Abstract
Free full text

Full-length prion protein incorporated into prion aggregates is a marker for prion strain-specific destabilization of aggregate structure following cellular uptake
Abstract
Accumulation of insoluble aggregates of infectious, partially protease-resistant prion protein (PrPD) generated via the misfolding of protease sensitive prion protein (PrPC) into the same infectious conformer, is a hallmark of prion diseases. Aggregated PrPD is taken up and degraded by cells, a process likely involving changes in aggregate structure that can be monitored by accessibility of the N-terminus of full-length PrPD to cellular proteases. We therefore tracked the protease sensitivity of full-length PrPD before and after cellular uptake for two murine prion strains, 22L and 87V. For both strains, PrPD aggregates were less stable following cellular uptake with increased accessibility of the N-terminus to cellular proteases across most aggregate sizes. However, a limited size range of aggregates was able to better protect the N-termini of full-length PrPD, with the N-terminus of 22L-derived PrPD more protected than that of 87V. Interestingly, changes in aggregate structure were associated with minimal changes to the protease-resistant core of PrPD. Our data show that cells destabilize the aggregate quaternary structure protecting PrPD from proteases in a strain-dependent manner, with structural changes exposing protease sensitive PrPD having little effect on the protease-resistant core, and thus conformation, of aggregated PrPD.
The native prion protein, PrPC, is not essential for mammalian survival (1) despite participating in protection from redox stress (2), nucleotide metabolism (3) and myelin formation (4). However, the prion protein can take on a range of non-native and infectious conformations (5,6), referred to collectively as PrPSc, for PrP scrapie, or PrPD, for disease associated PrP, the term used throughout this study. Prions propagate by converting native PrPC into the non-native conformation of PrPD(7,8). The main, if not sole, component of infectious prions is PrPD. As infectious prions propagate, they form protease-resistant PrPD aggregates that can deposit in tissues and interfere with vital cellular processes (9,10). Accumulation of PrPD faster than it can be cleared by cellular machinery is associated with the cell death and tissue degeneration that occurs in fatal and incurable prion diseases like chronic wasting disease in deer and elk, scrapie in sheep and Creutzfeldt–Jacob disease in humans. How PrPD eludes degradation long enough to successfully propagate is thus one of the most important questions in prion biology.
The properties of PrPD that govern infectivity, such as its ability to convert PrPC and its stability, may depend on the unique PrPD conformation associated with a given prion strain (11–14). However, each region and modification of the prion protein can contribute to the properties of aggregated PrPD in specific ways (15,16). The central and C-terminal regions of the prion protein comprise the protease-resistant core of PrPD from approximately amino acid residues 90 to 230 (17,18). This region generally adopts a β-sheet conformation unique to each prion strain and organizes into parallel in-register β-sheets in the quaternary structure of amyloid aggregate fibrils observed during prion infection (5,18–20). The ordered fibrillar cores of at least the 22L, RML and ME7 murine prion strains are not detectably affected by glycophosphatidyl-inositol (GPI) anchors and the extent of N-linked glycosylation (5,18,21). While the protease-resistant core of PrPD is structurally stable and capable of converting PrPC into PrPD, bound cofactors and posttranslational modifications can alter the properties of PrPD(22–24). The C-terminus of the prion protein is linked to a GPI anchor that helps PrPC and PrPD attach to membrane (24,25). In addition, the GPI anchor can be found integrated into large aggregates of PrPD(26) and may promote association between PrPD and membrane, which has been found to promote prion conversion (27–29). The prion protein can also be glycosylated at two different sites near the C-termini (30) and glycosylation can influence both the conversion and stability of PrPD in a strain and species specific manner (23,31,32).
The N-terminus of the prion protein can be naturally cleaved from PrPC at several different cleavage sites (reviewed in (33)), yielding N-terminally truncated fragments that have physiological importance not only for the function of PrPC(34) but also for the properties of PrPD(35). PrPC with N-terminal truncations or mutations are able to be converted into PrPD and form aggregates, but the presence of the full and unmutated N-terminus influences conversion and facilitates the formation of larger aggregates (22,36–39). Interprotein interactions between the N-terminus and other aggregate components may also contribute to the stability of prion aggregates, since binding sites for cofactors that promote the stability of prion aggregates are found in the N-terminal region of PrP (38,40–42). Importantly, the accessibility of the N-terminus of PrPD to protease digestion can vary by prion strain and is considered to be indicative of strain-specific differences in PrPD structure (43,44).
While the high structural stability and partial protease resistance of PrPD undoubtedly play roles in hindering clearance of PrPD by cellular machinery, prion strains with less stable PrPD are sometimes more capable of infecting cells than prion strains with more stable PrPD. For example, the mouse adapted prion strain 87V is more stable than other mouse adapted strains, such as 22L (45). However, 87V is unable to infect cell lines that can be infected by 22L even though de novo PrPD can be detected in cells following acute exposure to 87V prions (45,46). The fact that cellular PrPD formation induced by 87V is transient suggests that 87V PrPD may be more susceptible to cellular degradation and the activity of the proteostasis machinery than 22L PrPD. Indeed, by monitoring the size and protease resistance of 22L and 87V PrPD following cellular uptake we have recently shown that, while both strains undergo similar changes in aggregate size, protease resistance, trafficking and degradation, 87V PrPD was degraded more rapidly (47). This suggests that strain-specific structural differences between 87V and 22L PrPD aggregates may influence the stability of PrPD in the cell and thus its susceptibility to cellular degradation.
Since the accessibility of the PrPD N-terminus (amino acids ~ 23–89) to proteases varies with PrPD structure (43,44), we reasoned that tracking the ratio of full-length and N-terminally truncated prion protein in PrPD aggregates could provide insight into how the structure and stability of different prion strains change during cellular uptake. We therefore monitored over time both the percentage of PrP containing the N-terminus (i.e. full-length PrP) and the percentage of N-terminally truncated PrP following cellular uptake of 22L and 87V PrPD. Our data show that full-length PrP is part of the brain-derived PrPD aggregate, with the ratio of full-length PrP to N-terminally truncated PrP in both 22L and 87V PrPD being constant across a wide range of aggregate sizes. However, following cellular uptake and exposure to cellular proteases, the ratio of full-length PrP to N-terminally truncated PrP changes in a manner dependent upon lysosomal activity, aggregate size, and prion strain. Furthermore, we found that the N-termini of PrP can be protected from proteolytic degradation within large aggregates of PrPD, potentially allowing them to contribute to the properties of prion aggregates after proteolytic degradation. Our data suggest that during cellular processing the quaternary structure of PrPD aggregates is altered in a strain-specific manner, leading to changes in the exposure full-length PrPD with little impact on the protease-resistant core of PrPD.
23–89) to proteases varies with PrPD structure (43,44), we reasoned that tracking the ratio of full-length and N-terminally truncated prion protein in PrPD aggregates could provide insight into how the structure and stability of different prion strains change during cellular uptake. We therefore monitored over time both the percentage of PrP containing the N-terminus (i.e. full-length PrP) and the percentage of N-terminally truncated PrP following cellular uptake of 22L and 87V PrPD. Our data show that full-length PrP is part of the brain-derived PrPD aggregate, with the ratio of full-length PrP to N-terminally truncated PrP in both 22L and 87V PrPD being constant across a wide range of aggregate sizes. However, following cellular uptake and exposure to cellular proteases, the ratio of full-length PrP to N-terminally truncated PrP changes in a manner dependent upon lysosomal activity, aggregate size, and prion strain. Furthermore, we found that the N-termini of PrP can be protected from proteolytic degradation within large aggregates of PrPD, potentially allowing them to contribute to the properties of prion aggregates after proteolytic degradation. Our data suggest that during cellular processing the quaternary structure of PrPD aggregates is altered in a strain-specific manner, leading to changes in the exposure full-length PrPD with little impact on the protease-resistant core of PrPD.
Methods
Prion propagation and animal care
The 22L and 87V mouse-adapted prion strains were propagated in C57BL/10 and VMDK mice, respectively. Clinically positive mice were euthanized and the prion infected brains were removed and stored at −80°C until use. The recommended protocols in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health were stringently followed throughout the inoculation, care up to the clinical stage of prion disease and subsequent euthanasia of the mice that were used to propagate prions. All protocols used for animal experiments were reviewed and approved by the Rocky Mountain Laboratories Animal Care and Use Committee.
Preparation of 22L and 87V prion infected brain homogenate
Brains from mice infected with either 22L or 87V prions were homogenized in phosphate-buffered saline (PBS) using a Mini-BeadBeater-8 instrument (Biospec) at a concentration of 10% (wt/vol) as described previously (47). Cellular debris was pelleted from brain homogenates by centrifugation at 500g for 5 min before clarified supernatants were aliquoted and stored at −80°C until use.
Uptake of 22L and 87V prions by CF10 cells
CF10 cells, a neural cell line derived from mice lacking the PRNP gene (48), were grown to 95% confluency in complete media (Opti-MEM (Gibco) supplemented with 10% fetal bovine serum (FBS, ATLAS Biologicals), 100 U of penicillin (Gibco), and 100 μg of streptomycin (Gibco)), before being resuspended in saline-trypsin-versine (STV) solution, diluted into complete media, and added to 6-well plates at a concentration of 1.0 × 106 cells/well. Cells were then incubated at 37°C with 5% ambient carbon dioxide overnight in a humidified incubator. Prior to prion exposure, complete media was removed and CF10 cells were washed four times with warmed Opti-MEM, 1 mL/wash. Opti-MEM containing 100 U of penicillin, 100 μg of streptomycin and 0.25% brain homogenate from mice infected with 22L or 87V prions was then added to the cells (750 μL/well), and the cells incubated at 37°C for 4 h before the addition of 750 μL of pre-warmed complete medium containing 20% FBS. Samples taken after addition of complete medium constituted the 4 h time point, while the remaining cells were incubated at 37°C for an additional 20 h before samples were taken (24 h time point).
