Abstract
Free full text

Cross-protocol assessment of induction and durability of VISP/R in HIV preventive vaccine trial participants
Abstract
Candidate HIV vaccines are designed to induce antibodies to various components of the HIV virus. An unintended result of these antibodies is that they may also be detected by commercial HIV diagnostic kits designed to detect an immune response to HIV acquisition. This phenomenon is known as Vaccine-Induced Seropositivity/Reactivity (VISP/R). In order to identify the vaccine characteristics associated with VISP/R, we collated the VISP/R results from 8,155 participants from 75 phase 1/2 studies and estimated the odds of VISP/R by multivariable logistic regression and 10-year estimated probability of persistence in relation to vaccine platform, HIV gag and envelope (env) gene inserts, and protein boost. Recipients of viral vectors, protein boosts, and combinations of DNA and viral-vectored vaccines had higher odds of VISP/R compared to those who received DNA-only vaccines (odds ratio, OR = 10.7, 9.1, 6.8, respectively, p<0.001). Recipients of gp140+ env gene insert (OR = 7.079, p<0.001) or gp120 env (OR = 1.508, p<0.001) had higher odds of VISP/R compared to those participants who received no env. Recipients of gp140 protein had higher odds of VISP/R than those that did not receive protein (OR = 25.155, p<0.001), and recipients of gp120 protein, had lower odds of VISP/R than those that did not receive protein (OR = 0.192, p<0.001). VISP/R persisted at 10 years in more recipients of env gene insert or protein compared to those who did not (64% vs 2%). The inclusion of gag gene in a vaccine regimen had modest effects on these odds and was confounded by other covariates. Participants receiving gp140+ gene insert or protein were most often reactive across all serologic HIV tests. Conclusions from this association analysis will provide insight into the possible impact of vaccine design on the HIV diagnostic landscape and vaccinated populations.
Introduction
Nearly 40 years after the discovery of HIV, the virus continues to disproportionately burden vulnerable countries and groups. The World Health Organization (WHO) estimated that 1.5 million new infections occurred in 2020 [1], signifying that a safe and effective HIV vaccine remains a priority in curbing this pandemic.
More than three decades of HIV vaccine research has led to the clinical evaluation of many candidate regimens, though to date none have resulted in an efficacious product fit for licensure. These vaccines vary in platform (e.g., DNA or viral vector), gene insert (e.g., HIV gag or envelope [env]), protein boost (e.g., Env glycoprotein 120 [gp120] or gp140), and adjuvant (e.g., alum or MF59). Antibodies elicited to HIV antigens among participants receiving these candidate vaccine regimens may linger past the observation period of the clinical trial, which is typically only 6–12 months after the last vaccination [2]. Importantly, these antibodies may be detected by commercial HIV serologic diagnostic kits which aim to detect HIV-elicited antibodies and contain similar antigens to the vaccine regimens evaluated thus far: HIV p24, encoded by gag, and Env glycoprotein. Thus, these antibodies can confound the interpretation of serologic test results and subsequent diagnosis of an actual HIV acquisition, a phenomenon known as vaccine induced seropositivity/reactivity (VISP/R). Diagnostic tests that directly detect components of the HIV virus, such as nucleic acid amplification tests, are needed to differentiate a true HIV infection from VISP/R. VISP/R affects the trial participant’s ability to receive a timely and accurate diagnosis of their HIV status from healthcare providers unfamiliar with the diagnostic complications that arise from vaccine-induced antibodies. It may also result in detrimental social impacts to trial participants, such as complications obtaining health insurance, donating blood or organs, or serving in the military [3]. If an efficacious vaccine for HIV prevention also induces VISP/R, the successful deployment of this vaccine could be compromised unless diagnostic tests agnostic to VISP/R are concurrently developed, commercialized, and adopted globally.
A cross-sectional analysis of the prevalence of VISP/R in HIV vaccine recipients in 25 HIV Vaccine Trial Network (HVTN) studies was performed in 2010 [4]. The general findings of this analysis were that VISP/R varied by vaccine product, and that inclusion of env or gag inserts increased the rate of VISP/R in study participants. However, this analysis did not measure the correlation of specific vaccine characteristics with the increased the rate of VISP/R, nor the duration for which participants would need HIV testing that differentiates true HIV infection from VISP/R. Since 1999, the HVTN has evaluated the safety and efficacy of new vaccine regimens and employed up-to-date HIV diagnostic testing platforms. From 2011 to 2020, the HVTN conducted HVTN 910, a longitudinal observational study which measured the duration of VISP/R and its social impacts in participants who received a vaccine in preventative HIV vaccine trials. Therefore, further evaluation of which vaccine characteristics are associated with the occurrence of VISP/R and its duration in participants is now possible and warranted.
In this analysis, we assessed the occurrence of VISP/R across 75 HIV vaccine studies to identify vaccine characteristics associated with this phenomenon. We then estimated the duration of VISP/R in participants enrolled in HVTN 910, grouped by characteristics of the vaccine regimen they received in their parent protocol. We also assessed the relationship between vaccine-induced binding antibody titers and the occurrence of VISP/R. These analyses will help identify components of an experimental HIV vaccine regimen that may induce VISP/R in study participants and inform the impact an efficacious preventative HIV vaccine would have on the HIV diagnostic landscape.
Methods
Ethics statement
Participants received HIV testing during their participation in these trials in accordance with Centers for Disease Control and Prevention (CDC) and local guidelines. On-site testing results could be blinded when necessary. An end of study visit was usually performed 6–12 months after the last vaccination per study guidelines, or retrospectively at the last visit before loss-to-follow-up, to identify participants who had acquired VISP/R. HVTN provided participants with VISP/R post-study HIV testing until VISP/R was no longer detected.
All trials were approved by the institutional review board/ethics committees of each clinical research site (CRS) institution prior to participant screening and enrollment. All participants provided written informed consent for both the parent protocol and HVTN 910.
Study setting
We performed a longitudinal analysis of data collected from 75 HIV placebo-controlled, multicenter, double-blind, randomized vaccine trials from 1990 to 2020 in which participants were given a study product intended for preventing HIV acquisition: five phase 1 AIDS Vaccine Evaluation Group (AVEG) studies, 64 phase 1-2a HVTN studies, and 6 phase 2b HVTN studies (Table 1). Participants in these vaccine trials received preventative HIV vaccine regimens varying by DNA or viral vectors platform (VV; derived from alphavirus, adenovirus, poxvirus, or vesicular stomatitis virus), recombinant proteins, or a combination of the above. HIV-derived gene inserts primarily included env, gag, or both. Envelope gene insert and protein lengths were grouped into two categories: gp120 (sometimes linked to the gp41 transmembrane anchor peptide, gp41TM) and gp140+ (inclusive of gp120 with subunits of the gp41 ectodomain, gp140, gp145, gp150, gp160).
