Abstract
Free full text

H3N2 influenza A virus gradually adapts to human-type receptor binding and entry specificity after the start of the 1968 pandemic
Associated Data
Significance
Influenza A viruses originate from waterfowl. To become a human influenza virus, their hemagglutinin needs to adapt to bind human-type sialic acid receptors. Tight cell binding results from multiple, but individually weak, interactions. Low affinity enables crucial virus rolling over a cell surface but is poorly characterized at the molecular level. Systematic analysis of affinities for avian- and human-type receptors showed an unexpected plasticity in the receptor requirement for human H3N2 viruses isolated since its appearance in 1968. Neither rapid acquirement nor maintenance of high selectivity for human-type receptors is a strict necessity. Revealing changes of the kinetic parameters of receptor binding during 42 y of evolution improves our molecular understanding of the selection and maintenance of a human-to-human transmissible strain.
Abstract
To become established upon zoonotic transfer, influenza A viruses (IAV) need to switch binding from “avian-type” α2-3-linked sialic acid receptors (2-3Sia) to “human-type” Siaα2-6-linked sialic acid receptors (2-6Sia). For the 1968 H3N2 pandemic virus, this was accomplished by two canonical amino acid substitutions in its hemagglutinin (HA) although a full specificity shift had not occurred. The receptor repertoire on epithelial cells is highly diverse and simultaneous interaction of a virus particle with a range of low- to very low-affinity receptors results in tight heteromultivalent binding. How this range of affinities determines binding selectivity and virus motility remains largely unknown as the analysis of low-affinity monovalent HA–receptor interactions is technically challenging. Here, a biolayer interferometry assay enabled a comprehensive analysis of receptor-binding kinetics evolution upon host-switching. Virus-binding kinetics of H3N2 virus isolates slowly evolved from 1968 to 1979 from mixed 2-3/2-6Sia specificity to high 2-6Sia specificity, surprisingly followed by a decline in selectivity after 1992. By using genetically tuned HEK293 cells, presenting either a simplified 2-3Sia- or 2-6Sia-specific receptor repertoire, receptor-specific binding was shown to correlate strongly with receptor-specific entry. In conclusion, the slow and continuous evolution of entry and receptor-binding specificity of seasonal H3N2 viruses contrasts with the paradigm that human IAVs need to rapidly acquire and maintain a high specificity for 2-6Sia. Analysis of the kinetic parameters of receptor binding provides a basis for understanding virus-binding specificity, motility, and HA/neuraminidase balance at the molecular level.
Influenza A viruses (IAV) of waterfowl occasionally cross the species barrier to infect the respiratory tract of, for example, swine and humans. Only rarely they become established, causing a pandemic. Reassortment of gene segments between avian and mammalian IAVs can be one of the initial steps in the creation of a mammalian IAV followed by further adaptation to the new host. For the introduction of a HA of avian origin adaptation to “human-type” 2-6Sia receptors is a major hurdle to be taken (1). Two canonical mutations in the viral envelope–embedded hemagglutinin (HA) of the 1968 pandemic H3N2 virus cause a switch to human-type receptor specificity (2, 3). However, early isolates retain some avian-type 2-3Sia receptor binding (4–6) and a fully functional specificity switch likely requires mutations for fine-tuning of binding specificity, before or after establishment in the human host. Fluctuation of H3N2 virus-binding strength and selectivity over the years has been reported (6–9). Strong and long-lasting IAV binding to the cell surface requires multiple HA–Sia interactions. Individual HA–receptor interactions need to be weak to allow virus particles to roll over the cell surface. Such motility was shown to require neuraminidase (NA) activity and is essential for infectivity (10, 11). Association and dissociation rates of individual HA–Sia interactions are the primary determinants of IAV-binding strength and specificity, but technical difficulties have largely prohibited their determination. Here a method of analysis was developed and applied to investigate the evolution of binding and entry specificity of seasonal H3N2 viruses.
