser irradiation and then pollinated on a pistil. Because the N. benthamiana pistil is too thin for semi-in vivo assays, the N. tabacum pistil was used instead for pollination. The FDA fluorescent dye was not used because it may disturb pollination, whereas pollen morphology, that is, pollen shape under the bright field and pollen autofluorescence under the CFP bandpass filter (Nikon), were used as indicators of pollen grains. Unlike dust and other cells, N. benthamiana pollen shows strong blue autofluorescence, making pollen shape easy to detect. The tdTomato-expressing pollen was defined as the target pollen, and laser irradiation was omitted. A laser power of 145 mW at the focal plane and 10 ms irradiation time were used for the disruption of the wild-type pollen. After IR laser irradiation, the disrupted pollen bulk population was collected through the filter and pollinated to N. tabacum pistil (emasculated before 2 or 2 d). Immediately, the pollinated style was excised with a razor and placed horizontally on the pollen germination medium solidified with 1% (w/v) NuSieve GTG Agarose (Lonza, Switzerland), followed by incubation at 25–30°C for 18–24 hr under humid and dark conditions (Nagahara et al., 2021). Pollen tubes emerging from the cut end of the pistil were observed under an inverted fluorescence microscope (Eclipse Ti2-E; Nikon).
3. Results
3.1. Conditions of IR laser irradiation for selective disruption of non-target pollen
The FDA signal remained after 5 min of irradiation at 24 mW, even when the irradiation time was increased from 1 to 20 ms (Supplementary Table S1). In contrast, the FDA signal disappeared after irradiation at >48 mW. At an irradiation power of 48 mW, 165–225 s were required for the FDA fluorescence to disappear. The fluorescence signal disappeared within a short period of time after irradiation at >96 mW. At 194 mW, laser irradiation caused the pollen to jump and shift forward from the position observed before irradiation; such irradiation power was thus considered too strong for this study. For laser irradiation durations of 10 and 20 ms at 194 mW, the fluorescence disappeared simultaneously with laser irradiation (Supplementary Table S1); therefore, pollen was damaged during laser irradiation. To irradiate an increased number of non-target pollen grains without affecting the target cells, reducing the laser power and shortening the duration of irradiation is necessary to the maximum possible extent is necessary. Therefore, we determined the irradiation condition as the disappearance of fluorescence within 42.5 s, that is, a laser power of 145 mW at the focal plane and irradiation time of 10 ms, which was used for subsequent experiments.
3.2. Software-automated identification of pollen expressing exogenous genes is affected by pollen density and separation
First, the percentage of FDA-positive pollen in wild-type N. benthamiana was determined. As a result, 151 pollen grains were FDA-positive and 45 were FDA-negative, which indicated that 77.0% of wild-type N. benthamiana pollen grains were viable. Because a short laser irradiation period is preferred, FDA-negative pollen grains, which accounted for 23.0%, were excluded from subsequent laser irradiation. To identify the target pollen, a plasmid DNA vector containing the red fluorescent protein was introduced into the pollen. Gene expression was evaluated based on fluorescence signals at 18 hr after bombardment (Figure 1a). Each nucleus in the target pollen was labelled by expression of the introduced AtUBQ10p::H2B-tdTomato plasmid DNA. Five independent experiments on different days were used for the analysis (Table 1). Because it is necessary to irradiate many non-target pollen grains, the following five steps were executed and automatically performed by the graphically programmable software module: (a) image acquisition, (b) detection and definition of living pollen and their position on the microscope, (c) detection and masking of the target pollen, (d) IR laser irradiation of non-target pollen, and (e) image acquisition after irradiation (Figure 1b,c). The irradiation area is defined in step a. In step b, pollen viability was assessed by FDA staining. The FDA-positive pollen was considered viable and its position information was automatically registered. Pollen grains were suspended in pollen culture medium and stained with the FDA solution. After IR laser irradiation, the non-target pollen showed shrinkage and disappearance of the FDA signal (black arrows in Figure 1d,e), whereas the target pollen retained fluorescent signals derived from both FDA and H2B-tdTomato (Figure 1d). This indicated that the irradiated cells lost their viability. In contrast, non-target cells in the vicinity of the target cells (i.e., within the masked area) remained fluorescent because they were omitted from laser irradiation (Figure 1d). Of the five samples, 1,011–3,764 pollen grains, including 539–1,457 FDA-positive pollen grains, were counted (Table 1). The pollen density of each sample was 40.6–104.1 cells/mm2 (average 65.8 ± 25.4 cells/mm2; Table 1). Similarly, pollen showing a fluorescent signal derived from tdTomato expression was defined as the target cell for step c. The number of target pollens per experiment was 20–94, and the ratio of target pollen within viable pollen was 3.6%–9.4% (average 5.8 ± 2.4%, Table 1). Therefore, more than 90% of the pollen was defined as non-target pollen that needed to be disrupted by laser irradiation. Based on the x, y location of these defined cells, laser irradiation was performed at the centre of each non-target pollen in step d. Finally, the irradiated area was captured to assess the laser disruption of the non-target pollen. The average time required for laser irradiation was 68.4 ± 12.4 min for an irradiation area covering 80–120 tiles, when the average pollen density was 65.8 ± 25.4/mm2 (Table 1). Sample I, which had the lowest pollen density, had the highest cell-isolation efficiency. The proportion of tdTomato-expressing pollen after irradiation was 8.3-fold higher, increasing from 3.7% to 30.8% (Table 1). These results indicate that pollen density affects isolation efficiency.
Table 1
Number and proportions of pollen before and after laser irradiation in the five samples
| Irradiation area (mm2) | # of tiles covering area | # of the total pollen | Pollen (/mm2) | Before | After | Time irradiation (min) | Disruption rate %) | Viability pollen (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FDA positive | tdTomato expressing | Target pollen (%) | FDA positive | tdTomato expressing | Target pollen (%) | ||||||||
| I | 22.5 | 80 | 1011a | 44.9 | 539 | 20 | 3.7b | 39 | 12 | 30.8b | 51 | 94.8c | 60.0d |
| II | 24.3 | 99 | 1617a | 66.5 | 638 | 35 | 5.5b | 185 | 31 | 16.8b | 68 | 74.5c | 88.6d |
| III | 26.3 | 100 | 2739a | 104.1 | 918 | 86 | 9.4b | 202 | 43 | 21.3b | 77 | 80.9c | 50.0d |
| IV | 55.2 | 120 | 2241a | 40.6 | 1432 | 94 | 6.6b | 396 | 62 | 15.7b | 83 | 75.0c | 66.0d |
| V | 51.7 | 117 | 3764a | 72.8 | 1457 | 53 | 3.6b | 462 | 31 | 6.7b | 63 | 69.3c | 58.5d |
| Average | 65.8 ± 25.4e | 5.8 ± 2.4e | 18.2 ± 8.8e | 68.4 ± 12.4e | 78.9 ± 9.8e | 64.6 ± 14.6e | |||||||
To investigate the effects of other factors on the isolation efficiency, we performed a detailed analysis of the irradiation results for each tile, including at least one tdTomato-expressing pollen. Laser irradiation was performed on samples with pollen grains of one-fourth or one-sixth anthers suspended over an area of 25 mm2. First, the number of pollen grains in a single anther was calculated. The average pollen number in a single anther was 12,717 ± 2,814 (10 anthers from 5 flowers); thus, one-sixth of the anther was estimated to contain approximately 2,119 pollen grains, and the pollen density was calculated as 84.8/mm2. Similarly, one-fourth of anthers were estimated to contain approximately 3,179 pollen grains, and pollen density was calculated as 127.2/mm2. Next, the results for each tile were divided into two groups based on the presence or absence of pollen separation (Table 2). The number of pollen grains, pollen separation, and disruption success rate was counted for each tile (Table 2). When the pollen density was 84.8/mm2, the average disruption success rates were 87.2 ± 13.4% and 71.6 ± 14.1%. This was higher than the 72.0 ± 22.7% and 65.7 ± 19.9% for pollen density of 127.2/mm2. In contrast, when comparing the presence or absence of pollen separation, the disruption success rates were 87.2 ± 13.4% and 72.0 ± 22.7% for successful pollen separation, which were higher than that of unsuccessful pollen separation (71.6 ± 14.1% and 65.7 ± 19.9%). The maximum target pollen isolation rate 62.8 ± 35.5% and the disruption success rate 87.2 ± 13.4% were observed at the pollen density of 84.8 mm2 with pollen separation. This result suggests that the isolation efficiency was affected by both pollen density and pollen separation, and the lower pollen density achieved more success.