Table 1
| Vaccine Platform | Platform Details | env insert | Env Protein | Protocol numbers* |
|---|---|---|---|---|
| DNA | DNA | No Env | No Protein | HVTN 045 (NCT00043511), HVTN 060 (NCT00111605), HVTN 063 (NCT00115960), HVTN 119 (NCT03181789) |
| gp120+gp41TM | No Protein | HVTN 044 (NCT00069030), HVTN 052 (NCT00071851) | ||
| gp140 | No Protein | HVTN 092 (NCT01783977) | ||
| gp120 | HVTN 105 (NCT02207920), HVTN 108 (NCT02915016), HVTN 111 (NCT02997969) | |||
| gp140 | HVTN 049 (NCT00073216) | |||
| gp150 | No Protein | HVTN 070 (NCT00528489), HVTN 080 (NCT00991354), HVTN 098 (NCT02431767) | ||
| DNA.VV | DNA/Ad35 | gp140 | No Protein | HVTN 072 (NCT00472719), HVTN 077 (NCT00801697) |
| DNA/Ad5 | gp120TM/gp140 | No Protein | HVTN 068 (NCT00270218), HVTN 069 (NCT00384787), HVTN 082 (NCT01054872), HVTN 204 (NCT00125970) | |
| gp140 | No Protein | HVTN 057 (NCT00091416), HVTN 072 (NCT00472719), HVTN 076 (NCT00955006), HVTN 077 (NCT00801697) | ||
| gp140/gp145 | No Protein | HVTN 505 (NCT00865566) | ||
| DNA/MVA | No Env | No Protein | HVTN 065 (NCT00301184) | |
| gp150 | No Protein | HVTN 073 (NCT00574600), HVTN 086 (NCT01418235) | ||
| gp150/gp160 | No Protein | HVTN 094 (NCT01571960), HVTN 106 (NCT02296541), HVTN 205 (NCT00820846) | ||
| gp160 | No Protein | HVTN 114 (NCT02852005) | ||
| DNA/MVA/protein | gp150 | gp140 | HVTN 073 (NCT00574600), HVTN 086 (NCT01418235) | |
| gp160 | gp120 | HVTN 114 (NCT02852005) | ||
| DNA/NYVAC | gp140 | No Protein | HVTN 092 (NCT01783977) | |
| gp120 | HVTN 096 (NCT01799954) | |||
| DNA/VSV | gp140/gp160 | No Protein | HVTN 112 (NCT02654080) | |
| gp160 | No Protein | HVTN 087 (NCT01578889) | ||
| Protein | Protein | No Env | gp120 | AVEG 005B (NCT00000632), HVTN 041 (NCT00027365), HVTN 108 (NCT02915016), HVTN 110 (NCT02771730) |
| gp140 | HVTN 049 (NCT00073216), HVTN 073 (NCT00574600), HVTN 088 (NCT01376726) | |||
| gp145 | HVTN 122 (NCT03382418) | |||
| gp160 | AVEG 003 (NCT00000745), AVEG 004 (NCT00000968) | |||
| VV | Ad26/Ad35 | gp140 | No Protein | HVTN 091 (NCT01215149) |
| Ad26/protein | gp140 | gp140 | HVTN 117 (NCT02788045) | |
| HVTN 118 (NCT02935686) | ||||
| Ad35 | gp140 | No Protein | HVTN 083 (NCT01095224) | |
| Ad4/protein | No Env | gp120 | HVTN 110 (NCT02771730) | |
| gp150 | gp120 | HVTN 110 (NCT02771730) | ||
| Ad5 | No Env | No Protein | HVTN 050 (NCT00849732), HVTN 071 (NCT00486408), HVTN 084 (NCT01159990), HVTN 502 (NCT00095576), HVTN 503 (NCT00413725), HVTN 504 (n/a) | |
| gp140 | No Protein | HVTN 054 (NCT00119873), HVTN 068 (NCT00270218), HVTN 083 (NCT01095224), HVTN 084 (NCT01159990), HVTN 085 (NCT01479296) | ||
| Ad5/Ad35 | gp140 | No Protein | HVTN 072 (NCT00472719), HVTN 077 (NCT00801697), HVTN 083 (NCT01095224) | |
| Ad5/NYVAC | gp120/gp140 | No Protein | HVTN 078 (NCT00961883) | |
| Alphavirus | No Env | No Protein | HVTN 040 (NCT00063778), HVTN 059 (NCT00097838) | |
| ALVAC | gp120 | No Protein | HVTN 026 (NCT00011037), HVTN 039 (NCT00027261), HVTN 203 (NCT00007332) | |
| gp120 | HVTN 026 (NCT00011037), HVTN 097 (NCT02109354), HVTN 100 (NCT02404311), HVTN 107 (NCT03284710), HVTN 203 (NCT00007332) | |||
| gp120+gp41TM | No Protein | HVTN 042 (NCT00076063) | ||
| gp120 | HVTN 120 (NCT03122223), HVTN 702 (NCT02968849) | |||
| gp140 | gp160 | HVTN 034 (n/a) | ||
| p24 | HVTN 032 (n/a) | |||
| FPV | gp140 | No Protein | HVTN 055 (NCT00083603) | |
| MVA | gp140 | No Protein | HVTN 055 (NCT00083603) | |
| gp150 | No Protein | HVTN 065 (NCT00301184), HVTN 205 (NCT00820846) | ||
| gp140 | HVTN 086 (NCT01418235) | |||
| gp160 | No Protein | HVTN 114 (NCT02852005) | ||
| gp120 | HVTN 114 (NCT02852005) | |||
| MVA/FPV | gp140 | No Protein | HVTN 055 (NCT00083603) | |
| NYVAC/protein | gp140 | gp120 | HVTN 096 (NCT01799954) | |
| Vaccinia/Protein | gp160 | gp160 | AVEG 002 (NCT00000683), AVEG 002B (NCT00000631) | |
| VSV | No Env | No Protein | HVTN 090 (NCT01438606) |
*Italicized protocols contained gag genes or epitopes.
Cohort
From 2011 to 2020, a total of 21,578 participants from the Americas, Africa, and Southeast Asia were enrolled in the 75 HIV vaccine trials included in this analysis. Eligibility criteria for participants included having a low likelihood of HIV acquisition for most phase 1 and 2a studies and a high likelihood of HIV acquisition for phase 2b studies. Participants were healthy, HIV-seronegative adults between 18 and 60 years old at the time of enrollment. VISP/R testing was performed at or near the end of the protocol. To assess the vaccine characteristics associated with the occurrence of VISP/R, participants were excluded from this analysis if they received a placebo/control product (n = 8,397), did not undergo a VISP/R assessment in their parent protocol (n = 4,833), acquired HIV by end of study (n = 39), or received study product but terminated early or met other exclusion criteria (n = 154). Thus, this analysis was performed on a total of 8,155 participants (Fig 1).
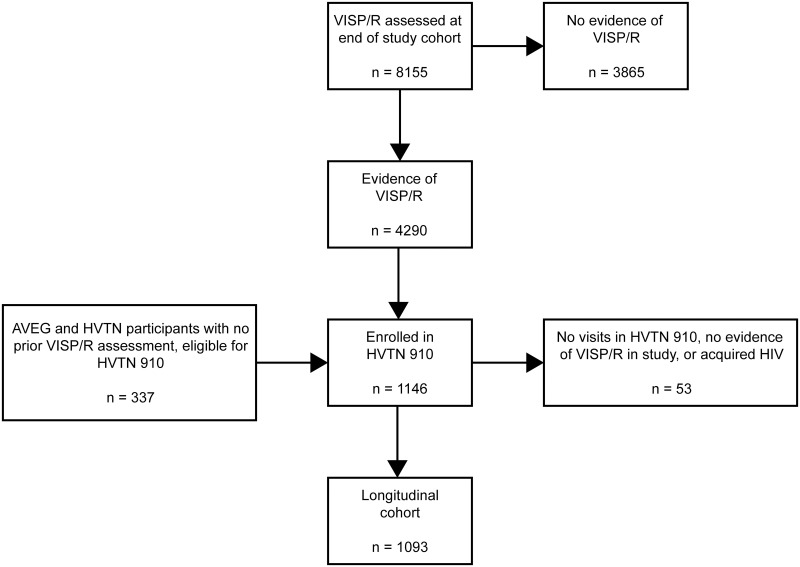
Participants were enrolled in HVTN vaccine trials and evaluated for VISP/R at the end of the study. Those that received vaccine product were included in the analysis to identify vaccine characteristics associated with the occurrence of VISP/R. Participants with VISP/R were eligible to enroll in the long-term observational trial HVTN 910 to observe their VISP/R status until it resolved. AVEG participants and those in HVTN 032, 034, 050, 057, 077, 502, 503, in which no previous VISP/R assessment was performed, were also eligible to enroll in HVTN 910. Participants that had detectable VISP/R in study comprised the longitudinal cohort analyzed to estimate the duration of VISP/R.
VISP/R was detected in 4,290 participants at the end of their parent protocols. These individuals were offered prospective, unscheduled HIV and VISP/R testing and were eligible to enroll in HVTN 910, a prospective observational trial designed to monitor of the persistence of VISP/R. Participants from previously completed AVEG protocols were also eligible to enroll in HVTN 910. To join HVTN 910, participants needed to have access to an active HVTN CRS, be willing to receive pre- and post-test HIV counseling and HIV results, demonstrate understanding of the study, and have no conditions that would be a contraindication to protocol adherence or the ability to give informed consent (in the judgement of the CRS investigator).
Of the eligible AVEG and HVTN participants, 1,146 were enrolled in HVTN 910. A total of 21 participants had no end of study VISP/R visit and their first record is within HVTN 910. Participants who had no detectable VISP/R during HVTN 910, acquired HIV while enrolled, or had no VISP/R visits after enrollment were excluded from the analysis of VISP/R persistence. Participants were offered follow-up HIV testing (recommended approximately every six months) until the resolution of VISP/R. HVTN 910 was closed on 01 October 2020 and all participants with persistent VISP/R are currently provided post-study testing though standard HVTN processes.
Laboratory testing
VISP/R testing was performed according to a preapproved HIV diagnostic algorithm at three Division of AIDS (DAIDS)-approved Good Clinical Laboratory Practice (GCLP)-compliant laboratories: the University of Washington Retrovirology Laboratory in Seattle, Washington, which performed testing of specimens from Europe, North America, and the Western Pacific (WP); the National Institute of Communicable Diseases in Johannesburg, South Africa, which performed testing of specimens from Africa; and the Asociación Civil Impacta Clinical Trials Unit (CTU) HIV Diagnostics Lab in Lima, Peru, which performed testing of specimens from South America. The Viral and Rickettsial Disease Laboratory at the California Department of Health Services (CL-Richmond) also performed diagnostic testing of specimens from North America prior to 2006.