After 4 and 24 h, media was removed and cells were washed four times (1 mL complete media/wash) before being washed once with 1 mL of STV. Cells were then incubated in 1 mL of STV for 1 min before being gently resuspended, mixed with 500 μL of complete media, and spun at 5400g for 5 min. After supernatant removal, cell pellets were resuspended in 1 mL of 1× PBS containing 2% n-octyl-β-d-glucopyranoside (NOG, EMD Milipore) and were allowed to lyse in the detergent over 2 min. Lysates were then spun at 5400g for 5 min before 950 μL of supernatant was pulled from each sample and mixed with 50 μL of 20% sarkosyl (MP Biomedicals) for a final concentration of 1% sarkosyl. The starting material (0 time point) was brain homogenate from mice infected with 22L and 87V prions mixed with NOG and sarkosyl to a concentration of 2 and 1% respectively. Samples were aliquoted and stored at −80°C until use. Where appropriate, samples were treated with proteinase K (PK; Novagen) at a concentration of 10 μg/mL for 1 h at 37°C, and then PK was inhibited by the addition of Pefabloc (Sigma-Aldrich) to 2 mM. Both PK treated and untreated samples were mixed 2:1 with a 5:1 2-butanol/methanol mixture before being centrifuged for 30 min at 18,300g in an F301.5 Beckmann rotor at 4°C to both precipitate and pellet protein from each sample.
Lysosomal activity assay
CF10 cells were prepared and exposed to prion infected brain homogenate as described above, except that the cells were placed in 12-well plates with 0.36 ×
× 106 cells/well and the media volumes used were halved. CF10 cells expressing mouse PrP bearing the 3F4 epitope that were or were not infected with 22L prions (49) were plated at the same concentrations as CF10 cells but were not exposed to brain homogenates. As a negative control, some wells were exposed to either media containing 0.25% normal brain homogenate (NBH) or media containing no brain homogenate. The lysosomal activity of CF10 cells was evaluated at 3 and 24 h post-exposure by quantifying the fluorescence of a self-quenched fluorescent peptide substrate (BioVision) that is un-quenched after uptake and degradation of the peptide in the lysosome. For the assay, cells were first washed with and then incubated in Opti-MEM not supplemented with FBS for 4 h. FBS was then added to the media and the cells incubated for 8 h. To assay for lysosomal activity, the self-quenched substrate was diluted to 0.02 mg/mL via a 66.6× dilution into cellular media 1 h before each time point, at which time cells were washed in complete media, resuspended with STV and analysed on an Accuri C6 flow cytometer (BD Biosciences) using the FL1/FITC channel. For each analysis, 30,000 to 35,000 cells were analysed. As a control for no lysosomal activity, 1 μM of bafilomycin (BioVision) was added to media that did not contain brain homogenate 1 h before the self-quenched dye was added.
106 cells/well and the media volumes used were halved. CF10 cells expressing mouse PrP bearing the 3F4 epitope that were or were not infected with 22L prions (49) were plated at the same concentrations as CF10 cells but were not exposed to brain homogenates. As a negative control, some wells were exposed to either media containing 0.25% normal brain homogenate (NBH) or media containing no brain homogenate. The lysosomal activity of CF10 cells was evaluated at 3 and 24 h post-exposure by quantifying the fluorescence of a self-quenched fluorescent peptide substrate (BioVision) that is un-quenched after uptake and degradation of the peptide in the lysosome. For the assay, cells were first washed with and then incubated in Opti-MEM not supplemented with FBS for 4 h. FBS was then added to the media and the cells incubated for 8 h. To assay for lysosomal activity, the self-quenched substrate was diluted to 0.02 mg/mL via a 66.6× dilution into cellular media 1 h before each time point, at which time cells were washed in complete media, resuspended with STV and analysed on an Accuri C6 flow cytometer (BD Biosciences) using the FL1/FITC channel. For each analysis, 30,000 to 35,000 cells were analysed. As a control for no lysosomal activity, 1 μM of bafilomycin (BioVision) was added to media that did not contain brain homogenate 1 h before the self-quenched dye was added.
Sucrose gradient fractionation
Discontinuous sucrose gradients were created by layering 10%, 15%, 20%, 25%, 30%, and 60% sucrose, made in a 1% sarkosyl and PBS buffer, in 750 μL volumes that were pre-chilled to 4°C. Pre-chilled samples were loaded onto the gradient and spun 1 h at 4°C and 200,00g in an SW55TI Beckmann bucket rotor. From the top of each gradient, 450 μL fractions were removed and mixed with 550 μL of 1× PBS and 1% sarkosyl, as described previously (47). Fractions were then split in half and one half was mixed with PK to a final concentration of 10 μg/mL and incubated at 37°C for 1 h. Pefabloc was then added to a final concentration of 2 mM and the solution incubated for 15 min at room temperature to inactivate PK. Subsequently, protein from both PK treated and untreated samples was precipitated by the addition of a 5:1 2-butanol/methanol mixture, which was mixed at a volumetric ratio of 1:2. Precipitated protein was then pelleted with a 30-min spin at 18,300g in an F301.5 Beckmann rotor at 4°C.
Guanidine hydrochloride stability assays
Infected brain homogenate and cell lysate samples were diluted to 1% brain homogenate or cell lysate material (wt/vol) in 1× PBS containing 1% sarkosyl and 2% NOG before being mixed 1:1 with guanidine hydrochloride (GdnHCl) and 1× PBS to final concentrations of 0, 1, 2, 3, and 4 M GdnHCl. Samples were denatured for 1 h on a shaking platform held at room temperature before being diluted 1:10 in lysis buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% sodium deoxycholate and 0.5% Triton X-100) containing enough GdnHCl to adjust the final concentration of GdnHCl to 0.4 M. Samples were split and either spun down over a 10% sucrose layer in lysis buffer for 45 min in an F301.5 Beckmann rotor at 18,300g or digested for 1 h at 37°C in 10 μg/mL PK before digestion was terminated by treatment with 2 mM Pefabloc and a 15 min incubation at room temperature. Samples treated with PK were diluted 2:1 with a 5:1 2-butanol/methanol mixture before being spun 30 min spin at 18,300g in an F301.5 Beckmann rotor at 4°C to precipitate the protein in each sample.
Western blot development
Pelleted samples from brain homogenate, guanidinium, sucrose gradient and uptake assays were resuspended in 25 to 100 μL of 2× NuPAGE sample buffer (Novex) before being heated at 95°C for 5 min. Samples from guanidinium stability assays that were not de-glycosylated were then vortexed, spun down and loaded onto a 15% BisTris 1.5 mM gel (Invitrogen). All other samples were de-glycosylated in sample buffer with PNGase F (NEB) by first mixing 12 μL of sample pre-diluted and denatured in NuPage sample buffer with 1 μL of glycoprotein denaturing buffer (NEB) and heating at 95°C for 5 min. Fully denatured samples were then mixed with 1 μL each of glycobuffer (NEB), NP-40 (NEB) and PNGase F (500,000 U/mL) before being incubated at 37°C overnight. Prior to loading on a gel, samples were again heated to 95°C for 5 min. All gels were run at a constant voltage of 75 V until the dye front reached the bottom of the gel (~3 h). The contents of each gel were transferred to polyvinylidene difluoride (PVDF, EMD Milipore) Immobilon-P membranes overnight with a NuPAGE Novex gel system run at a constant 33 V.
PVDF membranes were blocked with TBST (10 mM Tris HCl (pH 8.0), 150 mM NaCl and 0.05% Tween 20) containing 5% (wt/vol) dry milk before being rinsed and probed with anti-PrP monoclonal antibodies for 1 h at room temperature. The anti-PrP antibodies 6D11 (BioLegends, PrP epitope between amino acid residues 93–109), Saf32 (PrP epitope between amino acid residues 59 and 89) and Saf84 (PrP epitope between amino acid residues 159 and 169), both from Cayman Chemical Co., were diluted to 1:5000, 1:10,000 and 1:10,000, respectively. After incubation with the primary antibody membranes were rinsed four times with TBST over 30 min before being incubated with a 1:40,000 dilution of sheep anti-mouse horseradish peroxidase-conjugated secondary antibody (GE Amersham) for 1 h. The blots were coated in ECL Plus reagent (GE Healthcare Life Sciences) so they could be developed on photosensitive film, which was later scanned and quantified digitally using Un-Scan-IT software (version 7.1; Silk Scientific Corporation).
Blot analysis, calculations and statistics
Similarly to previous work (47), bands of PrP on scanned images of film were quantified in the Un-Scan-IT software using the ‘Segment Analysis’ tool. Bands in each lane were boxed in and quantified via summation of all pixels in the analysis region before the background value was subtracted from each analysis region. The background for each band was calculated by taking the average sum of pixels in four identically sized analysis boxes near each band. For data from guanidinium assays that were not PNGase F treated, the data were normalized to the 0 M guanidinium sample by dividing the background corrected pixel sums of each replicate by the average background corrected pixel sum of the 0 M guanidinium sample. The average and standard deviation of replicates for each guanidinium concentration was then calculated and each normalized average was then multiplied by 100 to convert each value into a percentage. For PNGase F treated samples, the percentage of the PrPD population was calculated by taking the background corrected pixel sum of either the 25 or 23 kDa PrPD bands and dividing it by the sum of all background corrected pixel sums for all PrPD bands in a lane. The ratio of 25 kDa PrPD to total PrPD from each lane was then multiplied by 100 in order to convert each ratio into a percentage before calculated percentages from each replicate were averaged and the standard deviation calculated. The data are thus normalized to the amount of PrPD within each lane, allowing the direct comparison of experimental samples between different experiments and gels. The consistency of the calculated percentages of PrPD across different gel exposures, individual gels, and PrPD concentrations was validated in a set of control experiments (Supplementary Figs S1–S3). Statistical analysis for each data set was done in Prism Graphpad Software (version 8.0.2). In figures, the asterisks represent P values within specific ranges with p* =
= 0.01–0.04, p**
0.01–0.04, p** =
= 0.005–0.009, p***
0.005–0.009, p*** =
= 0.001–0.004 and p****
0.001–0.004 and p**** =
= <
< 0.0009.