Table 2
Number and proportions of pollen before and after laser irradiation in each tile
| Pollen density (/mm2) a | Separation of pollen b | Tile | Before | After | Disruption success rate (%) d | Average of disruption success rate (%) e | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FDA positive | tdTomato expressing | Target pollen (%) c | Average of target pollen (%) | FDA positive | tdTomato expressing | Target pollen (%) c | Average of target pollen (%) | |||||
| 84.8 | True | 1 | 18 | 1 | 5.6 | 19.4 ± 15.3 | 3 | 1 | 33.3 | 62.8 ± 35.5 | 88.2 | 87.2 ± 13.4 |
| 2 | 14 | 2 | 14.3 | 5 | 2 | 40.0 | 75.0 | |||||
| 3 | 11 | 1 | 9.1 | 3 | 1 | 33.3 | 80.0 | |||||
| 4 | 10 | 1 | 10.0 | 4 | 1 | 25.0 | 66.7 | |||||
| 5 | 10 | 4 | 40.0 | 4 | 4 | 100.0 | 100.0 | |||||
| 6 | 9 | 1 | 11.1 | 3 | 1 | 33.3 | 75.0 | |||||
| 7 | 7 | 1 | 14.3 | 1 | 1 | 100.0 | 100.0 | |||||
| 8 | 5 | 1 | 20.0 | 1 | 1 | 100.0 | 100.0 | |||||
| 9 | 2 | 1 | 50.0 | 1 | 1 | 100.0 | 100.0 | |||||
| False | 1 | 31 | 2 | 6.5 | 13.9 ± 7.7 | 9 | 0 | 0.0 | 31.8 ± 24.4 | 69.0 | 71.6 ± 14.1 | |
| 2 | 25 | 4 | 16.0 | 12 | 3 | 25.0 | 57.1 | |||||
| 3 | 22 | 2 | 9.1 | 6 | 2 | 33.3 | 80.0 | |||||
| 4 | 21 | 1 | 4.8 | 10 | 1 | 10.0 | 55.0 | |||||
| 5 | 15 | 2 | 13.3 | 7 | 1 | 14.3 | 53.8 | |||||
| 6 | 15 | 3 | 20.0 | 4 | 2 | 50.0 | 83.3 | |||||
| 7 | 13 | 2 | 15.4 | 3 | 2 | 66.7 | 90.9 | |||||
| 8 | 10 | 3 | 30.0 | 3 | 2 | 66.7 | 85.7 | |||||
| 9 | 29 | 3 | 10.3 | 10 | 2 | 20.0 | 69.2 | |||||
| 127.2 | True | 1 | 26 | 2 | 7.7 | 15.1 ± 5.6 | 5 | 1 | 20.0 | 36.9 ± 28.3 | 83.3 | 72.0 ± 22.7 |
| 2 | 22 | 2 | 9.1 | 15 | 3 | 20.0 | 40.0 | |||||
| 3 | 14 | 3 | 21.4 | 6 | 3 | 50.0 | 72.7 | |||||
| 4 | 8 | 1 | 12.5 | 0 | 0 | 0.0 | 100.0 | |||||
| 5 | 8 | 1 | 12.5 | 2 | 1 | 50.0 | 85.7 | |||||
| 6 | 7 | 1 | 14.3 | 3 | 1 | 33.3 | 66.7 | |||||
| 7 | 6 | 1 | 16.7 | 3 | 1 | 33.3 | 60.0 | |||||
| 8 | 6 | 1 | 16.7 | 4 | 1 | 25.0 | 40.0 | |||||
| 9 | 4 | 1 | 25.0 | 1 | 1 | 100.0 | 100.0 | |||||
| False | 1 | 13 | 1 | 7.7 | 11.2 ± 14.7 | 10 | 1 | 10.0 | 23.8 ± 28.8 | 25.0 | 65.7 ± 19.9 | |
| 2 | 17 | 1 | 5.9 | 5 | 1 | 20.0 | 75.0 | |||||
| 3 | 15 | 1 | 6.7 | 7 | 1 | 14.3 | 57.1 | |||||
| 4 | 20 | 2 | 10.0 | 7 | 1 | 14.3 | 66.7 | |||||
| 5 | 46 | 1 | 2.2 | 13 | 1 | 7.7 | 73.3 | |||||
| 6 | 18 | 1 | 5.6 | 6 | 1 | 16.7 | 70.6 | |||||
| 7 | 25 | 2 | 8.0 | 12 | 2 | 16.7 | 56.5 | |||||
| 8 | 19 | 1 | 5.3 | 7 | 1 | 14.3 | 66.7 | |||||
| 9 | 2 | 1 | 50.0 | 1 | 1 | 100.0 | 100.0 | |||||
3.3. Pollen germination and pollination of the isolated pollen
In the case of bombardment, pollen germination occurs before the expression of the transgene; therefore, pollen germination and pollination of isolated pollen cannot be studied (Nagahara et al., 2021). Transgenic 35Sp::H2B-tdTomato was used for pollen germination analysis, and AtUBQ10p::tdTomato pollen grains were used for pollination analyses as target pollen expressing fluorescent proteins. To analyse pollen germination of the isolated pollen, the wild-type and 35Sp::H2B-tdTomato pollen grains were mixed, and pollen with nuclei labelled with tdTomato (white arrowheads in Figure 2) was defined as the target pollen and omitted from laser disruption. Pollen grains that did not show tdTomato fluorescence signals were disrupted using IR laser irradiation as a non-target pollen. After 1 hr of laser irradiation, germination of the target pollen grains with both the FDA signal and tdTomato expression was observed (white arrows in Figure 2), whereas it was not observed in the shrunken non-target pollen grains disrupted by laser irradiation (black arrows in Figure 2). The FDA fluorescent signals in several pollen grains disappeared, and these pollen grains showed bursts despite being protected from laser disruption (white dotted circles in Figure 2). Pollen tubes from the target pollen grains continued to elongate, and nuclei were transported to the pollen tube tips 3 hr after laser irradiation (yellow arrows in Figure 2).