Sialic acids (Sia) are a group of monosaccharides that terminate glycan chains attached to glycolipids and the N- and O-linked glycans of proteins. Members of this group are differentially distributed on epithelial tissues along the respiratory tract (12, 13). In general, “avian” IAVs preferentially attach to Siaα2-3Galβ receptors (“avian-type”) whereas “human” IAVs prefer binding to Siaα2-6Galβ/Siaα2-6GalNAc receptors (“human-type”) (2, 14). Linkage type-specificity determines the preferred infection sites, which are the 2-3Sia-rich avian enteric tract and the 2-6Sia-rich human upper respiratory tract.
The binding strength of individual HA–Sia interactions is called affinity and is measured by the equilibrium dissociation constant (KD). It is defined as the ratio of the dissociation rate constant (koff) and the association rate constant (kon) (KD=koff/kon) with molar (M) as a unit. A decrease in KD corresponds to an increase of affinity. The interaction of IAV particles with receptor-coated surfaces occurs by several simultaneous HA–Sia interactions of low affinity that collectively result in tight multivalent binding of high avidity (10, 15). A low affinity results in a fast alternation of HA–Sia dissociation and reassociation. Sia moieties can reassociate with the same HA or with a neighboring HA, thereby allowing the virus particle to wobble on the receptor surface. Alternatively, Sia can associate with the receptor-destroying NA, which removes Sia receptors by enzymatic cleavage. This creates a local Sia receptor gradient and induces directional motility of surface-associated IAV particles (11). Motility, which is essential for cell entry (11), is therefore critically dependent on HA–Sia binding and dissociation rates on the one hand and NA binding rate and activity on the other hand (10, 11, 16). The dynamics of HA–Sia interactions are determined by their kon and koff, which are the prime determinants of virus–cell surface interaction dynamics. However, virus-binding analysis has usually been limited to determination of binding avidity for homogeneously coated receptor surfaces (e.g., glycan arrays) whereas the question how affinity parameters for a specific receptor determine binding and entry efficiency into cells carrying such receptors is still largely unexplored.
Structural analysis has revealed crucial differences between the interaction of HA with an avian-type receptor and a human-type receptor. The Sia moieties of both receptor types are bound in a similar conformation to the receptor-binding site (RBS) (1, 15, 17). Most hydrogen bonds and van der Waals interactions are formed between conserved amino acids in the RBS and the Sia moiety, independent of Sia linkage-type. However, an α2-3 or α2-6 linkage-type determines the angle by which the subterminal glycan chain radiates from the RBS. Therefore, interactions between HA and the penultimate disaccharide depend on the linkage-type and largely determine binding specificity (1, 17). Unfortunately, binding affinities cannot be precisely estimated from HA-glycan structures, also because such static structures provide no insight into changes in entropy and ease of access of the Sia to the RBS (15).
The correlation between receptor binding and virus entry selectivity is only partially understood. Cells carrying major or minor amounts of a specific receptor can be infected with equal efficiency (8, 12). In accordance, we recently found that binding and entry of 2-3Sia-specific avian IAVs is strongly enhanced at low densities of 2-3Sia by mixed 2-6Sia receptors. Thus, low- and high-affinity interactions can collectively contribute to virus binding via heteromultivalent interactions (18). Understanding the adaptation of an IAV strain to the heterogeneous receptor repertoire of a novel host therefore requires knowledge on the evolution of the kinetic parameters for the different receptors present at the epithelial surface.
Here, we determined the evolution of the KD, koff and kon of HA–receptor interactions for H3N2 strains isolated between 1968 and 2009 using a biolayer interferometry (BLI)–based method. Changes in binding affinity were strictly dependent on changes of the association rate constant. We observe that the correlation of entry selectivity, i.e. the ratio of entry efficiency into genetically tuned 2-3Sia- versus 2-6Sia-receptor displaying HEK293 cells, with selectivity of binding affinity (KD) was better than correlation between binding avidity and affinity.
Results
Systematic Analysis of KD, kon, and koff for HA–Sia Receptor Interactions.