0.0009.
Results
PrPD from 22L and 87V contains a population of full-length PrP protected from proteases
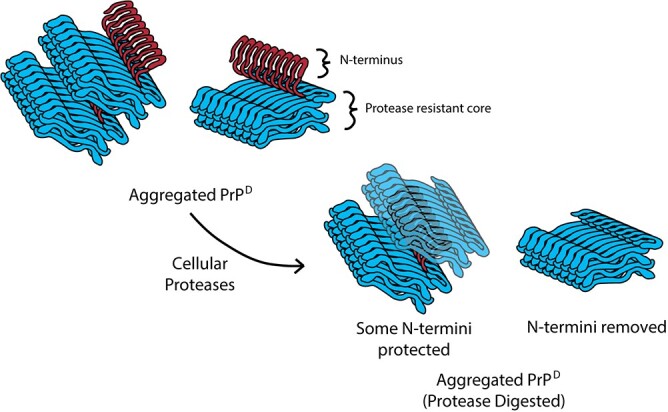
In order to understand how the cell alters aggregate structure following uptake of PrPD, we first characterized the sizes of PrP molecules in the brains of mice infected with either 22L or 87V prions. To simplify discussion of the data, we will continue to refer to PrP associated with prion infection as PrPD regardless of the protease-sensitivity or molecular mass of the PrP molecules. PrPD in brain homogenates from prion infected mice was de-glycosylated with PNGase F in sample buffer to prevent glycosylation from obscuring the actual size of the PrPD molecules. For those samples treated with PK to identify protease-resistant PrPD, de-glycosylation in SDS-PAGE sample buffer followed PK digestion. De-glycosylated untreated and PK treated samples were analysed by western blot (Fig. 1A) using the anti-PrP antibodies Saf32, 6D11 and Saf84. These antibodies recognize the N-terminal, central and C-terminal regions of PrP, respectively (Fig. 1, top graphic). In untreated brain homogenates from mice infected with either 22L or 87V prions, two sizes of PrPD could be observed: a 25 kDa band representing full-length PrPD that was seen with all three antibodies and a truncated form about 20 kDa in size that was predominantly visible only with the 6D11 and Saf84 antibodies (Fig. 1A). The absence of the 20 kDa band in blots probed with Saf32 indicates that the PrPD truncated by the brain lacks the N-terminus at least up to amino acid residue 89. After PK treatment, the majority of protease-resistant PrPD possessed a size of about 20 kDa representing N-terminally truncated PrPD. Unexpectedly, a small amount of the 25 kDa full-length PrPD was also protected from protease digestion and was still detectable in both the 22L and 87V prion samples (Fig. 1A). The N-terminus is not part of the protease-resistant core of PrPD, suggesting that the observed full-length PrPD was inaccessible to PK and could represent either full-length, protease-resistant PrPD or PrPC. In addition, since the PrPD in this experiment is from unfractionated samples, the percentage of full-length PrPD calculated from these blots likely represents an average across all potential PrPD sub-populations, which may possess similar or different percentages of 25 kDa PrPD.
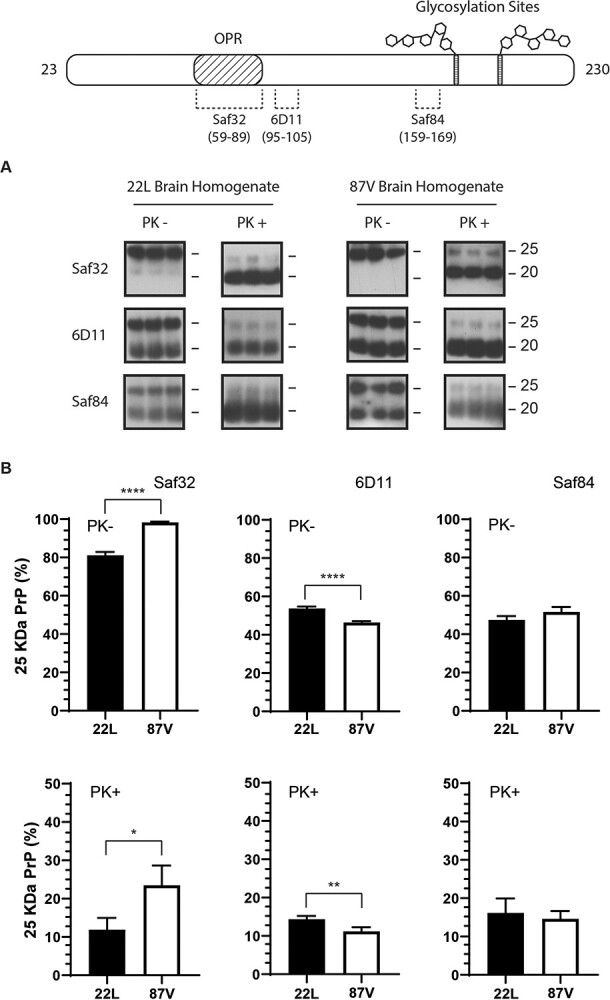
A small subpopulation of full-length PrP
D
is protected from truncation by PK. The molecular mass of PrPD in brain homogenates from mice infected with 22L and 87V prions was determined via immunoblot. (A) Immunoblots containing three replicates of PK treated and untreated PrPD from brain homogenates of 22L (left panels) and 87V (right panels) prion infected mice. All samples were de-glycosylated with PNGase F. Blots were developed using the anti-PrP antibodies Saf32 (upper panels), 6D11 (middle panels) and Saf84 (lower panels). Molecular mass markers representing 25 kDa (upper line) and 20 kDa (lower line) are shown to the right of each panel. Representative exposures from gel lanes within the same experiment are shown. (B) The percentage of 25 kDa PrP for PrPD in 22L brain homogenate (filled bars) or for PrPD in 87V brain homogenate (open bars) was determined for samples that were left untreated (upper plots) or digested with PK (lower plots), using immunoblots developed with either Saf32 (left plots), 6D11 (middle plots) or Saf84 (right plots). Percentages were calculated by quantifying the amount of full-length 25 kDa PrPD and dividing by the amount of total PrPD ((25 kDa PrPD/(25 kDa PrPD + 20 kDa PrPD)) x 100). Data were calculated from n
20 kDa PrPD)) x 100). Data were calculated from n =
= 3 for each time point and are given as mean
3 for each time point and are given as mean ±
± standard deviation. An unpaired one-tail t-test was used to calculate the statistical significance between 22L and 87V samples. Asterisks represent a range of P-values with p*
standard deviation. An unpaired one-tail t-test was used to calculate the statistical significance between 22L and 87V samples. Asterisks represent a range of P-values with p* =
= 0.02, p**
0.02, p** =
= 0.008 and p****
0.008 and p**** =
= <
< 0.0009. A map of PrP from residues 23–230 with the location of the epitopes for all 3 antibodies is shown at the top of the graph. The octapeptide repeat region (OPR) and glycosylation sites are indicated.
0.0009. A map of PrP from residues 23–230 with the location of the epitopes for all 3 antibodies is shown at the top of the graph. The octapeptide repeat region (OPR) and glycosylation sites are indicated.
Quantification of the relative amount of the 25 and 20 kDa PrPD bands in blots probed with the 6D11 and Saf84 antibodies, which recognize residues within the protease-resistant core of PrPD, confirmed that brains infected with 22L and 87V prions have similar percentages of full-length 25 kDa PrPD in both PK treated and untreated samples (Fig. 1B, 6D11 and Saf84 panels). However, in blots probed with Saf32, which binds the octapeptide repeat region (OPR) of PrP, the percentage of full-length PrPD in 87V was consistently higher than that in 22L irrespective of PK digestion (Fig. 1B, SAF32). The low relative intensity of the 20 kDa band before PK treatment in blots probed with Saf32, despite the 25 and 20 kDa PrPD bands having similar concentrations in blots probed with 6D11, indicates that the majority of PrPD truncated in the brain is missing the Saf32 epitope. The shift in the size of PrPD from 25 to 20 kDa after PK treatment for both strains in blots probed with Saf32, as opposed to a loss of PrPD, indicates that PK treatment does not remove the Saf32 epitope while proteolytic activity in the brain does. The data also suggest that a portion of full-length PrPD from brains infected with 22L or 87V is either partially cleaved or uncleaved PrPD, with 87V PrPD retaining more of the OPR than 22L PrPD following exposure to proteases in the brain.
Cells lose the ability to degrade full-length PrPD without a loss of lysosomal activity
We have previously shown that the protease resistance of 22L and 87V PrPD changes dramatically following cellular uptake (47), and we thought it likely that protection of the N-terminus of full-length PrPD could also change, thereby acting as a marker for cell-mediated structural changes in PrPD. We therefore incubated CF10 cells, which are ablated for PrP expression, with the same brain homogenates from mice infected with 22L or 87V prions used in Fig. 1. After 4 and 24 h, the cells were lysed and half of each lysate sample was treated with PK before all samples were denatured and de-glycosylated with PNGase F and analysed by immunoblot with the 6D11 antibody (Fig. 2A). While the amount of 25 kDa PrPD in brains infected with 22L or 87V detectable by the 6D11 antibody was about 50% of the total amount of PrPD (see Fig. 1B), it decreased to about 15% within the first 4 h of cellular uptake (Fig. 2A and andB,B, PK samples) and was barely or not at all detectable after PK treatment (Fig. 2A and andB,B, PK+ samples). The loss of PrPD containing the N-terminus and the change in its accessibility to proteases during initial cellular uptake suggests that PrPD aggregates were being partially disassembled and degraded by the cell, exposing the protected population of full-length PrPD to cellular proteases.