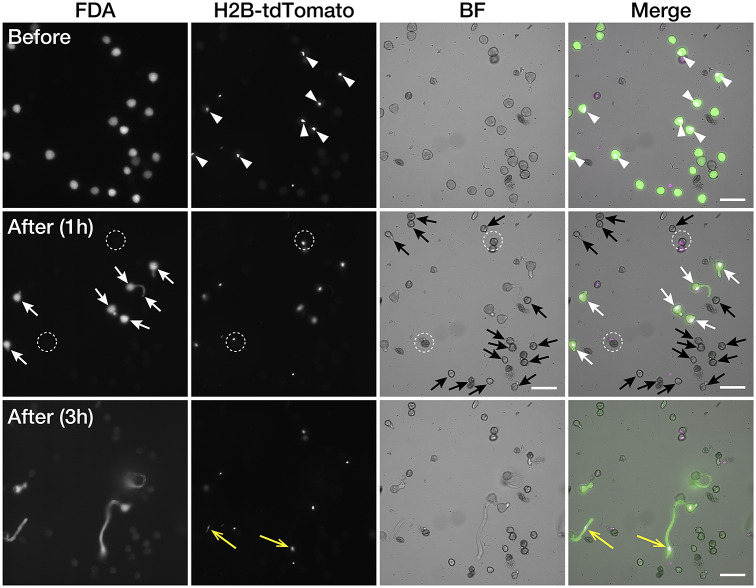
Automated target pollen detection and germination of the isolated pollen Fluorescence and brightfield images of pollen before and after infrared (IR) laser irradiation. Pollen germination in the media after laser irradiation was observed 1 and 3 hr after laser irradiation. Pollen with H2B-tdTomato-labeled nuclei was defined as the target pollen for cell isolation (white arrowheads). Fluorescein diacetate (FDA)-positive pollen grains without tdTomato expression were defined as non-target pollen. After laser irradiation, pollen tube germination from the target pollen was observed (white arrows). Pollen grains that disappeared from the FDA signal did not show pollen germination (white dotted-line circles). Pollen tube nuclei transported to the pollen tube tips were observed 3 hr after the laser irradiation (yellow arrows). Scale bars, 100 μm.
Pollination and pollen tube growth of the isolated pollen were analysed in the pistil using a semi-in vivo growth assay (Bosch & Hepler, 2006). Wild-type and AtUBQ10p::tdTomato pollen grains were mixed at a 1:1 ratio. The tdTomato-expressing AtUBQ10p::tdTomato pollen grains were defined as target pollen and omitted from the laser disruption. Although pollen morphology and autofluorescence, but not FDA fluorescence, were used as indicators, pollen grain detection and laser irradiation were successfully performed (Supplementary Video 1). After laser irradiation, the pollen bulk population (including disrupted non-target pollen and tdTomato-expressing target pollen) was pollinated with N. tabacum pistil (Figure 3a). A semi-in vivo growth assay revealed that pollen tubes emerged from the cut end of the pistil. After pollination, the number of pollen tubes that emerged was counted using each pistil (Figure 3b,c). A clear difference between irradiated (+) and non-irradiated (−) pollen pollination was observed in the proportion of pollen tubes expressing tdTomato (Figure 3b). In the case of the non-irradiated pistil #C1, 57 pollen tubes were observed, including 36 wild-type pollen tubes and 21 tdTomato-expressing pollen tubes (Figure 3c). The percentage of tdTomato-expressing pollen tubes was 36.8%. In the case of the irradiated pistil #IR1, there were 52 pollen tubes, including 17 wild-type pollen tubes and 35 tdTomato-expressing pollen tubes. Thus, the percentage of tdTomato-expressing pollen tubes was 67.3% (Figure 3c). The average percentage of tdTomato-expressing pollen tubes in the three laser-irradiated pistils was 77.2 ± 8.6%, which was more than approximately two times that of the control experiment #C1 (Figure 3c). The percentage of tdTomato-expressing pollen tubes was higher in the laser-irradiated bulk pollen population, suggesting that disruption of non-target pollen was effective.
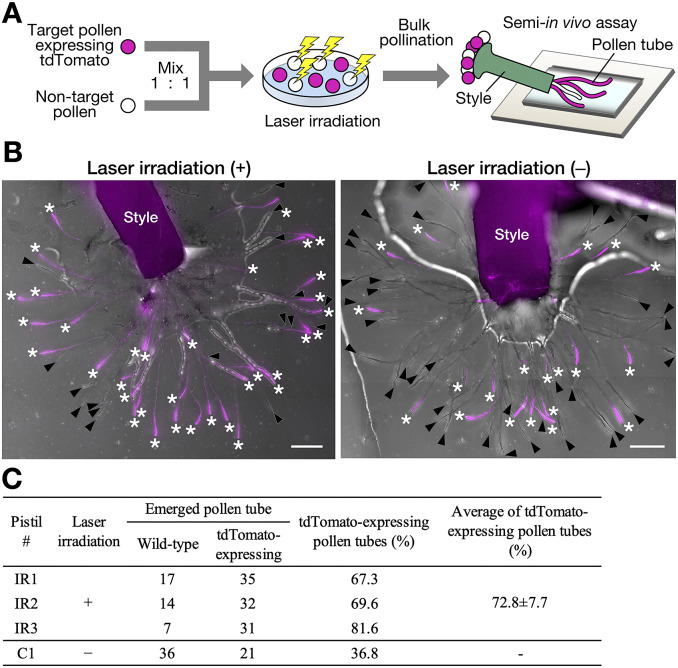
Pollination of the isolated pollen (a) Schematic representation of the semi-in vivo growth assay using the isolated pollen. Target pollen expressing tdTomato and non-target wild-type pollen were mixed at a ratio of 1:1 and infrared (IR) laser irradiation was performed. After irradiation, the bulk pollen population was pollinated to the pistil, and pollen tubes emerging from the cut end of the style were observed. (b) Semi-in vivo growth assay with bulk pollen population with (+) and without (−) laser irradiation. Red fluorescent and brightfield images were merged. The tdTomato signal is indicated in magenta. Asterisks, tdTomato-expressing pollen tube; arrowheads, tdTomato-negative pollen tube. Scale bars, 400 μm. (c) The number of pollen tubes that emerged in the semi-in vivo growth assay with the bulk pollen population. The laser-irradiated bulk pollen population was pollinated on three pistils (IR1–3). The bulk pollen population, without laser irradiation, was also pollinated on one pistil (c1).
These results suggest selective target pollen germination in vitro and in vivo by cell disruption of non-target pollen.