The evolution of the avidity and specificity of IAV–receptor surface interactions has been studied in detail. Still, at the nanoscale level, it remains unknown how affinity determines the dynamics and strength of these multivalent interactions. We therefore developed a BLI-based method (outlined in Fig. 1A) to determine the affinity of individual HA–receptor interactions by straightforward equilibrium binding measurements at a physiologically relevant temperature of 30 °C. By linking virus particles to the surface of BLI biosensors, the high HA density was obtained that is required for sensitive real-time recording of equilibrium binding and dissociation of small molecules. We measured the association and dissociation rates (Fig. 1B) of tri-saccharides that represent common avian-type (Siaα2-3Galβ1-4GlcNAc, 3SLN; Siaα2-3Galβ1-4Glc, 3SLac) and human-type IAV (Siaα2-6Galβ1-4GlcNAc, 6SLN; Siaα2-6Galβ1-4Glc, 6SLac) receptors. Association and dissociation rate constants for seasonal H3N2 isolates from 1968 to 2009, representing 10 consecutive antigenic clusters, were calculated [kon (SI Appendix, Table S1) and koff (SI Appendix, Table S2)]. Binding affinity (KD, Table 1) displayed a gradual decrease for α2-3Sia-glycans while binding affinity for α2-6Sia-glycans first increased, and decreased after 1992 (Fig. 1 C and D). This trend is similar for SLac and SLN. Interestingly, KD was mostly determined by changes in association rate (kon ~800 to ~50,000 M-1 min−1, Fig. 1E) with dissociation rate being stable over time for all virus–glycan combinations (koff ~50 min−1 to 70 min−1, Fig. 1F).
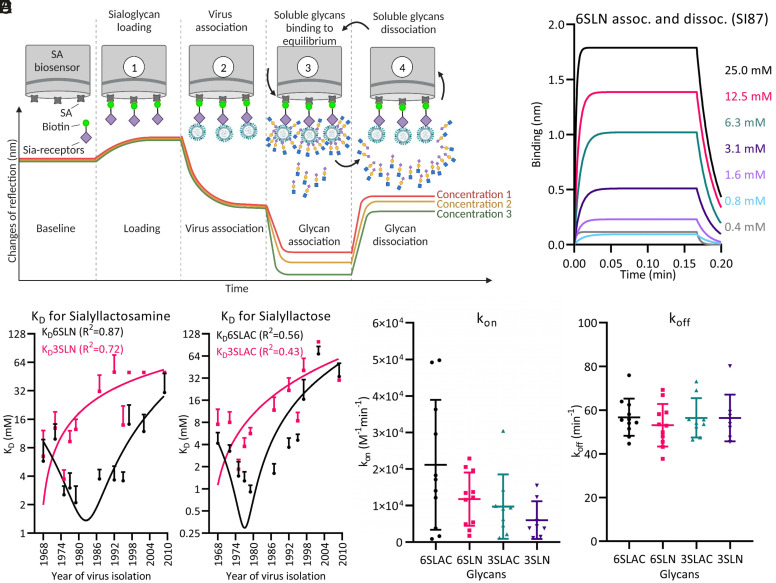
Equilibrium binding measurements of glycans in solution to IAV particles by BLI. (A) Biotinylated sialoglycans Siaα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc [2-6Sia-(LN)3] or 2-3Sia-(LN)3 are loaded to SA-coated biosensors followed by stable association of virus particles in the presence of 10 µM NA inhibitor OC. Sensors are immersed in an appropriate concentration range of soluble sialoglycans and after reaching equilibrium-binding levels, sensors were transferred to glycan-free buffer to record dissociation rate. (B) Binding and dissociation is recorded 5 times per sec at 30 °C at the indicated concentrations of 6SLN in the presence of OC to inhibit NA activity. Curves for SI87 are shown as an example. (C and D) Binding affinities (KD) for the indicated glycans were determined from multiple curves of two biological replicates and plotted against year of virus isolation. Error bars (SD) and R2 of trend curves (GraphPad prism) are indicated. (E and F) Box plot showing (E) association rate constant (kon) or (F) dissociation rate constant (koff) for 11 H3N2 strains with the four indicated glycans.