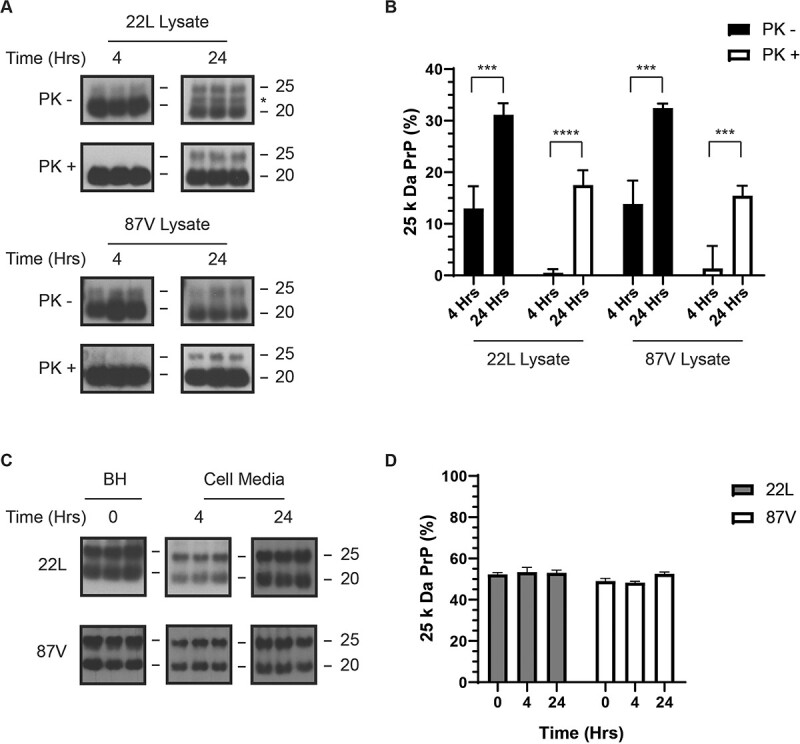
Full-length 22L or 87V PrP
D
is partially protected from cellular proteases 24 h after uptake. Media containing brain homogenate from either 22L or 87V prion infected mice was incubated with CF10 cells for 4 and 24 h and the percentage of full-length PrPD in cells and growth media was quantified. (A) Immunoblots of 22L (upper panels) and 87V (lower panels) PrPD from PK treated (PK+) and untreated (PK-) CF10 cell lysates. All samples were treated with PNGaseF and blots were developed with the 6D11 antibody. Molecular mass markers representing 25 kDa (upper line) and 20 kDa (lower line) are shown to the right of each panel while an asterisk indicates 23 kDa PrPD. (B) The average percentage of full-length PrPD in the total PrPD population from cell lysates of CF10 cells exposed to 22L or 87V prions for either 4 or 24 h. Samples were either PK treated (unfilled bars) or left untreated (filled bars). (C) Immunoblots of the starting brain homogenate (BH, representing time 0) or cell media incubated with 22L or 87V brain homogenate for 4 or 24 h. All samples were treated with PNGase F and the blots developed with the 6D11 antibody. (D) The average percentage of full-length PrPD in the total PrPD population of starting brain homogenate (time 0) or cell media incubated with 22L or 87V brain homogenate for 4 or 24 h. Gray bars represent 22L samples and white bars represent 87V samples. Percentages were calculated by quantifying the amount of full-length 25 kDa PrPD and dividing by the amount of total PrPD ((25 kDa PrPD/(25 kDa PrPD + 23 kDa PrPD +
23 kDa PrPD + 20 kDa PrPD)) x 100). Averages were calculated from n
20 kDa PrPD)) x 100). Averages were calculated from n =
= 3 for each time point and are given as mean
3 for each time point and are given as mean ±
± standard deviation. An unpaired one-tail t-test was used to calculate statistical significance between the 4 and 24 h time points. Asterisks represent a range of P-values with p***
standard deviation. An unpaired one-tail t-test was used to calculate statistical significance between the 4 and 24 h time points. Asterisks represent a range of P-values with p*** =
= 0.001–0.004 and p****
0.001–0.004 and p**** =
= 0.0003. Representative exposures from gel lanes within the same experiment are shown. For some panels, longer exposures are shown in order to clearly show the PrPD banding pattern.
0.0003. Representative exposures from gel lanes within the same experiment are shown. For some panels, longer exposures are shown in order to clearly show the PrPD banding pattern.
We have previously shown that up to 25% of PrPD in brain homogenate is taken up by CF10 cells, with the smallest aggregates taken up most rapidly (48), and uptake slowing by 15 h with a plateau at 24 h (47,48). Thus, it was unexpected that, after 24 h, the population of full-length PrPD increased for both 87V and 22L (Fig. 2A and andB),B), suggesting that the cells could be losing the ability to disassemble and degrade prion aggregates over time. Consistent with this interpretation, full-length PrPD in samples incubated with cells for 24 h comprised about 15% of total PrPD after PK treatment for both 22L and 87V, which is comparable to the amount of full-length PrPD protected from PK in the starting brain homogenates (see Fig. 1). Importantly, the percentage of full-length PrPD in cell media did not significantly decrease over 24 h, indicating that the increase of full-length PrPD in CF10 cells over 24 h was not likely the result of selective uptake (Fig. 2C and andDD).
The appearance of an intermediate sized band of approximately 23 kDa in 22L prion samples 24 h after uptake (Fig. 2A, asterisk) was also consistent with both the partial disassembly of prion aggregates and the exposure of the normally protease sensitive N-terminus to cellular proteases. We therefore further characterized the different populations of PrPD from cell lysate via western blot by probing with the Saf32, 6D11, and Saf84 antibodies (Fig. 3A and andB).B). For both strains before and after PK treatment, the banding pattern of 25 and 20 kDa PrPD in cellular lysate was similar to that of brain homogenate for all three antibodies (Fig. 1). By contrast, the 23 kDa band was visible with the Saf32, 6D11, and Saf84 antibodies, but only appeared in 22L associated samples prior to PK treatment (Fig. 3A, asterisk). The appearance of the 23 kDa band where there was not one previously and the presence of all three epitopes is consistent with it being an intermediate cleavage product of 25 kDa PrPD that was only partially truncated following exposure to cellular proteases. For both strains, the percentage of full-length PrPD observed without PK treatment was much lower after cellular uptake than in brain homogenate samples regardless of the antibody used (compare Fig. 1B with Fig. 3B). Analysis with all three antibodies showed that after PK treatment, cell lysate samples from the 24 h time point possessed similar percentages of full-length PrPD when compared to brain homogenate (compare Fig. 1B with Fig. 3B). However, the combined percentage of full-length and partially truncated 23 kDa PrPD is greater in 22L than 87V, indicating that more full-length PrPD is degraded or truncated in 87V samples following cellular uptake. The disappearance of the 23 kDa PrPD population in 22L associated samples after PK treatment indicates that this population is not protected from proteolytic degradation by PK the way it appears to be protected from cellular proteases, which may indicate either differences in the accessibility to different proteases or environmentally driven differences in PrPD structure. Taken together, the data are consistent with the hypothesis that as cells accumulate PrPD over time, their ability to disassemble and degrade prions becomes impaired.
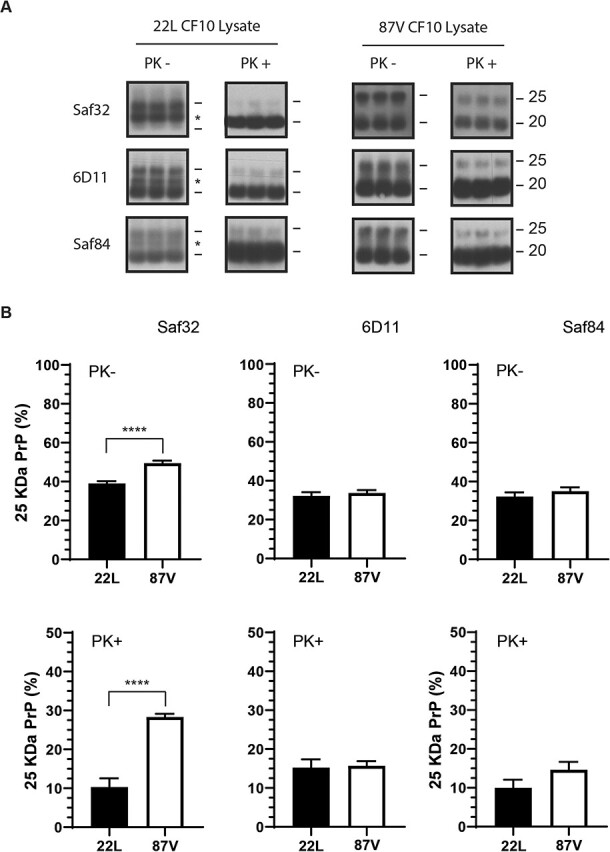
Full-length PrP
D
is partially accessible to cellular proteases in both 22L and 87V PrP
D
aggregates. (A) PrPD in lysates of CF10 cells exposed to 22L (left panels) and 87V (right panels) prion infected brain homogenates for 24 h. All samples were treated with PNGase F. Half of each cell lysate was PK treated and triplicate samples of digested and undigested lysates were run on the same gels. Blots were developed with the anti-PrP antibodies Saf32 (upper panels), 6D11 (middle panels), and Saf84 (lower panels). Molecular mass markers representing 25 kDa (upper line) and 20 kDa (lower line) are shown to the right of each panel while an asterisk indicates the presence of 23 kDa PrPD. Representative exposures from gel lanes within the same experiment are shown. (B) The percentage of 25 kDa PrPD in lysates of CF10 cells exposed to 22L brain homogenate (filled bars) or 87V brain homogenate (open bars) for 24 h. Samples were either not exposed to PK (upper plots) or PK treated (lower blots) and then analysed with the anti-PrP antibodies Saf32 (left plots), 6D11 (middle plots), and Saf84 (right plots). The percentage of full-length 25 kDa PrPD was calculated as in the legend to Figure 2. Data were calculated from n =
= 3 for each time point and are given as mean
3 for each time point and are given as mean ±
± standard deviation. An unpaired one-tail t-test was used to calculate the statistical significance between 22L and 87V PrPD samples. Asterisks represent a range of P-values with p****
standard deviation. An unpaired one-tail t-test was used to calculate the statistical significance between 22L and 87V PrPD samples. Asterisks represent a range of P-values with p**** =
= <
< 0.0009.