4. Discussion
Distinguishing cells of interest from other cells is important to better understand biological phenomena. With recent developments in molecular cell biology, there is an increasing need to study gene functions and phenotypes of particular cells in multicellular organisms (including the study of cell lineages, cell fates, cell differentiation, and cell-to-cell communication). Here, an IR laser was used for the disruption of cells; ultraviolet or visible light can be used as long as the laser is focused on the focal plane. Laser light can damage cells through thermal, mechanical, and photochemical effects, which, in turn, elicit cellular stress and lead to cell death. Among these laser light sources, IR lasers are suitable for inducing gene expression in living organisms at the single-cell level. IR lasers have a high cell-heating capacity because they operate at a wavelength of 1,480 nm, which can heat water with a 105-fold higher efficiency than 440-nm lasers can (Kamei et al., 2009). Therefore, IR lasers have been used to effectively induce gene expression using heat-shock promoters. In plants, only the female gametophyte in the ovule is stimulated and the heat-inducible Cre-loxP recombination system induces transgene expression (Hwang et al., 2019). In addition to inducing gene expression, IR lasers have been used in single-cell ablation and disruption experiments, because their high-power energy can be focused on a defined area. Laser disruption is used frequently in plant developmental studies. For example, laser ablation of the quiescent centre of Arabidopsis roots demonstrated that stem cells maintain their status by inhibiting the differentiation of the surrounding cells (van den Berg et al., 1995, 1997). Disrupted Arabidopsis embryo initial cells induce cell fate conversion of the suspensor cell to compensate for the embryo even after the cell fate has been specified (Gooh et al., 2015). These irradiation experiments have been performed on single cells in multicellular tissue but have rarely been applied for the selection of target cells. Cell damage depends on the IR-laser power and duration of exposure; however, the effective conditions differ among cell species (Kawasumi-Kita et al., 2015). Therefore, we first identified a suitable irradiation setting for the disruption of non-target pollen. In the current study, a method for the automatic irradiation of non-target cells was established to enable efficient selection of target cells. Our method can be applied not only to pollen but also to other isolated cells such as spores, algae, and fungal cells. It does not require specific culturing areas or dish shapes as long as the cell population is the focus. To ensure effective selection, the cells and background, as well as the target and non-target cells, should be binarised precisely. Target cells were binarised using dyes and fluorescent proteins. The number of non-target living pollen grains was recorded using FDA staining, which is commonly used to assess pollen viability (Heslop-Harrison et al., 1984). FDA is hydrolysed by cytoplasmic esterase to produce fluorescein, which accumulates in the cell and exhibits a strong green fluorescence. In contrast, dead cells are characterised by the absence of esterase activity, and thus do not exhibit fluorescence. Generally, mature wild-type anthers contain a proportion of undeveloped or dead pollen (Lee et al., 2008). Additionally, pollen viability is reduced depending on storage and culture time (Hashida et al., 2012). In Nicotiana, anthers contain normal pollen as well as undeveloped, abnormally shrunken, or dead pollen (Matoušek et al., 2020). Thus, only living pollen was counted using FDA staining. In our experiments, 77.0% of the N. benthamiana pollen grains showed FDA signals, which was considered viable. After laser irradiation, the FDA fluorescent signals in several target pollens disappeared despite being protected from laser disruption (Figure 2). This may be because of arrest during pollen development for endogenous reasons, or bombardment and FDA solution may have negatively affected pollen development. In addition to FDA, other indicators, such as cell state and morphology, can also be used to define target cells (Supplementary Video 1). An appropriate density of cells is also important for effective isolation because a high cell density responsible for more non-target pollen may lead to an overlap with the masking area, which reduces isolation efficiency. Once the conditions for isolation have been established, high-throughput studies will be possible through automated isolation.
Antibiotic-based selection based on resistance genes has generally been used to eliminate non-target cells (Southern & Berg, 1982). Antibiotic and herbicide-resistance genes have been widely used as selectable markers in transgenic plants and cells (Breyer et al., 2014). However, it is difficult to use antibiotics in some organisms and cell species that are already resistant or show varying selectivities at different concentrations (Angenon et al., 1994). In addition, physiological responses to antibiotics can be altered by their use. Because our method does not use antibiotic-based selection, it can be applied to select cells of interest. For example, it may be effective in the study of antibiotic resistance and drug discovery in multidrug-resistant bacteria (Lage et al., 2018). Moreover, the positional information before irradiation is completely maintained after laser irradiation, and our method facilitates continuous analysis, such as live imaging, for tracing individual cells. The method can be applied to other approaches, such as the analysis of cell-to-cell communication between disrupted and non-disrupted cells in scattered cells, similar to the known positive and negative pollen density effects on pollen tube germination (Boavida & McCormick, 2007).
Pollen germination is possible not only in the media but also on the pistil. We have previously reported that bombarded pollen can elongate pollen tubes on the pistil and reach the ovule (Nagahara et al., 2021). Our results suggest that pollination with isolated pollen may achieve efficient seed production from the target pollen (Figure 3). However, the proportion of pollen tubes expressing tdTomato was not 100%, which may need further improvement. A detailed analysis of the isolation results for each tile revealed that the disruption success rate showed a wide deviation, indicating the importance of both pollen density and separation (Table 2). For example, combining methods to spread pollen grains in pieces more reliably may improve the isolation efficiency. A high separation efficiency can be achieved by separating each tile with a plate or microfluidic device and collecting cells from each tile. This method may facilitate high-throughput studies at the single-cell level, such as gene expression analysis and cell dynamic imaging, in various organisms.
Acknowledgements
We sincerely thank Dr. Ueda and Dr. Kurihara for providing the plasmid vector and T. Shinagawa and E. Matsumoto for preparing the plant materials and performing the experiments. We also thank the members of the JST A-STEP project for helpful discussions.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/qpb.2022.24.
Financial support
This work was supported by the Japan Science and Technology Agency (JST) [Adaptable and Seamless Technology Transfer Program through Target-driven R&D (A-STEP, JPMJTR194H)], Japan Society for the Promotion of Science [Grant-in-Aid for Transformative Research Areas (20H05778, 20H05779)], and Program for Promoting the Enhancement of Research Universities (2022). This work was also supported by Advanced Bioimaging Support in MEXT/JSPS KAKENHI (22H04926).
Authorship contributions
Y.M. conceived and designed the study. I.K., M.I., and Y.M. collected data. Y.M., I.K., and T.H. wrote the manuscript.
Data availability statement
The data and processes of program that support the findings of this study are available from the corresponding author, Y.M., upon reasonable request.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/qpb.2022.24.