Table 1.
Equilibrium dissociation constant of human H3N2 strains
| KD (mM) | ||||||
|---|---|---|---|---|---|---|
| 6SLac | 6SLN | 3SLac | 3SLN | 3SLac/6SLac | 3SLN/6SLN | |
| HK68 | 4.2 ± 1.6* | 5.8 ± 4.0 | 7.5 ± 4.5 | 6.5 ± 5.6 | 1.8 | 1.1 |
| EN72 | 3.3 ± 0.7 | 9.9 ± 4.4 | 8.0 ± 3.1 | 12.9 ± 6.3 | 2.4 | 1.3 |
| VI75 | 1.5 ± 0.9 | 2.5 ± 0.6 | 1.8 ± 0.7 | 3.7 ± 0.9 | 1.2 | 1.5 |
| TX77 | 1.3 ± 0.4 | 3.0 ± 1.3 | 3.8 ± 1.1 | 9.3 ± 2.8 | 2.9 | 3.1 |
| BK79 | 0.9 ± 0.2 | 2.1 ± 1.0 | 5.6 ± 1.1 | 12.5 ± 3.5 | 6.2 | 6.0 |
| SI87 | 1.6 ± 0.6 | 3.7 ± 1.0 | 11.7 ± 5.8 | 31.5 ± 15.5 | 7.3 | 8.5 |
| BE92 | 3.7 ± 1.2 | 3.6 ± 1.4 | 21.7 ± 10.4 | 50.4 ± 26.2 | 5.8 | 14.0 |
| WU95 | 4.6 ± 0.9 | 3.6 ± 0.8 | 8.4 ± 2.5 | 13.9 ± 8.2 | 1.8 | 3.9 |
| SI97 | 16.5 ± 14.0 | 14.2 ± 8.4 | 40.7 ± 18.6 | 50† | 2.5 | 3.5 |
| FU02 | 68.4 ± 17.7 | 11.8 ± 6.1 | 100† | 50† | 1.5 | 4.2 |
| PE09 | 33.7 ± 17.1 | 30.6 ± 18.3 | 30† | 50† | 0.9 | 1.6 |
| H5N1 (HU02) | 7.1 ± 2.9 | ~30 | 0.8 ± 0.2 | 2.0 ± 1.1 | 0.1 | 0.1 |
6SLac (Siaα2-6Galβ1-4Glc); 6SLN (Siaα2-6Galβ1-4GlcNAc).
3SLac (Siaα2-3Galβ1-4Glc); 3SLN (Siaα2-3Galβ1-4GlcNAc).
*SD.
†Approximate KDs that could only be estimated from equilibrium binding levels.
A comparison of our systematic analysis with the scarce data from previous studies (19–26), obtained by various methods and mostly lacking data on kon and koff, shows similar KD ratios (3SLac/6SLac is 0.82 to 2.12) for HK68 or its closely related strain AI68 (SI Appendix, Table S3). Absolute KD values obtained here at 30 °C were twofold-to-threefold higher than previous determinations at lower temperatures. Remarkably, although KD values reported for AI68 by NMR (19) or microscale thermophoresis (23) at 24 °C were similar to those determined at 4 °C by end point binding (21), the KD for ME71 (similar to EN72), as determined by ellipsometry, was strongly temperature dependent (SI Appendix, Table S3). We conclude that kinetic parameters can be accurately determined by adapting straightforward equilibrium binding analysis to a BLI platform. The results show that H3N2 receptor-binding kinetics evolve over time but, in contrast to the common perception, rapid evolution to, and maintenance of, a high affinity and selectivity for human-type receptors did not occur.
Systematic Analysis of Receptor-Binding Rates of Human Seasonal H3N2 Strains.