0.0009.
Previous studies have indicated that PrPSc can interfere with the trafficking of endosomal vesicles to the lysosome by interfering with proteins that regulate the merger of endosomal and lysosomal vesicles, delaying the degradation of PrPSc(50). We thus considered that CF10 cells may lose the ability to traffic endosomal cargo to the lysosome after 24 h of exposure to prion infected brain homogenates, potentially impairing their disassembly and degradation. In order to determine if transport to the lysosome of CF10 cells changes as a result of exposure to prions, we employed a fluorescence-based assay to measure lysosomal activity. This assay utilizes a self-quenched fluorescent dye attached to a peptide that is trafficked to endo-lysosomal vesicles where it is degraded, with the level of intracellular lysosomal activity proportional to the amount of fluorescence detected. In CF10 cells exposed to 22L, 87V, or normal brain homogenate, lysosomal activity was reduced compared to an untreated control, with 22L infected brain homogenates causing the greatest loss of lysosomal activity (Fig. 4A, left panel). The impact of 22L infected brain homogenate was small but significantly different from that of uninfected brain homogenate (Fig. 4B), and the effect was the same at 3 and 24 h post-exposure to prions (Fig. 4A, left panel). By contrast, the effect of 87V prion infected brain homogenate on lysosomal activity did not differ significantly from that of normal brain homogenate (Fig. 4B). The reduced lysosomal activity of CF10 cells exposed to 22L infected brain homogenate may be a transient effect of acute exposure to prion infectivity as we found no difference in lysosomal activity in CF10 cells that were, or were not, chronically infected with 22L prions (Fig. 4A, right panel). Overall, the data indicate a minimal impact of prion infection on lysosomal activity, suggesting that the reduced ability of the cell to disassemble and degrade prion aggregates is not due to significant overall lysosomal impairment. However, the reduction of lysosomal activity by 22L PrPD may allow it to persist longer than 87V during initial cellular uptake, which in turn may facilitate its ability to infect cells that 87V cannot.
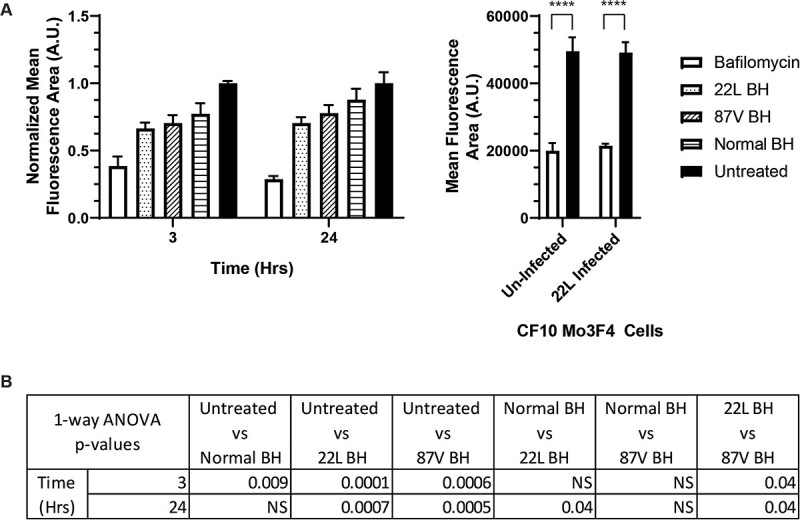
Changes in cellular lysosomal activity following PrP
D
uptake are limited. A protease sensitive, self-quenched fluorescent peptide was used to characterize the overall lysosomal activity of CF10 cells acutely exposed to 22L or 87V brain homogenates or CF10-Mo3F4 cells chronically infected with 22L prions. (A) Bar graphs of the average fluorescence intensities of CF10 cells exposed to 22L and 87V prions (left panel) or chronically infected with 22L prions (right panel). In the left panel, cells were acutely exposed to normal brain homogenate (Normal BH, bars with horizontal lines), 22L infected brain homogenate (22L BH, bars with spots), or 87V infected brain homogenate (87V BH, bars with diagonal lines). Samples in both panels were also treated with bafilomycin (white bars) or left untreated (black bars). Averages were calculated from the mean fluorescence area of cells from n =
= 3 separate experiments and are shown as mean
3 separate experiments and are shown as mean ±
± standard deviation. The average fluorescence values for CF10 cells taking up brain homogenate (left panel) were normalized to untreated cells while the values from CF10-Mo3F4 cells that were or were not chronically infected with 22L (right panel) were not normalized. (B) A one-way ANOVA Tukey's multiple comparisons test was used to calculate the statistical significance of the average fluorescence values of cells exposed to brain homogenate shown in (A), left panel. In the right panel, the statistical significance of untreated and bafilomycin treated CF10-Mo3F4 cells that were or were not chronically infected with 22L was calculated with a paired one-tail t-test. Asterisks represent a range of P-values with p****
standard deviation. The average fluorescence values for CF10 cells taking up brain homogenate (left panel) were normalized to untreated cells while the values from CF10-Mo3F4 cells that were or were not chronically infected with 22L (right panel) were not normalized. (B) A one-way ANOVA Tukey's multiple comparisons test was used to calculate the statistical significance of the average fluorescence values of cells exposed to brain homogenate shown in (A), left panel. In the right panel, the statistical significance of untreated and bafilomycin treated CF10-Mo3F4 cells that were or were not chronically infected with 22L was calculated with a paired one-tail t-test. Asterisks represent a range of P-values with p**** =
= <
< 0.0009.
0.0009.
Sensitivity of full-length PrPD to cellular proteases depends on prion strain and aggregate size
Aggregates of PrPD can be composites of many smaller aggregates (47,51) and normally protease sensitive material that is buried within aggregates may be inaccessible to proteolytic degradation. If concealment within aggregates prevents the proteolytic degradation of full-length PrPD, then 1) the amount of full-length PrPD protected from degradation could correlate with aggregate size and, 2) the cellular disaggregation machinery could influence the relationship between aggregate size and protease resistance.
In order to investigate the relationship between aggregate size and the protease resistance of full-length PrPD, we sucrose gradient fractionated brain homogenates from mice infected with 22L or 87V prions and characterized the percentage of full-length PrPD in each fraction before and after PK treatment (Fig. 5) via western blot analysis (Supplementary Figs S4-S7). In fractions 7 to 11, which have the highest percent sucrose and thus contain the largest aggregates, PrPD from both 22L and 87V infected brains consisted of 45% to 55% full-length PrPD in all fractions before PK treatment, indicating that the relative amount of full-length PrPD does not change with aggregate size (Fig. 5). However, after PK treatment, full-length PrPD was mostly concentrated in fractions 8 to 9 for 22L and fractions 8 to 10 for 87V (Fig. 5). The maximum amount of full-length PrPD after PK treatment was about 20% for 22L and 40% for 87V and localized to fractions 9 and 10, respectively (Fig. 5). The variability in the percentage of full-length PrPD across a wide size range confirms that the percentage in un-fractionated samples (Fig. 1) represents an average of multiple sub-populations that contain varying amounts of full-length PrPD. While the presence of full-length PrPD in large aggregates following PK treatment supports the possibility that protease sensitive PrPD is protected from degradation within larger aggregates, the largest aggregates of PrPD, found in fraction 11, had almost no full-length PrPD following PK treatment. These data suggest that, while a wide range of aggregate sizes contain equivalent proportions of full-length PrPD, only a specific population of large aggregates have a quaternary structure that both incorporates full-length PrPD and protects it from protease digestion.
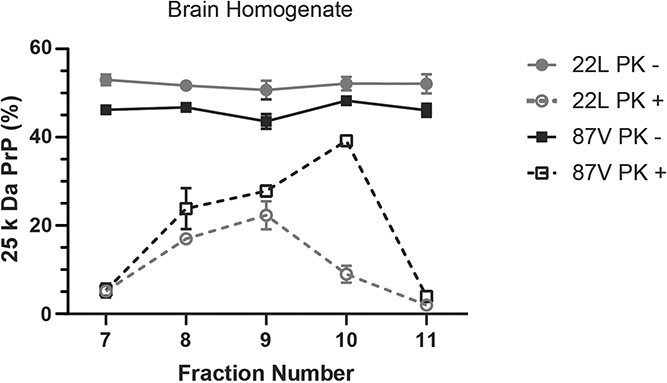
Full-length PrP
D
is uniformly present in aggregates across a wide size range but is partially inaccessible to proteases in intermediate sized aggregates. Brain homogenates from 22L (gray lines and circles) and 87V (black lines and squares) prion infected mice were fractionated via a 10% to 60% sucrose gradient. Half of each sucrose fraction was either PK treated (open shapes and dashed lines) or left undigested (filled shapes and solid lines). All samples were PNGase F treated and analysed via immunoblot using the 6D11 antibody. The average percentage of full-length PrPD in each sucrose fraction is shown. The percentage of full-length 25 kDa PrPD was calculated as in the legend to Figure 1, with the percentage of full-length PrPD in each fraction normalized to the amount of PrPD within each fraction. Thus, the percentage of 25 kDa PrPD is not weighted by concentration and will not equal the percentage of total 25 kDa PrPD calculated in the unfractionated samples in Figure 1. Averages were calculated from n =
= 3 for each fraction and are given as mean
3 for each fraction and are given as mean ±
± standard deviation.
standard deviation.