VISP/R was defined as a participant specimen resulting in a reactive result from a serological HIV laboratory screening or rapid test (i.e., antigen/antibody (Ag/Ab) combination or antibody enzyme immunoassay/chemiluminescent microparticle immunoassay (EIA/CMIA) test) and undetectable HIV-1 RNA by PCR. The VISP/R testing algorithm includes the performance of three or more independent U.S. Food and Drug Administration (FDA)-approved/Conformité Européene (CE)-marked HIV laboratory screening or rapid tests, followed by an HIV-1 RNA PCR if any positive or reactive laboratory screening or rapid results are obtained. To differentiate a possible HIV acquisition from VISP/R, participant specimens that elicited reactive laboratory screening or rapid and HIV-1 RNA negative results could also be tested by Western Blot or HIV-1/2 differentiation tests according to criteria by the CDC and the manufacturer’s package insert. The laboratory screening or rapid tests were selected by how commonly they were used in the participant’s regional diagnostic laboratories. The list of diagnostic tests for each laboratory is presented in Table 2. Testing was performed according to the manufacturer’s package insert.
Table 2
| gp120 env | gp140+ | No Envelope | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gp120 protein | No Protein | gp120 protein | gp140+ protein | No protein | gp120 protein | gp140+ protein | No Protein | |||||||
| Laboratory Screening Tests | Years of Use | HIV diagnostics lab | + gag | + gag | No gag | + gag | No gag | + gag | + gag | No gag | + gag | No gag | No gag | + gag |
| Abbott Architect HIV Ag/Ab Combo ** | 2011-present | UWVSL, NICD | 1% | 15% | 25%* | 99% | 79% | 91% | 0%* | 8% | 93% | 4% | ||
| Abbott Axsym HIV Ag/Ab Combo** | 2011–2014 | NICD | 93% | 53%* | 98%* | 0%* | ||||||||
| Abbott Murex HIV-1.2.O | 2008–2011 | NICD | 77%* | 70% | ||||||||||
| Abbott HIV AB HIV1/2 (rDNA) | 2006–2011 | UWVSL | 61% | 54% | 3%* | 100%* | 85% | 66% | 0% | 100%* | 60% | |||
| Abbott Prism Anti HIV-1/2, PSBC* | 2013–2021 | UWVSL | 0% | 16% | 0%* | 50%* | 63% | 84% | 0%* | 0%* | 86%* | 1% | ||
| bioMerieux Vironostika HIV Ag/Ab HIV 1/2 | 2011–2013 | NICD | 91% | 56%* | 98% | 0%* | ||||||||
| bioMerieux Vironostika HIV Uni-Form II + O | 2008–2011 | NICD | 64%* | 70% | ||||||||||
| bioMerieux Vironostika HIV-1 | 2006–2007 | UWVSL | 62% | 56% | 8%* | 100% | 87% | 0%* | 100%* | 17% | ||||
| BioRad GS HIV Combo Ag/Ab EIA | 2013-present | UWVSL, Impacta | 2% | 24% | 25%* | 98% | 76% | 82%* | 0%* | 13%* | 97% | 6% | ||
| BioRad GenScreen Ultra HIV Ag-Ab HIV 1/2 | 2011-present | NICD | 2% | 1% | 97% | 54%* | 98% | 0%* | 0%* | |||||
| BioRad Genetic Systems HIV 1/2 Plus O EIA | 2006–2013 | UWVSL | 43%* | 0%* | 100%* | 79% | 66% | 96% | 47% | |||||
| BioRad GenScreen HIV 1/2 | 2008–2011 | NICD | 79%* | 76% | ||||||||||
| BioRad Genetic Systems rLAV | 2007–2011 | UWVSL | 74% | 64% | 52% | |||||||||
| Rapid Tests | ||||||||||||||
| Alere Determine HIV-1/2 Ag/Ab Combo | 2016-present | UWVSL, NICD, Impacta | 2% | 7% | 100%* | 99% | 74% | 82%* | 7%* | 95%* | 6%* | |||
| Abbott Determine HIV Early Detect | 2022-present | NICD | 100%* | |||||||||||
| Alere HIV Combo | 2019–2021 | NICD | 1% | 0%* | 100%* | 0%* | ||||||||
| Biorad Multispot HIV-1/HIV-2 Rapid Test | 2009–2017 | UWVSL, NICD | 1% | 32% | 0%* | 93% | 79% | 92% | 0%* | 33%* | 92%* | 27% | ||
| SD Bioline HIV-1/2 3.0 | 2020–2020 | NICD | 0% | |||||||||||
| Western Blot/Differentiation tests | ||||||||||||||
| BioRad Geenius HIV 1/2 Confirmatory Assay | 2018-Present | UWVSL | 100%* | 81%* | 100% | 100%* | 100%* | 100%* | 75%* | |||||
| BioRad Genetic Systems HIV-1 | 2006–2016 | UWVSL, NICD | 86%* | 100%* | 100% | 93% | 82% | 100% | 64% | |||||
| Richmond In-House Western Blot | 2000–2006 | CL-Richmond | 95% | 97% | 100%* | 100%* | 100%* | |||||||
*VISP/R rate calculated from <50 participants. Color scale applied; red equals 0%, yellow 50%, and green 100%.
Statistical analysis
Participant demographics are summarized by each protocol and pooled. Geographic regions were consistent with WHO definitions, with Europe, the Americas, and WP grouped into one region since specimens were tested on comparable diagnostic platforms. Race categories were defined in accordance with United States Census race categories.
For each parent protocol, all enrolled vaccine recipients (i.e., those who received at least one dose of vaccine) were included in the analysis. VISP/R rates are summarized by demographics and vaccine characteristics, with 95% confidence intervals (CIs) computed using the Wilson score method [5]. Logistic regression models were applied to study the association between VISP/R response rates and vaccine characteristics, including vaccine product, gene insert, and protein. A multivariable logistic regression model was developed to examine the association between VISP/R status and vaccine characteristics, adjusted for age, region, trial phase and sex assigned at birth.
VISP/R duration analysis included all participants enrolled in HVTN 910 who had detectable VISP/R at the end of the parent protocol. Per-protocol VISP/R resolution in HVTN 910 was defined as having three consecutive seronegative tests over at least one year, followed by no subsequent positive tests. The per-protocol time to resolution was measured from the date of last vaccination in the parent protocol. For those whose VISP/R resolved, the event time was estimated as the midpoint between the date of last detection of VISP/R and first of three consecutive seronegative tests. Participants with only one or two seronegative tests were censored at the midpoint of the last positive and first negative test date. For participants who tested seropositive at their most recent HVTN 910 visit, time to VISP/R resolution was censored at the date of their last visit. Due to incomplete participant follow-up, we also considered a modified VISP/R resolution definition as having at least one seronegative test followed by no subsequent seropositive tests. The modified time to resolution was also measured from last vaccination, with event time defined as the midpoint between the date of the last detection of VISP/R and the first seronegative test.
Kaplan-Meier estimates of VISP/R persistence were calculated within study product categories. Median time to resolution and resolution rates at 2, 5, and 10 years were summarized within study product categories.
Assessments of the association between vaccine-induced immunogenicity, as measured by total IgG, to antibody reactivity to HIV diagnostic tests was performed using available binding antibody multiplex assay (BAMA) data previously collected from vaccine recipients in protocols HVTN 098, 100, 106, 107, 108, 111, 112, 114, 117, 118, 122, 205, and 505 [6–10]. Total IgG binding antibody responses were measured at two to four weeks following the last scheduled vaccination using matched antigen lots of gp120 Group B (Con 6 gp120/B), consensus gp140 (Con S gp140 CFI), variable loop 1 and 2 (V1/V2) (gp70_B.CaseA_V1_2), and gp41 antigens. Statistical analyses were performed using SAS (version 9.4, SAS Institute, Cary, NC) and R statistical software (version 3.5.3 R Foundation for Statistical Computing, Vienna, Austria).