References
- Alanis, A. J. (2005). Resistance to antibiotics: Are we in the post-antibiotic era? Archives of Medical Research, 36, 697–705. 10.1016/j.arcmed.2005.06.009. [Abstract] [CrossRef] [Google Scholar]
- Angenon, G. , Dillen, W. , & Van Montagu, M. (1994). Antibiotic resistance markers for plant transformation. In Gelvin S. B., & Schilperoort R. A. (Eds.), Plant molecular biology manual (pp. 125–137). Springer Netherlands. 10.1007/978-94-011-0511-8_9. [CrossRef] [Google Scholar]
- Boavida, L. C. , & McCormick, S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana . The Plant Journal, 52, 570–582. 10.1111/j.1365-313X.2007.03248.x. [Abstract] [CrossRef] [Google Scholar]
- Bosch, M. , & Hepler, PK . (2006). Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta, 223, 736–745. 10.1007/s00425-005-0131-x. [Abstract] [CrossRef] [Google Scholar]
- Breyer, D. , Kopertekh, L. , & Reheul, D. (2014). Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants – Scientific developments, current use, operational access and biosafety considerations. Critical Reviews in Plant Sciences, 33, 286–330. 10.1080/07352689.2013.870422. [CrossRef] [Google Scholar]
- Gooh, K. , Ueda, M. , Aruga, K. , Park, J. , Arata, H. , Higashiyama, T. , & Kurihara, D. (2015). Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Developmental Cell, 34, 242–251. 10.1016/j.devcel.2015.06.008. [Abstract] [CrossRef] [Google Scholar]
- Gross, A. , Schoendube, J. , Zimmermann, S. , Steeb, M. , Zengerle, R. , & Koltay, P. (2015). Technologies for single-cell isolation. International Journal of Molecular Sciences, 16, 16897–16919. 10.3390/ijms160816897. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hashida, S. , Takahashi, H. , Takahara, K. , Kawai-Yamada, M. , Kitazaki, K. , Shoji, K. , Goto, F. , Yoshihara, T. , & Uchimiya, H. (2012). NAD+ accumulation during pollen maturation in Arabidopsis regulating onset of germination. Molecular Plant, 6, 216–225. 10.1093/mp/sss071. [Abstract] [CrossRef] [Google Scholar]
- Hellens, R. P. , Edwards, E. A. , Leyland, N. R. , Bean, S. , & Mullineaux, P. M. (2000). pGreen: A versatile and flexible binary Ti vector for agrobacterium-mediated plant transformation. Plant Molecular Biology, 42, 819–832. 10.1023/A:1006496308160. [Abstract] [CrossRef] [Google Scholar]
- Heslop-Harrison, J. , Heslop-Harrison, Y. , & Shivanna, K. R. (1984). The evaluation of pollen quality, and a further appraisal of the fluorochromatic (FCR) test procedure. Theoretical and Applied Genetics, 67, 367–375. 10.1007/BF00272876. [Abstract] [CrossRef] [Google Scholar]
- Hoffmann, F. , Sibley, R. B. , & Tsay, S. (1988). Transgenic antibiotic resistance may be differentially silenced in germinating pollen grains. Plant Cell Reports, 7, 542–545. 10.1007/BF00272752. [Abstract] [CrossRef] [Google Scholar]
- Hu, P. , Zhang, W. , Xin, H. , & Deng, G. (2016). Single cell isolation and analysis. Frontiers in Cell and Developmental Biology, 4, 116. 10.3389/fcell.2016.00116. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hwang, D. , Wada, S. , Takahashi, A. , Urawa, H. , Kamei, Y. , & Nishikawa, S. I. (2019). Development of a heat-inducible gene expression system using female gametophytes of Arabidopsis thaliana . Plant & Cell Physiology, 60, 2564–2572. 10.1093/pcp/pcz148. [Abstract] [CrossRef] [Google Scholar]
- Kamei, Y. , Suzuki, M. , Watanabe, K. , Fujimori, K. , Kawasaki, T. , Deguchi, T. , Yoneda, Y. , Todo, T. , Takagi, S. , Funatsu, T. , & Yuba S. (2009). Infrared laser-mediated gene induction in targeted single cells in vivo . Nature Methods, 6, 79–81. 10.1038/nmeth.1278. [Abstract] [CrossRef] [Google Scholar]
- Kawasumi-Kita, A. , Hayashi, T. , Kobayashi, T. , Nagayama, C. , Hayashi, S. , Kamei, Y. , Morishita, Y. , Takeuchi, T. , Tamura, K. , & Yokoyama, H. (2015). Application of local gene induction by infrared laser-mediated microscope and temperature stimulator to amphibian regeneration study. Development, Growth & Differentiation, 57, 601–613. 10.1111/dgd.12241. [Abstract] [CrossRef] [Google Scholar]
- Lage, O. M. , Ramos, M. C. , Calisto, R. , Almeida, E. , Vasconcelos, V. , & Vicente, F. (2018). Current screening methodologies in drug discovery for selected human diseases. Marine Drugs, 16, 279. 10.3390/md16080279. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lee, Y. , Kim, E.-S. , Choi, Y. , Hwang, I. , Staiger, C. , Chung, Y.-Y. , & Lee, Y. (2008). The Arabidopsis phosphatidylinositol 3–kinase is important for pollen development. Plant Physiology, 147, 1886–1897. 10.1104/pp.108.121590. [Abstract] [CrossRef] [Google Scholar]
- Matoušek, J. , Steinbachová, L. , Drábková, L. Z. , Kocábek, T. , Potěšil, D. , Mishra, A. K. , Honys, D. , & Steger, G. (2020). Elimination of viroids from tobacco pollen involves a decrease in propagation rate and an increase of the degradation processes. International Journal of Molecular Sciences, 21, 3029. 10.3390/ijms21083029. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nagahara, S. , Higashiyama, T. , & Mizuta, Y. (2021). Detection of a biolistic delivery of fluorescent markers and CRISPR/Cas9 to the pollen tube. Plant Reproduction, 34, 191–205. 10.1007/s00497-021-00418-z. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sanford, J. C. (2000). The development of the biolistic process. Vitro Cellular & Developmental Biology – Plant, 36, 303–308. 10.1007/s11627-000-0056-9. [CrossRef] [Google Scholar]
- Southern, P. J. , & Berg, P. (1982). Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. Journal of Molecular and Applied Genetics, 1, 327–341. [Abstract] [Google Scholar]
- Tupý, J. , Řihová, L. , & Žárský, V. (1991). Production of fertile tobacco pollen from microspores in suspension culture and its storage for in situ pollination. Sexual Plant Reproduction, 4, 284–287. 10.1007/BF00200549. [CrossRef] [Google Scholar]
- van den Berg, C. , Willemsen, V. , Hage, W. , Weisbeek, P. , & Scheres, B. (1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature, 378, 62–65. 10.1038/378062a0. [Abstract] [CrossRef] [Google Scholar]
- van den Berg, C. , Willemsen, V. , Hendriks, G. , Weisbeek, P. , & Scheres, B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature, 390, 287–289. 10.1038/36856. [Abstract] [CrossRef] [Google Scholar]
February 18, 2022
Prof. Olivier Hamant
Editor-in-Chief
Quantitative Plant Biology
Dear Prof. Olivier Hamant,
We would like ask you to consider our manuscript entitled “Target pollen isolation using automated infrared laser‒mediated cell ablation” for publication in Quantitative Plant Biology as an original article.
In the plant biological sciences, various imaging techniques have been used to study molecular and cellular dynamics for understanding biological phenomena. In this study, we aimed to establish a method for target cell isolation from the isolated cell populations using automated infrared laser-mediated cell ablation systems. After isolation, target cells were retained at the same location as before the treatment and were enriched in the cell population. This method can facilitate high-throughput studies at the single-cell level, such as gene expression analysis and quantification of cell dynamics, independent of any specific microfluidic devices, cell species and skilled professionals.
-All the authors have read and approved this manuscript.
-The manuscript has not been previously published, nor is it under consideration for publication elsewhere, including the Internet.
-There is no one who has reviewed this paper.
-To the best of our knowledge, there are no potential conflicts of interest to this study.
We look forward to seeing valuable comments from you and referees.
We will appreciate your consideration in reviewing our report.
Sincerely,
Yoko
---
Yoko Mizuta, Ph.D.
Institute of Transformative Bio-Molecules (WPI-ITbM),
Nagoya University,
Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan
e-mail: [email protected]
Comments to Author: This manuscript is reporting a scheme comprised of “H2B-tdTomato gene transfer through particle bombardment targeting N. benthamiana pollen on a glass-bottom dish”, “FDA staining,” “position register of the living transgenic pollen,” and “IR-laser ablation targeting the pollen of not-interest.” The setting conditions on IR-laser ablation were examined based on the binarized FDA signal intensities, indicating the frequencies of the ablated pollen of not-interest and the living transgenic pollen.
I understood that the reported scheme is feasible for live-cell imaging because the position is precisely maintained. However, the authors performed the precise position control with the originally equipped system, and this report is not on the development of the system itself. Although the ablation of pollen except for living transgenic one succeeded in this study, it isn't easy to imagine the practical experimental design in high-throughput and single-cell analyses.