In previous studies, we showed that PrPSc from both 22L and 87V underwent a continual process of disassembly and degradation after uptake by CF10 cells (47). This process of disassembly and degradation could alter the structure of prion aggregates and make full-length PrP more accessible to cellular proteases. While the appearance of a 23 kDa PrP population after cellular uptake (see Fig. 2) supports the possibility that cellular interaction increases the accessibility of full-length PrPD to cellular proteases, this may not necessarily coincide with disaggregation. In order to determine whether or not proteolytic protection of full-length PrPD changes after cellular uptake, PrPD in cell lysates from CF10 cells exposed to 22L and 87V prion infected brain homogenates for 24 h were fractionated over a 10% to 60% sucrose gradient and analysed via western blot (Fig. 6, Supplementary Figs S8-S11). In contrast to the starting brain homogenate, prior to PK treatment full-length PrPD in lysate samples from both strains was predominately found in larger aggregate populations associated with fractions 8 to 11 and 10 to 11 for 22L and 87V, respectively (Fig. 6). The highest percentages of full-length PrPD per fraction were in lysates from cells exposed to 22L prions both before and after PK treatment when compared to 87V, suggesting that full-length PrPD is better protected in specific aggregate sizes of 22L during cellular uptake (Fig. 6). Consistent with this hypothesis, the 23 kDa PrPD population observed following uptake of 22L PrPD (see Fig. 3) was found in the largest PrPD aggregates in fractions 8 to 11 but was absent in fraction 7 and in all aggregate populations treated with PK (Fig. 6). The presence of this intermediate truncation product in the largest aggregate populations suggests that cellular processes can alter the structure of prion aggregates and increase the accessibility of full-length PrPD to cellular proteases without significant disaggregation. However, cellular proteases were unable to completely remove the N-terminus of PrPD in the largest aggregates, which required further digestion by PK for removal. Overall, the data suggest that PrPD aggregate structure is altered following cellular uptake with the previously protected N-terminus of full-length PrPD being at least partially exposed and accessible to cellular proteases.
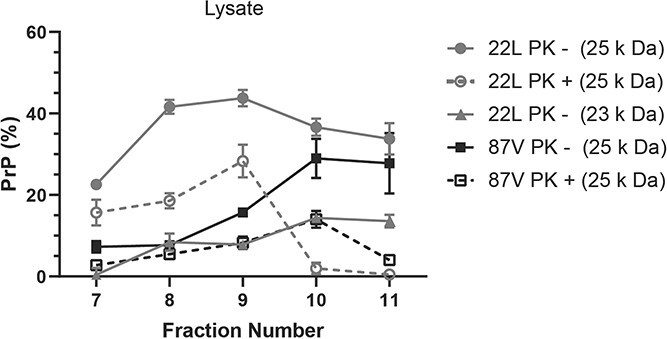
Strain specific protection of full-length PrP
D
in large aggregates following cellular uptake. CF10 cells were exposed to media containing brain homogenate from 22L and 87V prion infected mice for 24 h and then lysed. Cellular lysates were fractionated via a 10% to 60% sucrose gradient and half of each fraction was PK treated before all fraction samples were PNGase F treated and analysed by immunoblot using the 6D11 antibody. Plotted is the average percentage of 25 kDa (gray lines and circles) and 23 kDa (gray lines and triangles) PrPD in 22L PrPD and 25 kDa PrPD in 87V (black lines and squares), in cell lysates that have been either digested (open shapes and dashed lines) or left undigested (filled shapes and solid lines) with PK. Percentages of 25 kDa and 23 kDa PrPD in each population were calculated by dividing the amount of these bands by the total amount of PrPD as in the legend to Figure 2. Averages were calculated from n =
= 3 for each fraction and are given as mean
3 for each fraction and are given as mean ±
± standard deviation.
standard deviation.
Cells alter the stability of prion aggregates around protease sensitive PrPD
Given that the percentage and size distribution of full-length PrPD in PrPD aggregates from both 22L and 87V infected brains changed after cellular uptake (see Fig. 5), we wanted to determine if these changes correlated with changes in both PrPD aggregate structure (i.e. quaternary structure) and PrPD conformation (i.e. secondary/tertiary structure). Prior studies with 22L and 87V utilized incremental chemical denaturation with guanidinium hydrochloride (GndHCl) to characterize the differences in the structural stability of PrPD from both strains (45). We therefore applied the same methodology to investigate PrPD aggregate structure and conformation both before and after cellular uptake.
PrPD from brain homogenates of 22L and 87V infected mice was compared by western blot to PrPD taken up by CF10 cells after 4 and 24 h using incremental denaturation in 0, 1, 2, 3, and 4 M guanidinium hydrochloride (GndHCl) followed by either centrifugation (Fig. 7) or PK digestion (Fig. 8), to determine PrPD aggregate stability and protease resistance, respectively. Samples were not PNGase F treated and are thus normally glycosylated. At GndHCl concentrations greater than 1 M, PrPD from 22L samples showed a significant drop in the amount of aggregated PrPD as soon as 4 h after uptake by cells (Fig. 7A and B). The EC50 value of pelleted 22L in response to GndHCl exposure decreased from about 2.9 M for PrPD in brain homogenates to about 1.5 M for PrPD removed from cells after both 4 and 24 h of uptake (Fig. 7B, right panel). By contrast, the aggregate structure of PrPD from 87V samples was resistant to GndHCl denaturation even after cellular uptake, with an EC50 value that changed little within the first 4 h of uptake and only slightly dropped from 3.5 to 3.1 M after 24 h (Fig. 7B, lower panel). Compared to the change in aggregate stability, the degree to which cellular processes altered the PK resistance of PrPD after GndHCl exposure was minimal for both strains (Fig. 8A and B), with only 22L PrPD 24 h after cellular uptake showing a significant decrease in EC50 value when compared to PrPD in the starting brain homogenate (Fig. 8B). Taken together, these data suggest that, while the quaternary structure of PrPD aggregates can be destabilized following cellular uptake in a strain specific manner, the stability of the protease-resistant core, and thus the conformation of PrPD, changes very little.
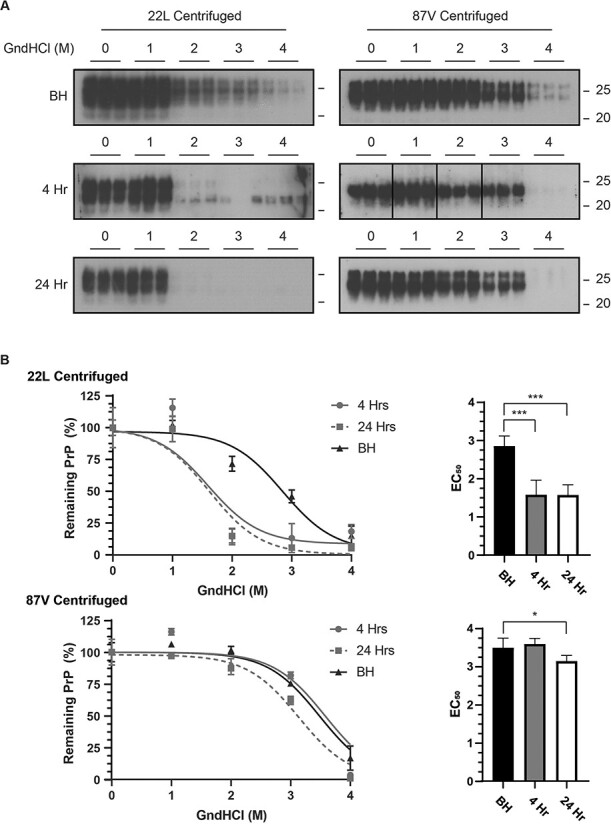
Strain specific changes in PrP
D
aggregate structural stability following cellular uptake. (A) Immunoblot analysis of 22L PrPD (left panels) or 87V PrPD (right panels) exposed to 0, 1, 2, 3, or 4 M GndHcl followed by centrifugation. A total of three separate samples are shown and all blots were analysed using the 6D11 antibody. Upper panel: PrPD in brain homogenate (BH); Middle panels: immunoblots of PrPD in cell lysate from CF10 cells exposed to prion infected brain homogenate for 4 h; Bottom panels: immunoblots of PrPD in cell lysate from CF10 cells exposed to prion infected brain homogenate for 24 h. Molecular mass markers representing 25 k Da (upper line) and 20 k Da (lower line) are shown to the right of each panel. Representative exposures from gel lanes within the same experiment are shown. (B) The total amount of insoluble PrPD remaining in the pellet was calculated from n =
= 3 for each GndHCl concentration and is given as mean
3 for each GndHCl concentration and is given as mean ±
± standard deviation. Curve fitting and EC50 values were calculated via a non-linear sigmoidal fitting model using GraphPad Prism. EC50 values calculated from each fit are shown in bar plots on the right side. Brain homogenate: black triangles, bars and lines; Cell lysate at 4 h: gray circles, bars and lines; Cell lysate at 24 h: gray squares, white bars and gray dashed lines. Significance values were calculated from the non-linear sigmoidal fit for each data set and asterisks represent a range of P-values with p*
standard deviation. Curve fitting and EC50 values were calculated via a non-linear sigmoidal fitting model using GraphPad Prism. EC50 values calculated from each fit are shown in bar plots on the right side. Brain homogenate: black triangles, bars and lines; Cell lysate at 4 h: gray circles, bars and lines; Cell lysate at 24 h: gray squares, white bars and gray dashed lines. Significance values were calculated from the non-linear sigmoidal fit for each data set and asterisks represent a range of P-values with p* =
= 0.03 and p***
0.03 and p*** =
= 0.001–0.004.
0.001–0.004.