Results
Risk of VISP/R in HIV preventative vaccine trial participants
Participant demographics including age, sex at birth, and race reflected the target populations enrolled for each geographic region (Table 3). Approximately 75% of these participants (6,205) were enrolled in the Americas, Europe, or the WP, of which less than 3% were enrolled in Europe and the WP. Of these 6,205 participants, 2,735 (44%) were under 30 years old, 3,863 (62%) were assigned male at birth, 3,873 (63%) identified as White, and 5,423 (87%) identified as non-Hispanic. Of the 1,950 participants enrolled in sub-Saharan Africa, 1,589 (81%) were under 30 years old, 991 (51%) were assigned male at birth, 1,869 (96%) identified as Black, and all identified as either non-Hispanic (1362, 70%) or were of unknown ethnicity (588, 30%). Over half of all participants across geographic regions were enrolled in phase 1-2a studies (58% in sub-Saharan Africa, 66% in Americas/Europe/WP).
Table 3
| Combined | Sub-Saharan Africa | Americas/Europe/ WP | |
|---|---|---|---|
| N = 8155 | N = 1950 | N = 6205 | |
| Age | |||
| 30 or above | 38% (3096) | 19% (361) | 44% (2735) |
| Less than 30 | 62% (5059) | 81% (1589) | 56% (3470) |
| Sex assigned at birth | |||
| Female | 40% (3301) | 49% (595) | 38% (2342) |
| Male | 60% (4854) | 51% (991) | 62% (3863) |
| Race/Ethnicity | |||
| American Indian or Alaska Native | 0% (36) | 0% (0) | 1% (36) |
| Asian | 2% (183) | 0% (2) | 3% (181) |
| Black or African American | 38% (3101) | 96% (1869) | 20% (1232) |
| Hispanic or Latino | 2% (157) | 0% (0) | 3% (157) |
| Multiracial | 6% (476) | 0% (0) | 8% (476) |
| Native Hawaiian or Other Pacific Islander | 0% (14) | 0% (0) | 0% (14) |
| Unknown | 4% (314) | 4% (78) | 4% (236) |
| White | 48% (3874) | 0% (1) | 62% (3873) |
| Ethnicity | |||
| Hispanic | 9% (767) | 0% (0) | 12% (767) |
| Non-Hispanic | 83% (6785) | 70% (1362) | 87% (5423) |
| Unknown | 7% (603) | 30% (588) | 0% (15) |
| Study Phase | |||
| Phase 1-2a | 64% (5208) | 58% (1123) | 66% (4085) |
| Phase 2b | 36% (2947) | 42% (827) | 34% (2120) |
Study phase (phase 2b vs. phase 1/2a), region (Americas/Europe/WP vs. SSA), vaccine platform (non-DNA vs. DNA), and the presence of a gp140 gene or protein were associated with increased odds of VISP/R in multivariate analyses (Table 4). Participants in the Americas, Europe, and WP had increased odds of VISP/R (OR = 2.559, p<0.001) compared to those from sub-Saharan Africa. Participants enrolled in phase 2b studies had increased odds of VISP/R (OR = 2.028, p = <0.001) compared to those in phase 1-2a. VV only, VV in combination with DNA, and protein-only regimens had increased odds of inducing VISP/R compared to DNA alone. The inclusion of a gp120 or gp140 gene insert in vaccine regimens increased the odds of VISP/R compared to participants who did not receive an env gene insert (OR = 1.508 and 7.079 respectively, p<0.001). The inclusion of gp140+ protein (OR = 25.155, p<0.001) in vaccine regimens, whether alone or in combination with DNA or viral vectors, increased the odds of VISP/R compared to participants who did not receive gp140+ protein. In contrast, the inclusion of a gp120 protein boost reduced the odds of VISP/R in participants (OR = 0.192, p<0.001). The inclusion of a gag gene insert decreased the odds of VISP/R (OR = 0.752, p = 0.017) despite a higher rate of VISP/R in participants with gag (60.4% VISP/R in those with gag versus 51.8% in those without gag). This is likely due to confounding by env and glycoprotein covariates since analysis of the inclusion of gag, adjusting for demographics and vaccine platform, demonstrated increased odds of VISP/R (OR = 1.634, p<0.001) (S1 Table). These observations confirm that regimens including gp140+ env gene inserts or protein boosts are more likely to induce specific seroreactive antibodies than regimens without these components.
Table 4
| Category | VISP/R rate | 95% CI | Multivariate Odds Ratio (95% CI) | P value |
|---|---|---|---|---|
| Age | ||||
| 30 or above | 55.1% (1706/3096) | (53.3%, 56.9%) | Ref | 0.026 |
| Less than 30 | 51.1% (2584/5059) | (49.7%, 52.4%) | 1.142 (1.016, 1.284) | |
| Sex assigned at birth | ||||
| Female | 47.0% (1550/3301) | (45.3%, 48.7%) | Ref | 0.646 |
| Male | 56.4% (2740/4854) | (55.0%, 57.8%) | 1.028 (0.914, 1.156) | |
| Study Phase | ||||
| Phase 1-2a | 48.0% (2502/5208) | (46.7%, 49.4%) | Ref | <0.001 |
| Phase 2b | 60.7% (1788/2947) | (58.9%, 62.4%) | 2.028 (1.747, 2.355) | |
| Region | ||||
| Sub-Saharan Africa | 26.8% (522/1950) | (24.9%, 28.8%) | Ref | <0.001 |
| Americas/Europe/WP | 60.7% (3768/6205) | (59.5%, 61.9%) | 2.559 (2.201, 2.975) | |
| Vaccine Platform | ||||
| DNA | 14.6% (177/1213) | (12.7%, 16.7%) | Ref | <0.001 |
| DNA.VV | 77.2% (1874/2426) | (75.5%, 78.9%) | 6.84 (5.443, 8.596) | |
| Protein | 47.7% (103/216) | (41.1%, 54.3%) | 9.126 (4.474, 18.614) | |
| VV | 49.7% (2136/4300) | (48.2%, 51.2%) | 10.693 (8.456, 13.521) | |
| Gag | ||||
| No gag | 51.8% (3853/7432) | (50.7%, 53.0%) | Ref | 0.017 |
| gag | 60.4% (437/723) | (56.8%, 63.9%) | 0.752 (0.595, 0.95) | |
| Env | ||||
| No Env | 40.8% (980/2401) | (38.9%, 42.8%) | Ref | <0.001 |
| gp120 env | 21.3% (302/1415) | (19.3%, 23.5%) | 1.508 (1.205, 1.887) | |
| gp140+ env | 69.3% (3008/4339) | (67.9%, 70.7%) | 7.079 (5.773, 8.68) | |
| Protein | ||||
| No protein | 60.3% (3563/5906) | (59.1%, 61.6%) | Ref | <0.001 |
| gp120 protein | 11.6% (197/1702) | (10.1%, 13.2%) | 0.192 (0.155, 0.238) | |
| gp140+ protein | 96.9% (530/547) | (95.1%, 98.0%) | 25.155 (13.981, 45.259) |
Estimating the risk of VISP/R by binding antibody titers to Env
To further characterize the relationship between vaccine Env antigens and seroreactive antibodies, we compared the occurrence of VISP/R for each vaccine platform or gene insert/protein combination to binding antibody responses. We chose antigens that were analyzed by BAMA for each study listed in the Methods: Con 6 gp120/B, Con S gp140, CaseA V1/V2, and gp41. Participants with VISP/R had higher binding antibody magnitudes than participants with no VISP/R for all antigens tested, regardless of vaccine platform or envelope gene insert/protein combination (Fig 2A). Multivariate logistic regression of participant age and sex assigned at birth, inclusion of gag, vaccine type, and binding antibody response magnitude indicate that responses to Con 6 gp120/B and CaseA V1/V2 result in a significant increase in the odds of VISP/R (S2 Table). There was no significant association between VISP/R and a response to gp140 antigen, likely due to insufficient gp140 antigen BAMA data from participants without VISP/R. A regression model to indicate if antibody response to gp41 affected the odds of VISP/R, adjusting for envelope/glycoprotein combinations, was unstable, likely due to lack of data for all combinations.