After having determined the binding affinities of a range of H3N2 virus strains, we can now systematically analyze how monovalent binding affinity determines the avidity of multivalent IAV binding to receptor-coated surfaces. We thereto measured virus-binding rates at 30 °C using BLI sensors coated with 1) synthetic glycans or, representing natural sialoglycoprotein receptors, 2) the LAMP1 glycoprotein. LAMP1 was produced in a HEK293 cell–based system that offers the major advantage of the possibility for a direct comparison of virus-binding avidity to virus entry using the same receptor repertoire (see below). In short, HEK293 cells that are genetically Sia-deficient (HEK293ΔSia) were transiently transfected with sialyltransferases ST3GAL4 (HEK2932-3Sia) or ST6GAL1 (HEK2932-6Sia) to display, respectively, only 2-3Sias or 2-6Sias on its cell surface and secreted proteins. Dual binding specificity for 2-3Sia and 2-6Sia (Fig. 2A) was observed for H3N2 strains isolated until 1975 (HK68, EN72 and VI75). Remarkably, glycan chain length appeared to affect binding rate as VI75 and BK79 prefer binding to the synthetic 2-3Sia glycans with three LacNAc repeats (LN3) over LN2. While more limited 2-3Sia binding was retained by TX77 and BK79, later strains strictly bound to 2-6Sias. Still, after 40 y of evolution, PE09 regained dual specificity. Avian H5N1 strain HU02 displayed specific binding to 2-3Sia.

Analysis of virus-binding rate to biosensor surfaces coated with synthetic glycans or sialoglycoprotein LAMP1. (A) Sialylated LAMP1 carrying 2-3Sia (grey bars) or 2-6Sia (red bars) or biotinylated synthetic glycans consisting of two [(LN)2] or three [(LN)3] LacNAc repeats carrying 2-3Sia (grey bars) or 2-6Sia (red bars) were loaded to maximum capacity on biosensors. Binding rates of the indicated virus strains were recorded and plotted in bar diagrams. Mean values are indicated. Error bars show SD of three biological replicates. (B and C) Virus binding rate (left yaxis) 2-6Sia-LAMP1 (B) or 2-6Sia-(LN)3 (C) and affinity (1/KD; right yaxis) are plotted against year of virus strain isolation. R2 for trend curves (GraphPad prism): 1/KD 6SLN = 0.7, 2-6Sia-LAMP1 = 0.78, 2-6Sia-(LN)3 = 0.12. Correlation (and significance) between binding rate and 1/KD: (B) 0.83 (P= 0.0017); (C) −0.11 (P= 1). (D) BLI recordings of 2-6Sia-specific lectin SNA binding to LAMP1 produced in HEK293ΔSia cells transfected with ST6Gal1 (ST6) with or without B3GNT2 (B3) and B4GALT1 (B4) as indicated (E) BLI recordings of H3N2 strains FU02, SY97 and VI75 binding to 2-6Sia-LAMP1 produced in HEK293ΔSia cells transfected with ST6Gal1 (ST6) with or without B3GNT2 (B3) and B4GALT1 (B4) are shown.
Binding affinity for 6SLN (Fig. 2 B and C) correlated well (0.82; P = 0.0017) with binding rate to 2-6Sia-LAMP1 but not (−0.11) to 2-6Sia-(LN)3, unless strains SY97, FU02, and PE09 were excluded (0.50; P = 0.21). Remarkably, SY97, FU02, and PE09 bound well to 2-6Sia-(LN)3 and thereby resemble H3N2 viruses isolated after 1990s that preferred binding to elongated glycan chains (7, 9, 27). As N-glycans produced in HEK293 cells carry short glycan chains (28, 29), we cotransfected HEK293ΔSia cells with β3GlcNAc-transferase B3GNT2 and β4Gal-transferase B4GALT1 in addition to LAMP1 and ST6Gal1. Collectively, B3GNT2 and B4GALT1 elongate glycan chains with Galβ1-4GlcNAc repeats. 2-6Sia-LAMP1 with or without elongated chains displayed similar sialylation as was shown by staining with 2-6Sia-specific lectin SNA (Fig. 2D). Efficient 2-6Sia-LAMP1 binding by SY97 or FU02 was dependent on chain elongation by B3GNT2/B4GALT1 in a dose-dependent way (Fig. 2E). In contrast, binding of early isolate VI75 was negatively affected. We conclude that binding affinity logically correlates with virus-binding rate but additional factors, such as chain length, can further affect binding rate.