It is critical to show an analytical example of an experiment using isolated pollen. The “isolated” pollen has remained on the glass-bottom dish. How are these living transgenic pollen moved from here and used? For example, are the ablated pollen population collected as bulk and available for the following analysis, such as pollination?
The technical novelty is hardly found in the current content. If a kind of analytical example using the “isolated” living transgenic pollen is indicated, the manuscript would be acceptable.
<Minor points>
(1) Some of the non-interest pollen escaped from ablation because such pollen is located in the vicinity of the living transgenic pollen. How about examining the masking region size (diameter) or adjusting the pollen density in a glass-bottom dish?
(2) The last sentence in section 3.1., “…140 mW laser power…” is 145 mW correctly?
(3) Second sentence from the last in section 3.2, “No pollen showing FDA-positive signals expressed red fluorescent protein” is correct?
(4) The words “bombarded pollen” (ex. Table 3) are a bit confusing because bombarded pollen does not always synthesize the transgene product, actually. How about using “tdTomato-positive pollen”?
Comments to Author: In this study, the authors aimed to isolate target pollen by laser ablation. Usually, cell isolation was conducted by selecting target cells, but this has several disadvantages, such as damage to cells and low throughput. Cell isolation by ablation of non-target cells is reverse thinking, and it is safer than using the target cell method. Furthermore, implementing automatization turns the ablation system into a high-throughput method. Overall, I agree with the author’s claims in this study and the novelty of this study. However, while the manuscript is written understandably, there are sentences that are difficult to read or sometimes hard to grasp. I commented on several points as follows.
#1 Introduction
“The methods used to individually recognize and separate cells of interest from other cells facilitate studying their biology, physiology, biochemistry, and taxonomy at the single-cell level (Gross et al., 2015).”
Finally, I could recognize “facilitate” is the actual verb in this sentence (not “used”, “recognize”, and “separate”). It was really confusing to read this sentence. This is a typical example; I strongly recommend authors carefully check each sentence and improve them throughout the manuscript.
#2 Introduction
“Laser capture microdissection, which was recently combined with image analysis algorithms (computer-assisted microscopy isolation) (Brasko et al., 2018), facilitates capture of individual target cells. However, accurate definition is complicated when cells show complex shapes. Micromanipulation, such as manual cell picking with micropipettes using a microscope, is a simple approach for single-cell isolation; however, its throughput is limited, and skilled professionals are required. Additionally, the above methods are not suitable for sequential analysis to evaluate the effects of environmental factors under constant conditions because cell location and environment are altered during cell isolation.”
I finally recognized that these sentences are explained as category (1) after repeated reading of this manuscript.
First, I thought that the explanation of category (1) is completed in this sentence.
“Fluorescence- activated cell sorting (FACS) and microfluidic droplet-based platforms are methods of category (1), which have been widely used for high-throughput isolation; however, this requires a large number of cells, and these harvesting techniques interfere with the cell location and phenotypic profile.”
And then the explanation of category (2) started after several sentences.
“Regarding methods of category (2), probably the most commonly…”
I was confused about what the intermediate sentences are (Laser capture microdissection…).
I strongly recommend the authors to mention these sentences belong to category (1).
#3 Introduction
In this manuscript, the authors developed a high-throughput ablation system with Nicotiana benthamiana pollen. However, there was almost no explanation about why the authors chose pollen and Nicotiana benthamiana in this study in the introduction section. Thus, I could not sufficiently catch the significance of this study from the introduction section. Part of these explanations seems to be mentioned in the discussion section. I think it is better to move these sentences to the introduction.
#4 Pollen vs. Pollens vs. Pollen grains
The grammatically correct plural of “pollen” is pollens. It is not largely used. If you refer to pollen in plural, but you do not want to use “pollens” then you can use “pollen grains”. Check this throughout the manuscript.
For example, page 12: “In this study, the pollen from one anther were suspended in pollen culture medium and were stained using FDA solution.”
#5 section 3.1
In this study, pollen viability was only defined by the FDA signal. It is possible that IR-irradiation disappears the FDA signal but pollen still may be viable. If the authors confirm pollen viability after irradiation by other evidence (such as by pollen germination ratio), it will support the authors claim. Or, it is better to cite it if the authors observed apparent morphological change after irradiation (for example, bursting pollen or leakage of pollen content, etc.). Even though the authors observed no morphological difference, it is important to cite them in the text.
In section 3.4
“The non- target pollen showed no FDA signal and shrank, whereas the target pollen retained both fluorescent signals derived from FDA and H2B-tdTomato (Figure 2).”
If this “shrank” indicate shrunk pollen grain, I recommend to explain it in more detail with a figure containing microscopic observations.
In the discussion, the authors wrote as follows,
“Not only the FDA, but other indicators such as cell state and morphology can also be used. “
This information should be placed in the result section and should be directly shown by evidence with a figure.
#6
Related to #5, it is important to show pollen viability about successfully bombarded pollen. I thought this was one of the important results of this study. (i.e., Isolated pollen is available (or not available?) for a subsequent experiment and making progeny.)
#7 section 3.2
“Pollen occur as isolated cells when collected from the anther, which is a simple collection method.”
This was a very confusing sentence for me.
#8 section 3.2
“After bombardment, gene expression was evaluated based on expression of fluorescent proteins at 18 hab (Figure 1).”
If the abbreviation “hab” is brought back to formal wording, it becomes as follows.
“After bombardment, gene expression was evaluated based on expression of fluorescent proteins at 18 h after bombardment (Figure 1).”
I feel strange about two “after bombardment” appearing in a single sentence. Is it OK for you?
#9 section 3.2
“A total of 20−94 introduced pollen grains were obtained per bombardment (the average efficiency of gene introduction was 57.6 ± 31.9 pollen).”
I could not understand why authors cite the average value here. What is “average efficiency”? I feel this is redundant because 57.6 ± 31.9 is 25.7-89.5, and these values are close to 20-94 and felt that this is meaningless. The authors should remove it or explain it in more detail.
I was also confused about reading this sentence.
“3.6%−9.4% (mean 5.8 ± 2.4%, Table 2).”
What does “mean” mean? Different statistic was applied with “average efficiency”?
#10 Fig1
No explanation for white circles in A1-3 and black circles A4. I assumed that they are aborted pollen (or non-stained pollen?). It should be explained in the Figure.
#11 Fig2
I think it is better to put symbols to ablated pollen grains in Figure2 “after”. I assumed that authors want to show this system can ablate non-target pollen efficiently. Currently, only failed pollen grains were emphasized by arrows.
#12 Page 5 – Plant materials and growth conditions
“Two-weeks-old seedlings were transferred to soil and were grown in a green room at 25–30 °C under long-day conditions (16 h light/8 h dark) in a greenhouse at 25–30 °C.”
Here the authors mention the green room and greenhouse. Is there any difference? If yes, please explain these differences or remove one.
#13 Page 12
“In step (1), the region in interest within the field of view was defined.”
It should be “the region of interest”.
#14 Discussion
“This indicates that the fluorescent signal derived from the FDA of approximately one third of the pollen was below the threshold. Target bombarded pollen is masked and omitted from laser irradiation, the cause of which may be photobleaching of the FDA dye to under the threshold. The fluorescent proteins and dyes used for target cell visualization should thus be photostable. This method may facilitate high-throughput studies at the single- cell level, such as gene expression analysis and cell dynamic imaging in various organisms.”