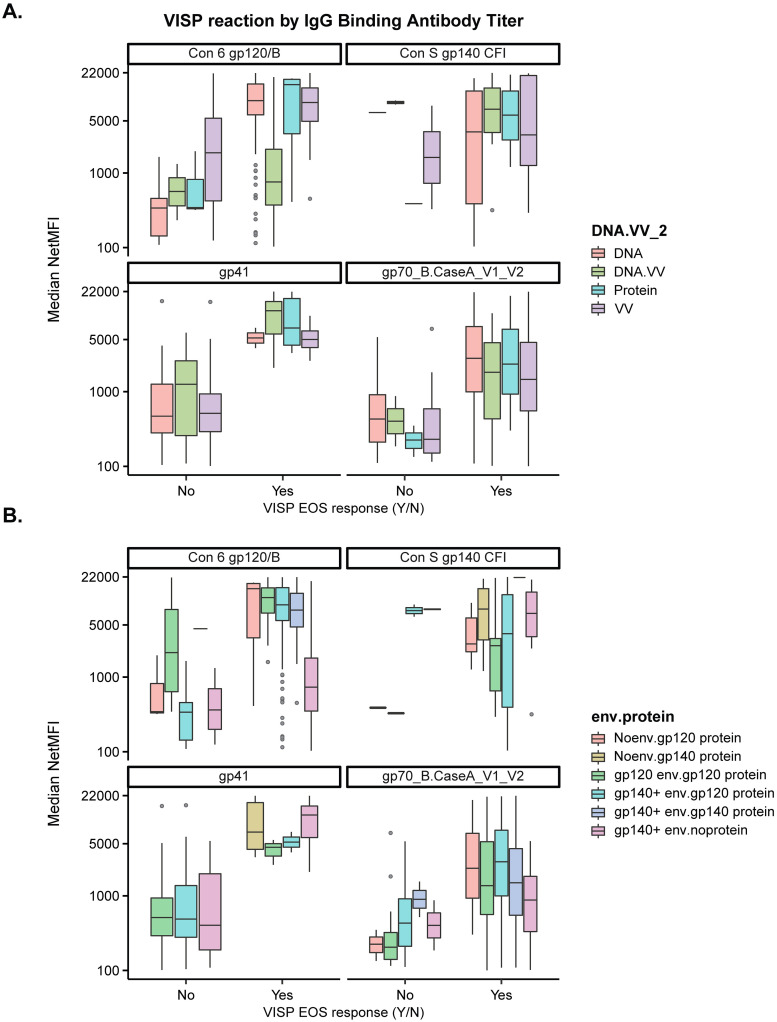
Participants enrolled in HVTN 098, 100, 106, 107, 108, 111, 112, 114, 117, 118, 122, 205, 505 clinical trials have available IgG binding antibody magnitudes to antigens Con 6 gp120/B, Con S gp140 CFI, gp41, gp70_B.CaseA_V1_V2. Median IgG binding antibody (netMFI) was assessed at the peak time point, two to four weeks post last scheduled vaccination, by (A) product type and (B) envelope gene/protein combination. Gray circles represent individual BAMA netMFI results greater than 1.5x the interquartile range away from the median.
Duration of VISP/R post-vaccination
Participants who had VISP/R detected in their parent protocol or were previously enrolled in an AVEG study were offered enrollment into the long-term follow-up study: HVTN 910. In total, 1,093 participants completed at least one visit in which VISP/R was identified, enrolled in HVTN 910, and were not later diagnosed as having acquired HIV (Fig 1 and S1 Fig). Demographics represent regional participant characteristics (S3 Table). Participants from all regions were enrolled in HVTN 910, with comparable years of follow-up since last vaccination, follow-up time from enrollment in HVTN 910, and median number of VISP/R assessments (Table 5). Of these participants, 1% from the Americas, Europe, and the WP and 4% from sub-Saharan Africa terminated from the study due to acquiring HIV. More participants from the Americas, Europe, and the WP terminated from the study due to study closure (75%) rather than resolution of VISP/R (10%). More participants from sub-Saharan Africa terminated from the study due to resolution of VISP/R (45%) than from study closure (40%). Due to incomplete follow-up of participants, we measured the frequency at which participants met a modified definition of VISP/R resolution as described above. For all groups, more participants had met a modified VISP/R resolution than met the original per-protocol definition of VISP/R resolution (S2 Fig).
Table 5
| Combined | Sub-Saharan Africa | Americas/Europe/WP | ||
|---|---|---|---|---|
| N* | N = 1093 | N = 230 | N = 863 | |
| Years follow-up since HVTN 910 enrollment (Median) [min, 25%, 75%, max] | 1085 | 2.0 [0.0, 1.0, 4.0, 8.4] | 1.9 [0.0, 1.0, 3.2, 7.7] | 2.0 [0.0, 0.9, 4.2, 8.4] |
| 0** | 11% (120/1085) | 13% (29/230) | 11% (91/855) | |
| >0, <1 | 15% (161/1085) | 10% (22/230) | 16% (139/855) | |
| ≥1, <2 | 25% (273/1085) | 30% (68/230) | 24% (205/855) | |
| ≥2, <4 | 24% (259/1085) | 31% (71/230) | 22% (188/855) | |
| ≥4, ≤10 | 25% (272/1085) | 17% (40/230) | 27% (232/855) | |
| Years since parent protocol VISP/R assessment (Median) [min, 25%, 75%, max] | 1084 | 4.3 [0.0, 2.4, 7.0, 15.2] | 3.6 [0.0, 2.1, 5.8, 12.9] | 4.6 [0.1, 2.5, 7.1, 5.2] |
| 0 | 1% (13/1084) | 6% (13/230) | 0% (0/854) | |
| >0, <1 | 3% (36/1084) | 5% (11/230) | 3% (25/854) | |
| ≥1, <2 | 14% (147/1084) | 13% (31/230) | 14% (116/854) | |
| ≥2, <4 | 27% (298/1084) | 33% (76/230) | 26% (222/854) | |
| ≥4, <10 | 50% (537/1084) | 40% (92/230) | 52% (445/854) | |
| ≥10, <30 | 5% (53/1084) | 3% (7/230) | 5% (46/854) | |
| Years since last vaccination (Median) [min, 25%, 75%, max] | 1093 | 6.6 [0.8, 3.5, 8.3, 28.9] | 5.6 [1.0, 3.0, 7.7, 13.4] | 6.7 [0.8, 3.8, 8.5, 28.9] |
| >0, <1 | 0% (2/1093) | 0% (0/230) | 0% (2/863) | |
| ≥1, <2 | 7% (78/1093) | 11% (26/230) | 6% (52/863) | |
| ≥2, <4 | 22% (239/1093) | 26% (60/230) | 21% (179/863) | |
| ≥4, <10 | 62% (679/1093) | 59% (135/230) | 63% (544/863) | |
| ≥10, <30 | 9% (95/1093) | 4% (9/230) | 10% (86/863) | |
| Terminated from HVTN 910 | 1093 | 100% (1091/1093) | 100% (230/230) | 100% (861/863) |
| Death | 0% (5/1093) | 0% (1/230) | 0% (4/863) | |
| Early Termination | 2% (19/1093) | 1% (2/230) | 2% (17/863) | |
| HIV infection | 2% (19/1093) | 4% (10/230) | 1% (9/863) | |
| Relocated | 5% (51/1093) | 2% (4/230) | 5% (47/863) | |
| Resolution of VISP/R | 17% (189/1093) | 45% (104/230) | 10% (85/863) | |
| Site Closure | 2% (19/1093) | 0% (0/230) | 2% (19/863) | |
| Study Closure | 67% (735/1093) | 40% (92/230) | 75% (643/863) | |
| Unable to contact participant | 5% (56/1093) | 7% (17/230) | 5% (39/863) | |
| Median number of VISP/R assessments [25%, 75%] | 1093 | 5 [3, 8] | 5 [3, 6] | 5 [3, 9] |
| Median number of VISP/R reactive [25%, 75%] | 1093 | 4 [2, 7] | 2 [1, 4] | 4 [2, 8] |
| One negative test | 1093 | 25% (276/1093) | 63% (146/230) | 15% (130/863) |
| Two consecutive negative tests | 1093 | 21% (225/1093) | 52% (120/230) | 12% (105/863) |
| Three consecutive negative tests | 1093 | 18% (195/1093) | 46% (106/230) | 10% (89/863) |
| Three consecutive negative tests over 1 year | 1093 | 12% (132/1093) | 33% (76/230) | 6% (56/863) |
*Some parameters have totals <1093 resulting from missing data (e.g., evaluation of VISP/R date)
**0 years follow-up indicates that there was only one VISP/R-related visit, and no follow-up
We attempted to perform survival analysis of the HVTN 910 data to assess the duration of VISP/R in this population. By 10 years of follow-up, 47.4% of participants enrolled in HVTN 910 saw VISP/R resolution (Fig 3A). When grouped by vaccine platform, the 10-year probability of VISP/R persistence was 63% (95% CI: 46–86%) for DNA vaccine recipients, 23% (95% CI: 18–29%) for VV vaccine recipients, 69% (95% CI: 64–76%) for DNA/VV vaccine recipients, and 97% (95% CI: 90–100%) for protein recipients (Fig 3B). Wide confidence intervals were typically a result of irregular participant visits and the uneven follow-up time.