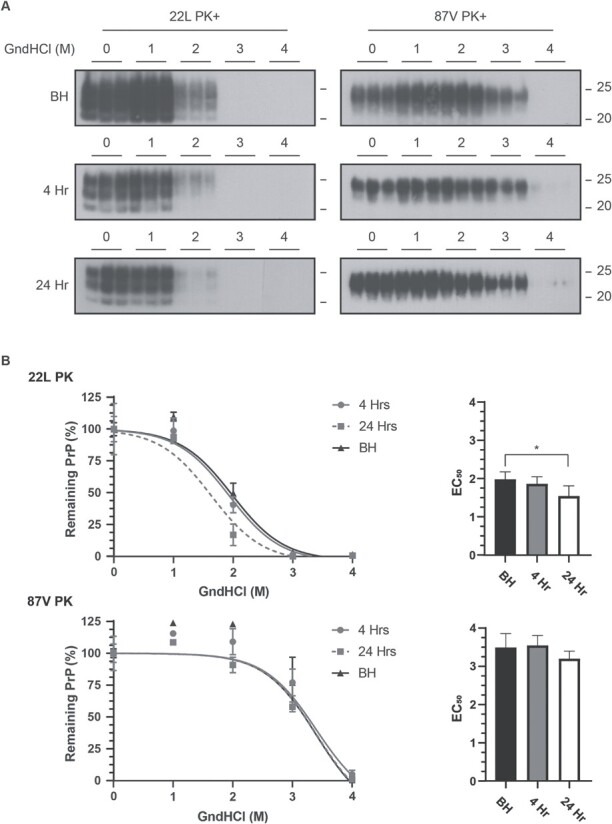
The stability of PK-resistant 22L and 87V PrP
D
is minimally altered following cellular uptake. (A) Immunoblot analysis of 22L PrPD (left panels) or 87V PrPD (right panels) exposed to 0, 1, 2, 3, or 4 M GndHcl followed by PK treatment. A total of three separate samples are shown and all blots were analysed using the 6D11 antibody. Upper panel: PrPD in brain homogenate (BH); Middle panels: immunoblots of PrPD in cell lysate from CF10 cells exposed to prion infected brain homogenate for 4 h; Bottom panels: immunoblots of PrPD in cell lysate from CF10 cells exposed to prion infected brain homogenate for 24 h. Molecular mass markers representing 25 k Da (upper line) and 20 k Da (lower line) are shown to the right of each panel. Representative exposures from gel lanes within the same experiment are shown. (B) The total amount of PrPD remaining after protease digestion was calculated from n =
= 3 for each GndHCl concentration and is given as mean
3 for each GndHCl concentration and is given as mean ±
± standard deviation. Curve fitting and EC50 values were calculated via a non-linear sigmoidal fitting model using GraphPad Prism. EC50 values calculated from each fit are shown in bar plots on the right side. Brain homogenate: black triangles, bars and lines; Cell lysate at 4 h: gray circles, bars and lines; Cell lysate at 24 h: gray squares, white bars and gray dashed lines. Averages were calculated from n
standard deviation. Curve fitting and EC50 values were calculated via a non-linear sigmoidal fitting model using GraphPad Prism. EC50 values calculated from each fit are shown in bar plots on the right side. Brain homogenate: black triangles, bars and lines; Cell lysate at 4 h: gray circles, bars and lines; Cell lysate at 24 h: gray squares, white bars and gray dashed lines. Averages were calculated from n =
= 3 and are given as mean
3 and are given as mean ±
± standard deviation. Fits in each graph were calculated with a non-linear sigmoidal fitting model in GraphPad Prism, which was used to calculate the EC50 values, shown as bar plots (right side), and determine their significance values. Asterisks represent a P-value with p*
standard deviation. Fits in each graph were calculated with a non-linear sigmoidal fitting model in GraphPad Prism, which was used to calculate the EC50 values, shown as bar plots (right side), and determine their significance values. Asterisks represent a P-value with p* =
= 0.03.
0.03.
Our data are consistent with protection of full-length PrPD by the aggregate, with loss of protection correlating with a change in aggregate structure but not in the protease-resistant core conformation of PrPD. Within the first 4 h of uptake, the overall PK resistance of PrPD changed very little after exposure to a range of GndHCl concentrations despite the N-terminus of full-length PrPD from both strains almost completely losing its resistance to PK (see Figs 2 and and7).7). Cellular processes may thus be altering aggregate structure such that full-length PrPD is exposed to cellular proteases while still maintaining the protease-resistant core of PrPD. To gain insight into whether cells can alter the structure of prion aggregates around full-length PrPD, brain homogenate and post-exposure 24 h cell lysates for both strains were denatured in 0, 1, 2, 3, and 4 M GndHCl before being either centrifuged or treated with PK. All samples were then PNGase F treated and analysed via western blot for the presence of full-length PrPD (Fig. 9). In brain homogenates from both strains, full-length PrPD resisted degradation by PK at higher concentrations of GndHCl when compared to full-length PrPD from cell lysate (Fig. 9, PK samples). With fitting and analysis, the GndHCl EC50 values for full-length PrPD treated with PK were found to greatly vary for 22L PrPD, with brain homogenate and cell lysate samples having values of 3.84 and 1.96 respectively (P = 0.0001). By a wide margin, 87V showed the largest change in EC50 value after cellular uptake with brain homogenate and cell lysate respectively showing values of >
0.0001). By a wide margin, 87V showed the largest change in EC50 value after cellular uptake with brain homogenate and cell lysate respectively showing values of > 4 and 0.72. The EC50 value of 87V from brain homogenate was not able to be accurately calculated as the remaining full-length PrPD never got below 50% in this sample. However, fitting comparisons for the lines showed that they were significantly different (P >
4 and 0.72. The EC50 value of 87V from brain homogenate was not able to be accurately calculated as the remaining full-length PrPD never got below 50% in this sample. However, fitting comparisons for the lines showed that they were significantly different (P > 0.0001). By contrast, samples that were centrifuged showed no significant differences in the percentage of full-length PrPD with increasing GndHCl concentration (Fig. 9, centrifuged samples). The constant percentage of full-length PrPD in both strains with increasing GndHCl concentration is consistent with full-length PrPD and protease-resistant, truncated PrPD being part of the same aggregate structure both before and after cellular uptake. Furthermore, the data show that the stability of PrPD aggregates from both 22L and 87V change following cellular uptake, exposing the N-terminus of PrPD, and suggest that 87V may be more sensitive to this destabilization than 22L.
0.0001). By contrast, samples that were centrifuged showed no significant differences in the percentage of full-length PrPD with increasing GndHCl concentration (Fig. 9, centrifuged samples). The constant percentage of full-length PrPD in both strains with increasing GndHCl concentration is consistent with full-length PrPD and protease-resistant, truncated PrPD being part of the same aggregate structure both before and after cellular uptake. Furthermore, the data show that the stability of PrPD aggregates from both 22L and 87V change following cellular uptake, exposing the N-terminus of PrPD, and suggest that 87V may be more sensitive to this destabilization than 22L.
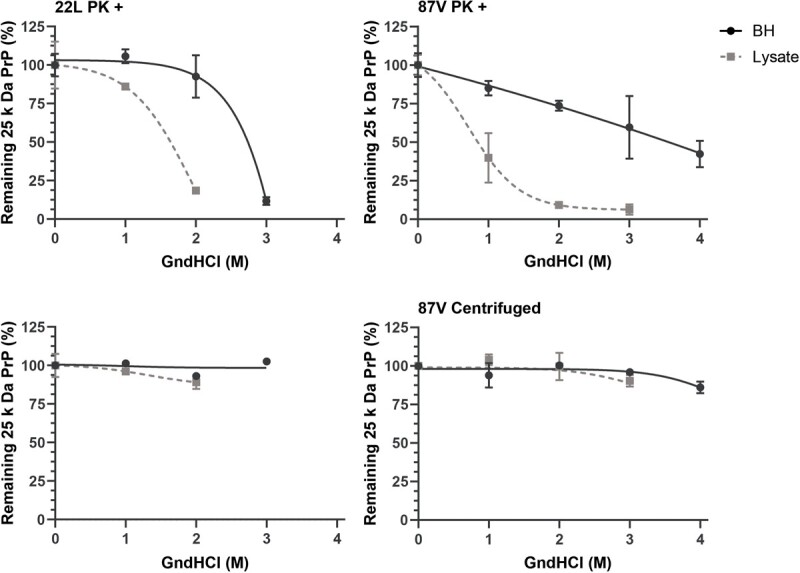
Aggregate structure protecting full-length PrP
D
is destabilized following cellular uptake. The GndHCl stability of full-length 25 kDa PrPD in populations of 22L (left panels) and 87V PrPD (right panels) before exposure to cells (BH, brain homogenate) and 24 h after cellular uptake (Lysate) was tested by exposure to 0, 1, 2, 3, and 4 M GndHCl followed by either treatment with PK (PK+, upper panels) or centrifugation with no PK treatment (lower panels). All samples were PNGase F treated and analysed by immunoblot using the 6D11 antibody. The percentage of 25 kDa PrPD in each population was calculated as in the legend to Figure 1. Shown is the average percentage of 25 kDa PrPD in each sample. Averages were calculated from n =
= 3 for each GndHCl concentration and are given as mean
3 for each GndHCl concentration and are given as mean ±
± standard deviation while fits and EC50 values were calculated via a non-linear sigmoidal fitting model in GraphPad Prism. Brain homogenate: black circles and lines; CF10 cell lysate 24 h post-exposure: gray squares and dashed lines.
standard deviation while fits and EC50 values were calculated via a non-linear sigmoidal fitting model in GraphPad Prism. Brain homogenate: black circles and lines; CF10 cell lysate 24 h post-exposure: gray squares and dashed lines.
Discussion
The N-terminus of PrP contributes significantly to prion biology despite being susceptible to degradation by cellular proteases (15). In this work, we tracked the ratio of full-length and N-terminally truncated PrPD during cellular uptake of prions. We found that while brain-derived PrPD from the mouse prion strains 22L and 87V exhibited a constant ratio of full-length to N-terminally truncated PrPD over a broad size range of aggregates (Fig. 5), only full-length PrPD in the largest aggregate populations was protected from degradation by the cell over 24 h (Fig. 6). Following extracellular PK digestion, full-length PrPD was almost completely absent from the smallest and largest PrPD aggregate populations analysed but was protected from protease digestion in an intermediate-sized aggregate population, regardless of whether it was trafficked through cells (Figs 5 and and6).6). As a result of its proteolytic protection, it is difficult to discern whether the full-length PrPD that survives both cellular proteases and PK degradation is fully protease sensitive, meaning that it can be degraded after exposure to proteases, or protease-resistant PrPD with an un-cleaved N-terminus. Regardless, our data suggest that either the cellular ability to efficiently degrade aggregates following uptake is aggregate-size dependent or that PrPD aggregates may have size-dependent differences in structure. In the latter case, mid-sized aggregates could have a more compact structure that better protects the N-terminus of PrPD and possibly other prion aggregate components, like PrPC and non-PrP proteins, from proteolysis.