I could not understand what the authors would like to claim here.
Do these sentences imply that FDA is not suitable for cell isolation?
Comments to Author: Dear Dr. Mizuta and colleagues,
We have just received the comments of two expert reviewers on your manuscript submission. Their detailed comments are attached.
As you will see, a reviewer suggests the changes that could be addressed by rephrasing and modulating the figure. But the other reviewer asks to add the data showing the analytical example using the isolated pollen.
It would be ideal if actual experimental data could be added. But if this is not possible, I think it would help if you could propose several examples of a feasible analysis.
We would be happy to receive a corrected version of your manuscript when it is ready.
I thank you again for having submitted your excellent manuscript to Quantitative Plant Biology.
Best regards,
Minako Ueda
No accompanying comment.
No accompanying comment.
Not Approved
Comments to Author: The revised manuscript has been improved by showing the germination ability of pollen isolated by IR-laser ablation. In addition, overall descriptions have been more understandable than in the previous version. I agree with these improved points; however, the present content seems not to meet the level for publication yet. This is because the structure of the content is still hard to understand, and the use of inappropriate values is shown to some extent. I strongly recommend being subjected to English editing by the main expert. If the authors want to claim the feasibility of this study, more quantitative data is required. Also, it is recommended to reconsider the claim possible from the present results.
In Fig. 2B, a higher proportion (67.3%) of pollen tubes germinated from the target pollen were indeed observed; however, at least 3 biological repeats should be performed to obtain the quantitative data statistically. Moreover, the proportion is less than 2 times that of the control experiment (36.8%). Although the enrichment of target pollen succeeded, it seems not enough as the feasible level for the future analyses raised by the authors.
The authors added the reason why they chose Nicotiana pollen for this study and the possible application in “Introduction” in response to reviewer 2. I agree with this point because the result in Fig. 2B coincides with this claim. Therefore, please revise the “Discussion” section in accordance with the “Introduction” section. If authors insist on “high-throughput studies at single-cell level” as the future application, a specific example should be raised. What can be evaluated by using bombarded and isolated pollen, even if the introduced gene is not expressed before pollen tube germination?
The critical point of this study is how to increase the purity (rate of content) of the target pollen among the samples after the laser ablation loops. That’s why I requested to evaluate the density as one of the previous comments. I meant that pollen density can be adjusted quantitatively by calculation because the pollen is suspended with a liquid medium before being mounted on the glass-bottom dish.
In the revised manuscript, why did the authors use UV excitation (for CFP image) for indication of the pollen grains? I wonder it because pollen viability seemed to be easily judged by the morphology in bright field. Please add the explanation.
<Other specific points>
<b>Section 3.1</b>
The value 165-190s (line 3 in this section) should be revised to 165-225s. The authors finally set the irradiation time at 10 ms. Please add the reason why 20 ms was not employed in spite of the required time for irradiation is shorter than that at 10 ms.
<b>Table 1<b>
● Please add a caption explaining “N. D..”
● Please move the item words “Laser power at the focal plane (mW)” to the upper side of the values.
● How about rephrasing the title “Time required for disappearance of FDA signals in irradiated pollen.”
<b>Section 3.2 and Table 2<b>
● Regarding Table 2 and the part after the descriptions from “Thereafter, the number of non-target pollen……”, the numbers of FDA-positive pollen in the population from a single anther (ca. 20000 pollen grains) are indicated? Also, I wonder why the values as the number of FDA-positive pollen is the product of the multiplication of “Irradiation area” by “Density.” Does it mean that all cells mounted on a glass-bottom dish were FDA-positive? Please confirm if these values are appropriate.
● In the text, the authors mentioned the frequency and the average. It would be helpful if you add these items to Table 2.
<b>Section 3.3<b>
● The values in the sentence “Using multiple fields of view……” (lines 12-15 from the start of this section) are not included in both Tables 2 and 3. Please indicate the source and process to obtain these values. If this description corresponds to my question regarding FDA-positive pollen in Table2, this part should be moved to section 3.2.
● At line 18 in the same section, the values 24.0-28.2 should be revised to 24.0-34.9.
● Overall, it is difficult to understand the correlation between the “Irradiation area,” “The xy tiling,” and “fields of view.” Similarly, the mixture use of “processing” and “irradiation” is confusing. Please clarify the definition of these words, or try to unify the word with the same meaning. The item words in tables also should be minded.
● In the last part of this section, the authors described the time required for a single loop. Please add this information in Table 2. I couldn’t calculate these values from the present information. It would be helpful for readers to indicate the source information.
● In the same part, the author expressed the time using “minutes.” Please unify the expression in the text and Table 2.
<b>Section 3.4<b>
● Indeed the result described in the title was obtained, but the correlation between the FDA-positive pollen density and the target isolation efficiency was not seen in the indicated results. I understand this type of study tends to show wider deviation. It might be good to mention the range of the obtained efficiency, in addition to the descriptions of the best result.
● As for another point, the present descriptions focus only on the FDA-positive pollen density. I think the overall density, including both viable and non-viable pollen, influences the efficiency more because it influences the effectiveness of masking. Therefore I’ve requested the evaluation of pollen density. Moreover, it seems better to combine the present Tables 2 and 3.
<b>Section 3.5<b>
● The 35Sp::H2B-tdTomato transgenic tobacco was not mentioned in the materials and methods. Please add the related information.
● Regarding the sentence “The FDA fluorescent signals in several pollens disappeared, and these pollen grains showed bursts despite being kept from laser ablation” (from line 12th in this section), please add discussion. What is considered to cause this? And I recommend replacing “pollens” with “pollen grains.”
● From lines 18 to 19, which transgenic line was used (35Sp::H2B-tdTomato or AtUBQ10p::tdTomato)?
<b>Fig. 2A<b>
Please indicate the cyan circle lines also on the H2B-tdTomato picture after the irradiation.
<b>Legend of Sup. Video 1<b>
The size of the masking area is described as “30 µm”, but “60 µm” in the legend of Fig. 1. Please revise to the correct value.
Comments to Author: In this study, the authors aimed to isolate the target pollen by laser ablation. This version of the manuscript has really improved readability with additional data about pollen tube elongation in a culture medium and semi-in vivo assay. I think that the current version of the manuscript provides enough novelty and data to be accepted. I put some comments to improve readability as follows.
#1 Low selectivity for target pollen grains.
The authors claimed increased selectivity of target pollen as follows(page 17),
“The highest cell isolation efficiency occurred in sample I, and the proportion of tdTomato-positive pollen after irradiation was 8.3-fold higher, increasing from 3.7 % to 30.8 %.”
For me, the ratio (30.8%-3.7%, Table 3) of targeted pollen/total pollen does not seems to be a high selectivity to me.
In this point, the authors stated (page20-21),
“The appropriate density of cells is also important for effective isolation because a high cell density responsible for more non-target pollen may lead to an overlap with the masking area, which reduces isolation efficiency (Figure 2).”
But I thought alternative idea of improving the selectivity ratio would be possible. For example,
ex1. Improving area definition
Now it seems if non-target pollen overlapped with the masking region, it will be not ablated; however, improving ablation condition by imaging program(or other method?), I think selectivity will be drastically improving.
ex2. Two times ablation after physically moving pollens
If the addition of one more ablation step after (gently) shaking the media after the 1st ablation, the selectivity ratio will be improved.