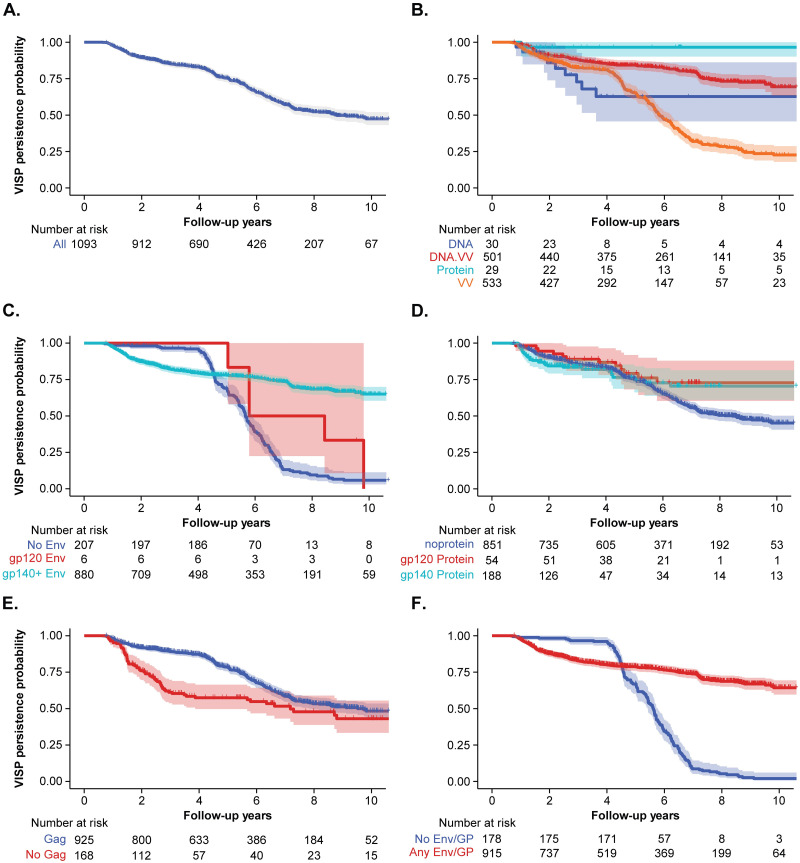
Kaplan-Meier (KM) estimates of HVTN 910 participants; shaded regions represent 95% CIs. KM estimates for (A) all HVTN 910 participants; and segregated by (B) vaccine platform, (C) Env gene, (D) Envelope protein, (E) Gag gene, and (F) Envelope gene or glycoprotein (GP). Participants were censored at time of their modified VISP/R resolution. “At risk” refers to number of participants observed at each timepoint that can contribute to the estimation of VISP/R persistence.
Due to the association between gag, gp140+ gene insert, and protein boost with VISP/R, we also assessed the duration of VISP/R in participants with or without these vaccine components. The 10-year VISP/R persistence rate was 6% (95% CI: 3–11%) for those receiving regimens containing no env and 65% (95% CI: 60–70%) for those receiving gp140+ env (Fig 3C). Of the few gp120 env recipients enrolled in HVTN 910, the 9-year VISP/R persistence rate was 33% (95% CI: 11–100%). When grouped by protein boost, the 10-year probability of VISP/R persistence was 45% (95% CI: 41–50%) for those receiving no glycoprotein, 73% (95% CI: 60–88%) for gp120 protein recipients, and 70% (95% CI: 61–81%) in all others (Fig 3D). The 10-year VISP/R persistence rate was 48% (95% CI: 44–53%) for those receiving regimens containing gag and 43% (95% CI: 33–56%) for those who did not (Fig 3E). Since the durations of VISP/R for gp120 and gp140+ gene and protein recipients were comparable, we assessed the persistence of VISP/R for participants who did not receive envelope gene or glycoprotein (gag only) to those that received any envelope or glycoprotein (with or without gag) (Fig 3F). The 10-year VISP/R persistence rate was 2% (95% CI: 0.7–6%) for participants receiving no envelope or glycoprotein and was 64% (95% CI: 60–70%) for those that received any combination of envelope and/or glycoprotein.
Assessing the VISP/R rate per diagnostic platform
Most serological HIV diagnostic tests are currently designed to detect Gag- and Env-specific antibodies. We measured the rate of VISP/R for each test used in the 75 HIV vaccine clinical trials in order to identify which assays most commonly resulted in the identification of VISP/R (Table 2). HIV diagnostic tests had variable rates of VISP/R in participants who only received gag gene inserts (1–70%). Most laboratory screening and rapid HIV tests had VISP/R rates >50% in participants who received gp140+ gene with or without gp140 protein (64–100%). None of these HIV tests had VISP/R rates >50% in participants who received gp140+ gene and gp120 protein (1–32%). The Abbott HIV AB HIV1/2 (rDNA) and bioMerieux Vironostika HIV-1 were the only laboratory screening or rapid tests that had VISP/R rates >50% in participants who received gp120 envelope or proteins. Although Western Blot/differentiation tests are not considered in the definition of VISP/R, 64–100% participants were reactive in these assays, regardless of the regimen received.
Discussion
Researchers continue to evaluate and advance experimental preventative HIV vaccines that vary in platform, HIV antigens, and adjuvants. While primary analyses in this study evaluated 10-year persistence, it is noteworthy that we observed candidate HIV vaccines inducing VISP/R persistent in study participants nearly 30 years after trial participation. The probability of developing long-lasting VISP/R is mostly related to the vaccine regimen and HIV antigens included in the study product(s), with minimal impact from participant characteristics. This analysis showed that participants receiving viral vectors, viral vectors in combination with DNA vaccines, or proteins were more likely to have VISP/R at the end of the parent protocol than those receiving DNA vaccine regimens. In contrast, more participants who received DNA or DNA/viral vector combination vaccines still had VISP/R 10 years after vaccination than those only receiving viral vector vaccines. Although participant characteristics like geographic location and study phase were associated with increased odds of VISP/R in our analysis, these observations are likely due to operational decisions for study conduct, such as clinical research site selection and progression of immunogenic HIV vaccines from phase 1/2a to phase 2b studies.
The effect of gag in a vaccine regimen on inducing VISP/R was difficult to discern when administered in combination with regimens containing envelope or glycoprotein. It was noteworthy that a portion of participants who received regimens with only gag and no envelope or glycoprotein did have VISP/R at the end of their parent protocol, but only 2% of these participants had VISP/R 10 years after vaccination. Gag is therefore capable of inducing VISP/R, consistent with the presence of Gag antigens in commercial diagnostic tests, but Gag-induced VISP/R appears to be shorter-lived relative to envelope- or glycoprotein-induced VISP/R.
Participants receiving regimens with gp140+ gene inserts or proteins had higher rates of VISP/R at the end of their participation in a clinical trial, and higher persistence of VISP/R at 10 years than those who received any other product. Inclusion of gp120 envelope in a regimen increased the odds of VISP/R in recipients, but a gp120 protein boost, regardless of the type of env gene insert, reduced these odds. A hypothesis for this seemingly contradictory observation could be that vaccine regimens with a gp140+-containing prime induces the presence of anti-envelope antibodies detected by common diagnostic tests, but a gp120 protein boost then diverts the immune response away from generating the cross-reactive antibodies that cause VISP/R.
Participants with VISP/R had high levels of antibody responses to gp120, gp140, V1V2 and gp41 antigens for all subsets of vaccine platforms. Uneven availability of BAMA data for every env/glycoprotein combination made inferences of antibody response by envelope gene insert of protein received difficult. However, results from Palli et. al. provide evidence that while participants receiving DNA and/or viral vector (ALVAC versus modified vaccinia Ankara) combinations with gp120 or gp140+ gene and protein components resulted in IgG and IgG3 antibody responses to these antigens, the gp140 and gp41 IgG responses had longer half-lives than gp120 responses [2]. These results suggest that the persistence of VISP/R in recipients of regimens containing gp140+ genes or proteins is related to the characteristics of the antibodies targeting gp140 and gp41 antigens, as generated by these vaccine regimens.
The induction and persistence of HIV antibodies is an unavoidable (and in fact, desirable) aspect of a successful vaccine candidate. The occurrence of VISP/R associated with these antibodies, however, is equally and fundamentally dependent on the design of commercial HIV tests in use. Many commercial HIV tests indicate HIV acquisition by detecting antibodies to the immunodominant epitopes in gp41 [11–13]. Reliance on this immunodominant domain improves the performance of these assays in detecting acute HIV acquisitions, but complicates the diagnoses of HIV status in former vaccine trial participants [6, 14, 15]. Since these antibody assays are cheaper and simpler to perform than assays that detect HIV nucleic acid (a more direct indication of HIV acquisition in a participant specimen), diagnostic laboratories continue to rely on detection of these antibodies despite the potential complications in assay interpretation. If HIV vaccine developers continue to include gp140 in candidate vaccine regimens, many more study participants may develop VISP/R and then require careful management of their diagnostic testing until a new paradigm in HIV testing is widely adopted in public health settings.