While the percentages of full-length PrPD in samples of 22L and 87V 24 h after cellular uptake were similar, the combined percentage of full-length PrPD and the 23 kDa partial truncation product is greater in samples of 22L than 87V. Furthermore, the presence of a 23 kDa partial truncation product in samples of 22L indicates that the N-terminus may be better protected from cellular proteases in aggregates of 22L than in 87V. This possibility is supported by our GndHCl stability assays, which showed that cells were better at destabilizing the area within 87V aggregates where full-length PrPD was buried compared to 22L (Fig. 9), despite 87V PrPD having a more stable protease-resistant core than 22L PrPD (Fig. 8). These observations suggest that large PrPD aggregates may form from the accretion of smaller PrPD aggregates, and that each strain may have a different binding strength between layers within aggregates where protease sensitive full-length PrPD is protected from proteases. Within the PrPD aggregate population of a single prion strain there also appears to be a sub-population of mid-sized ‘Goldilocks’ aggregates. In these aggregates, PrPD is more tightly packed and full-length PrPD is better protected from extracellular proteolytic digestion via PK than the largest and smallest aggregate sub-populations, even though the largest aggregates are resistant to cellular proteases. The observation that the largest aggregates of both strains lose the ability to protect full-length PrPD outside the cell suggests that either aggregation of PrPD occurs through multiple unique pathways with different capacities to protect full-length PrPD or that the largest aggregate populations are unstable in different chemical environments.
The degree to which full-length PrPD is protected from proteases varies with size, but the ratio of full-length and N-terminally truncated PrPD from un-digested samples of both 22L and 87V infected brains did not vary significantly with aggregate size or after denaturation with GndHCl. This strongly suggests that full-length and truncated PrPD are part of the same aggregate populations as the N-terminus contributes significantly to the properties of PrP (15) and thus aggregates composed entirely of either full-length or truncated PrPD are unlikely to have similar stabilities and sizes. Finally, the constant percentage of full-length PrPD across a wide range of aggregate sizes indicates that full-length and truncated PrPD associate with each other at a fixed ratio that could be determined when smaller aggregates form and is maintained as small aggregates coalesce into larger aggregates. A similar model was proposed in another study which found that prion fibers from multiple prion strains combine lengthwise into paired strands with a double helical structure that later coalesced with other paired strands of PrPSc into larger aggregates (51). In previous work, we found that cells removed all protease sensitive PrPD during disassembly and degradation of prion aggregates (47). This may leave behind individual strands of truncated PrPD that form helical pairs with newly converted and full-length PrPD. While 22L and 87V showed an almost 1:1 ratio of full-length and truncated PrPD in brain homogenate, earlier work with the 127S prion strain showed the percentage of full-length PrPD was 70% and 5% in brain and spleen homogenate respectively (52). The different percentages of full-length PrPD may result from how frequently specific strains of PrPD are taken up by different cell types and exposed to proteases.
Exposure to the harsh environment of the lysosome likely facilitates the structural changes in aggregated PrPD that allow protease sensitive full-length PrPD and its un-cleaved N-termini to be exposed to lysosomal proteases and degraded. While we observed that cells were initially able to degrade full-length 22L and 87V PrPD, with some full-length PrPD that survived cellular proteases becoming more accessible to extracellular PK digestion (Fig. 2), cells lost the ability to effectively expose and degrade full-length PrPD over 24 h. Furthermore, the appearance of a 23 kDa partial truncation product 24 h after uptake of 22L prions (Figs 2 and and3)3) and in cell lines infected with 127S prions (52), but not after uptake of 87V prions (Figs 2 and and3),3), indicates that cells can lose the ability to fully degrade PrPD in a strain specific manner. However, we observed that cells lost the ability to effectively degrade PrPD without an overall loss of lysosomal activity, suggesting that changes in how cells alter prion aggregates over time may result from PrPD disrupting the trafficking of endosomal vesicles to the lysosome (50) rather than disrupting lysosomal activity directly. Thus, partial truncation products like the 23 kDa band may not be the product of lysosomal failure, but rather a reduction in either the rate at which PrPD is cycled in and out of the lysosome or a failure to send PrPD back to the lysosome after an initial passage. In either case, the lack of a 23 kDa band in samples of 87V PrPD and the significantly lower lysosomal activity in cells exposed to 22L suggest that 22L may be better at interfering with cellular trafficking systems, which may explain why 22L can persistently infect cells that 87V cannot.
While proteolytic protection of full-length PrPD may result from its burial within large aggregates, other biomolecules, like lipids, could also contribute to the protease resistance of full-length PrPD. Cell membranes promote prion conversion and the lipids that comprise membranes may be incorporated into prion aggregates (27–29) where they could either block access to full-length PrPD directly or alter the structure of aggregates in a way that makes full-length PrPD less accessible to proteases. Lipids may contribute to the structure of aggregates by promoting the coalescence of aggregates, a process that may be assisted by the ability of the N-terminus of PrP to interact with membrane components (40). Support for the possibility that membrane may participate in proteolytic protection can be found by a comparison of how well proteases inside the cell degrade full-length PrPD versus extracellular digestion with PK. Both full-length PrPD and the 23 kDa PrPD truncation product resisted degradation by cellular proteases at the 24 h time point despite being very susceptible to degradation by PK. While differences in the chemical environment between cells and buffered solutions may influence the degree to which N-termini are protected from proteases, the presence of the detergent sarkosyl during PK treatment may have disrupted interactions between PrPD and bound membrane, increasing the accessibility of PrPD to PK. Previous work has found that sarkosyl concentrations of ≥2% completely disrupt the neurotoxic properties of prions without influencing infectivity (53) and proteolytic protection of the N-terminus may be similarly disrupted by detergent. This may explain why studies that isolated 22L using detergents such as sodium deoxycholate that were not used in this study, did not see any protection of the N-terminus after PK treatment (54).
Recent high-resolution cryo-EM structures of PrPSc from several rodent prion strains do not appear to contain the N-terminus of PrP (19). However, these cryo-EM structures of the protease-resistant core of PrPSc contain unstructured regions that could be remnants of the N-terminus that survive the PK digestion of PrPSc prior to freezing (5). Visualization of intact N-termini in PrPSc by cryo-EM may be difficult as high quality cryo-EM structures often require large numbers of regularly structured objects and protease treatment removes most of the protease sensitive N-termini in smaller PrPD aggregates (Fig. 5) that are likely best suited for high quality cryo-EM images. This means that the bundles of fibers and globular aggregate structures that most likely contain protected N-termini with more regular secondary and tertiary structure are less likely to be used for calculating the final cryo-EM structure. Finally, intrinsically disordered regions of proteins, like the N-terminus of PrP, can often adopt a range of secondary conformations with structures that depend upon the molecules to which they bind (55). Thus, the disordered nature of the N-terminus as well as the enzyme treatments used during PrPSc purification may make it especially difficult to visualize any N-termini in PrPSc using cryo-EM.
Acknowledgments
The authors would like to thank Drs. Byron Caughey and Simote Foliaki for critically reviewing the manuscript. This research was supported by a grant from the National Institutes of Health, National Institute of Allergy and Infectious Diseases Division of Intramural Research.
Funding
This research was supported by a grant (AI-000752-27) from the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Division of Intramural Research.
Contributor Information
Daniel Shoup, Laboratory of Persistent Viral Diseases, Rocky Mountain Laboratories, National Institute of Allergy & Infectious Diseases, National Institutes of Health, 903 S. 4th Str, Hamilton, MT 59840 USA.
Suzette A Priola, Laboratory of Persistent Viral Diseases, Rocky Mountain Laboratories, National Institute of Allergy & Infectious Diseases, National Institutes of Health, 903 S. 4th Str, Hamilton, MT 59840 USA.
REFERENCES
 Å cryo-EM structure of ex vivo RML prion fibrils. Nat. Commun. 13, 4004. [Europe PMC free article] [Abstract] [Google Scholar]
Å cryo-EM structure of ex vivo RML prion fibrils. Nat. Commun. 13, 4004. [Europe PMC free article] [Abstract] [Google Scholar]Articles from Journal of Biochemistry are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jb/mvad032
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jb/advance-article-pdf/doi/10.1093/jb/mvad032/50100842/mvad032.pdf
Citations & impact
Impact metrics
Article citations
Grp78 destabilization of infectious prions is strain-specific and modified by multiple factors including accessory chaperones and pH.
J Biol Chem, 300(6):107346, 06 May 2024
Cited by: 0 articles | PMID: 38718859 | PMCID: PMC11176782
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The Size and Stability of Infectious Prion Aggregates Fluctuate Dynamically during Cellular Uptake and Disaggregation.
Biochemistry, 60(5):398-411, 26 Jan 2021
Cited by: 3 articles | PMID: 33497187
A specific population of abnormal prion protein aggregates is preferentially taken up by cells and disaggregated in a strain-dependent manner.
J Virol, 87(21):11552-11561, 21 Aug 2013
Cited by: 8 articles | PMID: 23966386 | PMCID: PMC3807341
Pathogenic mutations within the hydrophobic domain of the prion protein lead to the formation of protease-sensitive prion species with increased lethality.
J Virol, 88(5):2690-2703, 18 Dec 2013
Cited by: 11 articles | PMID: 24352465 | PMCID: PMC3958101
Properties and pathogenicity of prion-derived peptides.
Protein Pept Lett, 16(3):230-238, 01 Jan 2009
Cited by: 6 articles | PMID: 19275735
Review
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z01 AI000752