I recommend the authors discuss the improvement of the selectivity ratio.
#2 Table 1
#2-1
The column “laser power at…” should be located above the mW number (i.e., “24”)
#2-2
What “N.D.” means? I could not find any explanation for this abbreviation.
(“No data”? If it is yes, all columns of 24mW are unnecessary, “Not disappeared”? If so, the authors should explain it).
#3 Table 3
“Viability of bombarded pollen (%)” may not be correct.
“Viability of tdTomato-positive pollen before and after IR-laser irradiation (%)” may be correct.
#4 Fig1B
From this figure, this method seems to be able to select target pollen with 100% selectivity. However, the current method is far from 100% (max. 30.8%). I recommend putting at least one FDA positive pollen (non-target pollen) in “After irradiation” figure to avoid overestimating/misunderstanding for the readers.
#5 Fig 2B
I am really confused after reading this sentence (page 18).
“The tdTomato- expressing pollen tubes were 35 in number, i.e., 67.3 %, (out of a total of 52 tubes that underwent laser irradiation) (Figure 2B), whereas there were 21 out of a total of 57 pollen tubes that did not undergo laser-irradiation (36.8 %, Figure 2B).”
#5-1
Please distinguish Figure 2B legend using such as “2B left/right” or “2B -Laser irradiation/+Laser irradiation”.
#5-2
I also could not catch where the numbers “35, 52, 21, 57” came from.
I recommend author cite this data somewhere (in Fig2B?, making a new plot or table?).
#5-3
I could not understand why tdTomato signal was colored green. First, I misunderstood this green signal as FDA. I recommend that the authors should use a uniform color code throughout the manuscript. But if it is difficult technically, I can accept this figure.
#6
“Pollen grains obtained from flower buds were used for biolistic delivery of plasmid vectors into cells”
I think that it would increase the reproducibility if the authors could state in which developmental stage the flower buds were.
#7
“We previously reported that bombardment pollen…”
It should be “We previously reported that bombarded pollen…”
Comments to Author: Dear Dr. Mizuta and colleagues,
We have just received the comments of two expert reviewers on your revised manuscript. Their detailed comments are attached.
As you will see, a reviewer requested the evaluation of pollen density and the biological repeats of pollen tube germination experiment. I think both of these are feasible and would be very important for publication in Quantitative Plant Biology.
We would be happy to receive a corrected version of your manuscript when it is ready.
I thank you again for having submitted your excellent manuscript to Quantitative Plant Biology.
Best regards,
Minako Ueda
No accompanying comment.
Dear Editor,
We wish to thank you and the peer reviewers for very positive reviews and for very useful and insightful comments regarding how to improve the manuscript. We are sorry that our response and edits are so late because we had to conduct additional experiments. Now, I am pleased to inform you that all of the requests have been met, and we not only highlight places in the text where edits were made, but also provide a detailed response to peers’ requests below.
All authors have seen and assisted with the editing of the revised manuscript and approve the re-submission to Quantitative Plant Biology. There are no conflicts of interest.
Please feel free to let us know if any requests remain and we look forward to seeing or findings published in Quantitative Plant Biology.
Sincerely,
Yoko
Comments to Author: The revised manuscript contains newly added quantitative data enough to claim the future feasibility of the reported scheme. The present data satisfies the level for publication in QPB. In minding reader-friendly construction, it might be better to revise the order of descriptions in sections 3.2 and 3.3 because some sentences are repeated.
1) It seems good to combine sections 3.2 and 3.3. My suggestions are below;
>Move the sentences at lines 1-6 from the bottom of page 14 (First, 151 pollen grains……., were excluded from subsequent laser irradiation.) to the top of section 3.2.
>Remove the sentences from line 20 of p13 to line 9 of p14 (The FDA solution was added at 18h, ………Using the above five samples, we isolated the target pollen by cell disruption of non-target pollen with IR laser irradiation.).
>Move and place the sentences from lines 2-7 of p16 after the sentence “Pollen grains were suspended in pollen culture medium and stained with the FDA solution” (p14).
>Replace the sentence “As the number of pollen grains increased……Irradiation time depended on the area and number of pollen grains.” (lines12-14 in p15) with the sentence “Sample I, which had the lowest pollen density……These results indicate that pollen density affects isolation efficiency” (lines 17-21 in p16).
>Remove the remained part in section 3.3, retitling the combined section.
2) Other minor points;
>Section 3.3 and Table 2; Authors evaluated the separation of pollen, but I couldn’t well imagine the state of “pollen separation” from the caption. In the case of “True”, does it means that all pollen grains in a single tile are independently dispersed without merging or contacting each other? It would be helpful for readers if more detailed explanations were added.
>Legend “a” in Table 2: culturated -> calculated
>Line 6 of p17: How about adding “(10 anthers from 5 flowers)” after “12,717 ± 2,814”.
>Lines 20-21 of p21: How about if you mention the actual counted data of pollen FDA-staining?
>Lines 10-12 (p13) and Table S1: Although it could be my misunderstanding, 1 minute is 60,000 milliseconds. If the unit in Table S1 is ms, all the indicated times are less than 1 minute. Please confirm the unit for time.
Comments to Author: This version of the manuscript has improved tables (i.e., clear and informative) and improved figures (i.e., intuitive and close to results). Furthermore, the authors added some tables, which are quantitative and well-support the data. The authors replied to my comments properly.
Comments to Author: We are pleased to inform you that your manuscript has finally been accepted. We would appreciate it if you correct the minor points pointed out by Reviewer1.
No accompanying comment.
Dear Editor,
It is a great pleasure to accept our manuscript.
We corrected the minor points pointed out by Reviewer 2 in the revised manuscript R3 as a final version.
We look forward to seeing publishing in Quantitative Plant Biology.
Sincerely,
Yoko
No accompanying comment.
No accompanying comment.
Articles from Quantitative Plant Biology are provided here courtesy of Cambridge University Press and the John Innes Centre
Full text links
Read article at publisher's site: https://doi.org/10.1017/qpb.2022.24
Read article for free, from open access legal sources, via Unpaywall:
https://www.cambridge.org/core/services/aop-cambridge-core/content/view/E7CE0CCEA634D80020A95567ED0800F8/S2632882822000248a.pdf/div-class-title-target-pollen-isolation-using-automated-infrared-laser-mediated-cell-disruption-div.pdf
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/140380553
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa.
BMC Genomics, 11:338, 28 May 2010
Cited by: 92 articles | PMID: 20507633 | PMCID: PMC2895629
Doubly guaranteed mechanism for pollination and fertilization in Ipomoea purpurea.
Plant Biol (Stuttg), 22(5):910-916, 08 Jun 2020
Cited by: 2 articles | PMID: 32270571
Detection of a biolistic delivery of fluorescent markers and CRISPR/Cas9 to the pollen tube.
Plant Reprod, 34(3):191-205, 19 Jun 2021
Cited by: 7 articles | PMID: 34146158 | PMCID: PMC8360903
Synchronicity of pollination and inoculation with Claviceps africana and its effects on pollen-pistil compatibility and seed production in sorghum.
Fungal Biol, 114(4):285-292, 03 Mar 2010
Cited by: 1 article | PMID: 20943138
Review

 1
,
5
1
,
5