This study has several key limitations. Meta-analysis of these HIV vaccine trials resulted in a dataset that lacked VISP/R rates for all possible combinations of vaccine candidate characteristics. Variable enrollment numbers in vaccine trial arms also meant varied depth of data available for each vaccine characteristic combination, particularly for associations between VISP/R and antibody response. This, in addition to the uneven and incomplete participant follow-up, impacted the precision to which VISP/R persistence could be measured in HVTN 910. This analysis also did not identify the epitopes within gp140 that result in VISP/R, and could not differentiate how the conformation of the gp41 region in the gp140+ env or protein within the vaccine regimen affects immunogenicity. For example, if vaccinated study participants receive gp140+ env or protein in which the immunodominant domain of gp41 is exposed, we hypothesize that their immune system would direct antibodies to this region, and these antibodies would in turn result in VISP/R. As vaccine developers generate products that express stable glycoprotein trimers in which this same region is modified or occluded, the antigenicity of the immunodominant domain (and resulting likelihood of inducing VISP/R) may be lower, but this hypothesis remains untested. Lastly, although all presented analyses tried to adjust for confounding factors that could potentially influence the effect of the demographics and vaccine characteristics on the rate and durability of VISP/R, there could be other unmeasured factors that may contribute to the observed patterns in these data.
Despite these limitations, this study represents the most comprehensive analysis of VISP/R to date. Analysis of 75 HIV vaccine trials and long-term observation of VISP/R in HVTN 910 has indicated that HIV envelope antigens from gene inserts or protein boosts increase the rate of VISP/R in study populations. This effect is modulated in part by other vaccine characteristics (e.g. vaccine platform, gag inserts). As long as HIV antibody-based diagnostic methods are in use, participants and study teams must be prepared to manage this VISP/R potentially for many years after product administration as long as HIV-antibody based diagnostic methods are in use. Vaccine and diagnostic test developers should use this analysis to identify strategies to mitigate this impact on study participants through long-term diagnostic testing support and the development of VISP/R-agnostic testing methods.
Supporting information
S1 Fig
Follow up time of HVTN 910 participants from last vaccination.Participants enrolled in HVTN 910 were monitored for HIV infection during the active study period and monitored for HIV infection and VISP/R during the HVTN 910 active study period. Time of first VISP/R is assumed as date of last HIV vaccination, and participants were followed up until VISP/R resolution, loss to follow up, or study termination, whichever comes first.
(TIF)
S2 Fig
HVTN 910 Kaplan-Meier analysis of persistence of VISP/R.Kaplan-Meier (KM) estimates of all HVTN 910 Participants; shaded regions represent 95% CIs. Participants were censored at time of their per-protocol VISP/R resolution. “At risk” refers to number of participants observed at each timepoint that can contribute to the estimation of VISP/R persistence.
(TIF)
Acknowledgments
We would like to thank the participants and study staff participating in AVEG and HVTN trials. We would also like to thank the HIV diagnostic testing laboratories for performing the HIV diagnostic testing analyzed here. We would like to thank Janine Maenza and Mary Allen for their participation in the HVTN 910 study team and the ongoing follow-up of participants with VISP/R. We would also like to thank Gail Broder, Michelle Andrasik, Mindy Miner, Sam Robinson, and the HVTN Scientific Review Committee for helpful editing.
Funding Statement
This work was supported by the National Institutes of Health (UM1 AI068614 to GDT; UM1 AI068618 to JM; UM1 AI068635 to XH, SG, YH; UM1 AI069412 to SRW; UL1 RR025758 to SRW; P30 AI064518 to GDT). MA and PD The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Data used from this analysis is available on the ATLAS data portal (https://atlas.scharp.org/cpas/project/HVTN%20Public%20Data/begin.view?) and can be provided upon request.
References
Decision Letter 0
16 Mar 2023
PGPH-D-22-02087
Cross-protocol assessment of induction and durability of VISP/R in HIV preventive vaccine trial participants
PLOS Global Public Health
Dear Dr. Hural,
Thank you for submitting your manuscript to PLOS Global Public Health. After careful consideration, we feel that it has merit but does not fully meet PLOS Global Public Health’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
The reviewers find that the manuscript is overall well written but needs minor revisions. For example, the reviewers suggest clarifying which vaccination arms/study products are being compared in your study, improving the clarity of the cohort description, and discussing the choice of antibody titer analyses in the Introduction section. The detailed comments are appended below.
Please submit your revised manuscript by Apr 14 2023 11:59PM. If you will need more time than this to complete your revisions, please reply to this message or contact the journal office at gro.solp@htlaehbuplabolg. When you're ready to submit your revision, log on to https://www.editorialmanager.com/pgph/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the editor and reviewer(s). You should upload this letter as a separate file labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. You should upload this as a separate file labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. You should upload this as a separate file labeled 'Manuscript'.
Guidelines for resubmitting your figure files are available below the reviewer comments at the end of this letter.
We look forward to receiving your revised manuscript.
Kind regards,
Alex Schaefer, PhD
Associate Editor
PLOS Global Public Health
Journal Requirements:
1. Please review your reference list to ensure that it is complete and correct. If you have cited papers that have been retracted, please include the rationale for doing so in the manuscript text, or remove these references and replace them with relevant current references. Any changes to the reference list should be mentioned in the rebuttal letter that accompanies your revised manuscript. If you need to cite a retracted article, indicate the article’s retracted status in the References list and also include a citation and full reference for the retraction notice.
2. We do not publish any copyright or trademark symbols that usually accompany proprietary names, eg ©, ®, ™ (e.g. next to drug or reagent names). Please remove all instances of trademark/copyright symbols throughout the text, including ® on page 24.
Additional Editor Comments (if provided):
[Note: HTML markup is below. Please do not edit.]
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. Does this manuscript meet PLOS Global Public Health’s publication criteria? Is the manuscript technically sound, and do the data support the conclusions? The manuscript must describe methodologically and ethically rigorous research with conclusions that are appropriately drawn based on the data presented.
Reviewer #1: Yes
Reviewer #2: Yes
**********
2. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: Yes
**********
3. Have the authors made all data underlying the findings in their manuscript fully available (please refer to the Data Availability Statement at the start of the manuscript PDF file)?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception. The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: Yes
**********
4. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS Global Public Health does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: Yes
**********
5. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: TITLE:
Cross-protocol assessment of induction and durability of VISP/R in HIV preventive
vaccine trial participants
AIM:
The study assessed the impact of HIV vaccine characteristics (e.g. vaccine platform, gag inserts, adjuvants etc) on the induction and persistence of VISP/R among HIV vaccine recipients. The authors also identified HIV vaccine components that are more likely to induce VISP/R and assessed the detection rate of VISR by antibody based diagnostic assays routinely used in diagnosis of HIV infection.
Minor comments
1. Line 36-39: It’s not clear which vaccination arms/ study products are being compared.
a) gp140+ env gene insert or protein Vs gp120 env or protein components?
b) gp140+ env gene insert or protein Vs participants who received no env?
c) Vs gp120 env or protein components Vs participants who received no env?
2. Lines 46-47: The sentence would read better if it started with “Nearly 40 years after the discovery of HIV, the virus continues to disproportionately burden vulnerable countries and groups ….…
3. Line 48…add two words;
a) that before 1.5 million
b) occurred before in 2020
4. Line 113: define CDC since it is the first time it appears in the manuscript.
5. To improve clarity, I would suggest moving the text between lines 118-127 to appear under the description of the cohort. Merge the information in that paragraph with the cohort description
6. Lines 133-137: Since a total of 8,155 participants were tested for VISP/R out of 21,578 participants initially included in the analysis, it would be good to know how many of the 21,578 were placebo recipients, how many acquired HIV infection during follow up and how many did not complete the studies. This would help to understand why only about a third of the participants were tested for VISP/R at the end of their parent protocols.
7. The text in lines 140-145 does not match with what is presented in Fig 2 (the flow diagram). Fig 2 shows that 4290 participants had evidence of VISR but of these only 1146 were enrolled into HVTN910. What happened to the 3144 participants (4290-1146)? Why weren’t they enrolled into HVTN910? Moreover, the 4290 are not mentioned anywhere in the cohort description, they just pop up in Fig 2.
8. Lines 148-152: The fig 2 legend does not match the information in fig 2. The AVEG, HIVNET and HVTN 032, 034, 050, 057, 077, 502, 503 participants are not shown in Fig 2.
9. Line 156: DAIDS and GCLP appear for the first time in the manuscript. They need to be defined.
10. Line 160: CTU appears for the first time in the manuscript. They need to be defined.
11. Line 168; FDA and CE appear for the first time in the manuscript. They need to be defined.
12. Table 3: The probability of developing VISP/R is not related to participant characteristics. It’s highly influenced by the vaccine characteristics and the immunization schedule. I wonder why participants’ results have been split into two groups; sub-Saharan Africa vs the rest of the World. It would be good if the authors had provided justification for splitting the analysis of the data into those two groups. Additionally, I would suggest the authors add a column for overall (combined) results in the table, so the reader sees an overall picture first before going into the subgroups.
13. Table 4: add “s” to the word odd in the column containing multivariate odd ratio
14. Line 262: Add “the” before risk of VISP/R by binding antibody titers to Env
15. Line 287: EOS appears for the first time. It should be defined.
16. Table 5:
a) Provide overall (combined) results first
b) Provide justification for splitting the results into two groups
c) There should be an explanation below the table explaining why for some parameters the total number of volunteers is less than 1093 (i.e 1085 and 1084)
d) HVTN910 should NOT be written as 910
e) Median years since 910 enrollment [IQR].. what does it stand for? follow-up time since enrollment in HVTN 910? It’s not clear
f) Median years since 910 enrollment (%)… same comment as above
g) What is the difference between 0 and 0-1? Why shouldn’t the authors present the data as <1 year?
h) The content would be clearer if both actual number and % were used in the table 5 i.e 4 (X%).
i) Years since EOS [IQR]… Same comments as in e above
j) Years since last vaccination [IQR]… Same comments as in e above
k) Median N of VISP assessment. what does N mean? Also add (IQR)
l) Three consecutive Ns over 1 yr… What does Ns and Yr mean?
m) Ensure uniformity in presentation of data. The numbers and percentages in the table should either be centred, aligned to the right or to the left.
17. Lines 308-334: Maintain consistency. Indicate 95% CI inside the parenthesis of all VISP/R results. i.e 63% (95% CI: 46-86%), 23% (18-29%), 69% (64-76%), 97% (90-100%).
18. Lines 353-355: it’s written that the study “found that some HIV vaccine candidates have induced VISP/R in study participants lasting between 10-30 years”. But the participants were only followed up for a maximum of 10 years only. The authors should rephrase the statement to reflect the results in their manuscript.
19. Line 407: an ‘a’ is missing in the word metanalysis.
Reviewer #2: The manuscript reports the long-term results of induced seropositivity in HIV vaccine recipients in 75 studies in several continents. This issue may not be known to the PLOS Global Public Health readership but is addressed here by the largest study ever conducted.
Overall, the results are quite complex but well presented. However, there are some points that the authors need to address.
Minor comments:
Figure 1: Sorry to say that it is not very informative. The right area of the graph with the rapid test image does not describe the VISP algorithm and is quite confusing.
Line 106: Do the authors mean that gp120 and gp41 were grouped together? Please specify as the groupings are not apparent in Table 1.
Line 119: Unscheduled testing, but when were they recommended ?
Line 179: Antibody titer data appear at this point in the manuscript. What are the objectives in this study? Please discuss in the Introduction and briefly justify the choice of specific IgG antibodies instead of just giving references.
Line 244-245: A single p-value does not allow to conclude that the 3 platforms are different from DNA platform. My suggestion is to add the confidence intervals of the multivariate odds ratios in Table 4.
Line 256: OR=1.634 in the text but OR=0.752 in SI Table 1. Please clarify.
Figure 3: Gray circles are outside 1.5 x interquartile range. Please check the legend.
Table 5: For times since vaccination and since EOS, the 4+ year class is not very informative. We would have seen more extreme classes and the range in addition to the IQR.
Line 354: ‘10-30 years of VISP/R’, this is not shown in the results (see Table 5 comment)
Line 369: ‘98% no longer had VISP/R 10 years after” , not in the Results , except in Figure S1 but only graphically
Figure S2: Individual points are beyond the “outliers” in Figure 3. Please check or justify.
**********
6. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
Do you want your identity to be public for this peer review? If you choose “no”, your identity will remain anonymous but your review may still be made public.
For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
**********
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email PLOS at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 0
28 Apr 2023
Attachment
Submitted filename: Response_to_Reviewers.docx
Decision Letter 1
17 May 2023
Cross-protocol assessment of induction and durability of VISP/R in HIV preventive vaccine trial participants
PGPH-D-22-02087R1
Dear Dr. Hural,
We are pleased to inform you that your manuscript 'Cross-protocol assessment of induction and durability of VISP/R in HIV preventive vaccine trial participants' has been provisionally accepted for publication in PLOS Global Public Health.
Before your manuscript can be formally accepted you will need to complete some formatting changes, which you will receive in a follow up email. A member of our team will be in touch with a set of requests.
Please note that your manuscript will not be scheduled for publication until you have made the required changes, so a swift response is appreciated.
IMPORTANT: The editorial review process is now complete. PLOS will only permit corrections to spelling, formatting or significant scientific errors from this point onwards. Requests for major changes, or any which affect the scientific understanding of your work, will cause delays to the publication date of your manuscript.
If your institution or institutions have a press office, please notify them about your upcoming paper to help maximize its impact. If they'll be preparing press materials, please inform our press team as soon as possible -- no later than 48 hours after receiving the formal acceptance. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information, please contact gro.solp@htlaehbuplabolg.
Thank you again for supporting Open Access publishing; we are looking forward to publishing your work in PLOS Global Public Health.
Best regards,
Julia Robinson
Executive Editor
PLOS Global Public Health
***********************************************************
Reviewer Comments (if any, and for reference):
Reviewer's Responses to Questions
Comments to the Author
1. If the authors have adequately addressed your comments raised in a previous round of review and you feel that this manuscript is now acceptable for publication, you may indicate that here to bypass the “Comments to the Author” section, enter your conflict of interest statement in the “Confidential to Editor” section, and submit your "Accept" recommendation.
Reviewer #1: All comments have been addressed
Reviewer #2: All comments have been addressed
**********
2. Does this manuscript meet PLOS Global Public Health’s publication criteria? Is the manuscript technically sound, and do the data support the conclusions? The manuscript must describe methodologically and ethically rigorous research with conclusions that are appropriately drawn based on the data presented.
Reviewer #1: Yes
Reviewer #2: Yes
**********
3. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: Yes
**********
4. Have the authors made all data underlying the findings in their manuscript fully available (please refer to the Data Availability Statement at the start of the manuscript PDF file)?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception. The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: Yes
**********
5. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS Global Public Health does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: Yes
**********
6. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: No further comments
Reviewer #2: (No Response)
**********
7. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
Do you want your identity to be public for this peer review? If you choose “no”, your identity will remain anonymous but your review may still be made public.
For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
**********
Articles from PLOS Global Public Health are provided here courtesy of PLOS
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/149767672
Article citations
HIV Diagnostics and Vaccines: It Takes Two to Tango.
J Infect Dis, 229(6):1919-1925, 01 Jun 2024
Cited by: 1 article | PMID: 38451247
Review
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Vaccine-induced HIV seropositivity/reactivity in noninfected HIV vaccine recipients.
JAMA, 304(3):275-283, 01 Jul 2010
Cited by: 35 articles | PMID: 20639561 | PMCID: PMC3086635
Comparison of shortened mosaic HIV-1 vaccine schedules: a randomised, double-blind, placebo-controlled phase 1 trial (IPCAVD010/HPX1002) and a preclinical study in rhesus monkeys (NHP 17-22).
Lancet HIV, 7(6):e410-e421, 17 Feb 2020
Cited by: 14 articles | PMID: 32078815 | PMCID: PMC7297076
Inclusion of a CRF01_AE HIV envelope protein boost with a DNA/MVA prime-boost vaccine: Impact on humoral and cellular immunogenicity and viral load reduction after SHIV-E challenge.
Vaccine, 30(10):1830-1840, 09 Jan 2012
Cited by: 13 articles | PMID: 22234262 | PMCID: PMC3324265
Exploring a community's understanding of HIV vaccine‑induced seropositivity in a South African research setting.
S Afr Med J, 113(1):36-41, 20 Dec 2022
Cited by: 1 article | PMID: 36537546
Funding
Funders who supported this work.
Center for AIDS Research, Duke University (1)
Grant ID: P30 AI064518
NCRR NIH HHS (1)
Grant ID: UL1 RR025758
NIAID NIH HHS (3)
Grant ID: UM1 AI068618
Grant ID: UM1 AI068635
Grant ID: P30 AI064518
National Center for Research Resources (1)
Grant ID: UL1 RR025758
National Institute of Allergy and Infectious Diseases (4)
Grant ID: UM1 AI068614
Grant ID: UM1 AI068635
Grant ID: UM1 AI069412
Grant ID: UM1 AI068618

 1
,*
1
,*