The short half-life time (~0.8 s) of monovalently bound virus particles limits detection by BLI, which mainly records multivalent virus–receptor interactions that provide long-lasting association of high avidity. Multivalent binding depends, among others, on receptor density which determines the number of HA–Sia interactions that can be formed with a virus particle. We determined the receptor density required for supporting a binding rate that is 50% of the maximally achievable binding rate (SI Appendix, Fig. S1 andTable S4), which appeared to correlate well with the KD (0.84; P = 0.0012). We conclude that at a higher affinity, less monovalent HA–Sia interactions are required for stable multivalent binding.
Avian-type 2-3Sia receptors display a 1.5 to 5.9 lower affinity than 2-6Sia receptors for the H3N2 strains (SI Appendix, Table S4). Remarkably, they were shown (18) to contribute to binding of human strains by reducing the density of 2-6Sia receptors that is required for half-maximal binding rate. Clearly (SI Appendix, Table S4), positive heteromultivalent binding effects of avian-type receptors on binding of human IAVs are not restricted to a narrow affinity range for 3SLN (3.7 to ~50 mM) nor do the ratios of the KDs for avian-type and human-type (1.5 to 5.9) match precisely with the magnitude of the heteromultivalent binding effects (1.4-fold to 2.1-fold). In conclusion, the density at which human-type receptors support multivalent binding of human H3N2 viruses correlates strongly with the KD. Avian-type receptors display a wide range of affinities for these human H3N2 viruses and support their binding without a strong correlation with their absolute KD.
Entry Selectivity of H3N2 Strains into HEK293 Cells with a Specific Receptor Repertoire.
Receptor density and complexity on cells is much higher than on the artificial surfaces used for virus-binding studies (10). As a consequence, virus strains displaying high receptor binding specificity in vitro, can enter cells with different receptor repertoires at similar efficiency. Also, binding strength often hardly affected entry efficiency (8, 12). To address this issue, we studied virus entry into cells presenting less sialoglycan diversity by limiting sialylation to Galβ1–4GlcNAc termini. These are abundantly present on N-linked glycans with only minor levels having been identified on O-glycans and glycolipids (30). HEK2932-3Sia and HEK2932-6Sia cells display a Sia-linkage type-specific receptor surface, and entry into these cells was compared to entry into wild-type HEK293WT cells and Sia-deficient HEK293ΔSia cells. In parallel, LAMP1 produced in the four cell types was used as a reporter to confirm the expected sialylation status of these cells by binding of 2-3Sia- specific or 2-6Sia-specific lectins (SI Appendix, Fig. S2). To determine virus infection efficiency, the different HEK293 cell lines were transfected with a plasmid encoding a Gaussia luciferase reporter that becomes transcribed by the viral replication machinery upon infection. Entry efficiency was plotted against virus particle numbers (Fig. 3), which were determined for each strain by nano particle tracking analysis. A infection dose based on virus particle numbers enables unbiased comparison of binding and entry in contrast to, for instance, infectious titer-based doses obtained on MDCK-II (Madin–Darby canine kidney) cells displaying a heterogeneous, cell type-specific, receptor repertoire. Here, for example, the ratio of particle number over infectious titer was on average ~60 but with large differences between virus strains (SI Appendix, Fig. S3). Virus entry efficiency could be recorded over a ~4 log dynamic range and reached saturation at doses over 100 particles/cell for most strains in HEK293WT. Entry of early isolates HK68, EN72, and VI75 into HEK2932-6Sia and HEK2932-3Sia was equally efficient followed by increasing, and after 1992 decreasing, selectivity for entry into HEK2932-6Sia cells. Selectivity was completely lost again in 2009. In conclusion, entry selectivity gradually evolves between 1968 and 2009 from nonselective to selective for 2-6Sia and back to nonselective. This pattern resembles the pattern obtained for the KD for 6SLN (Fig. 2C).
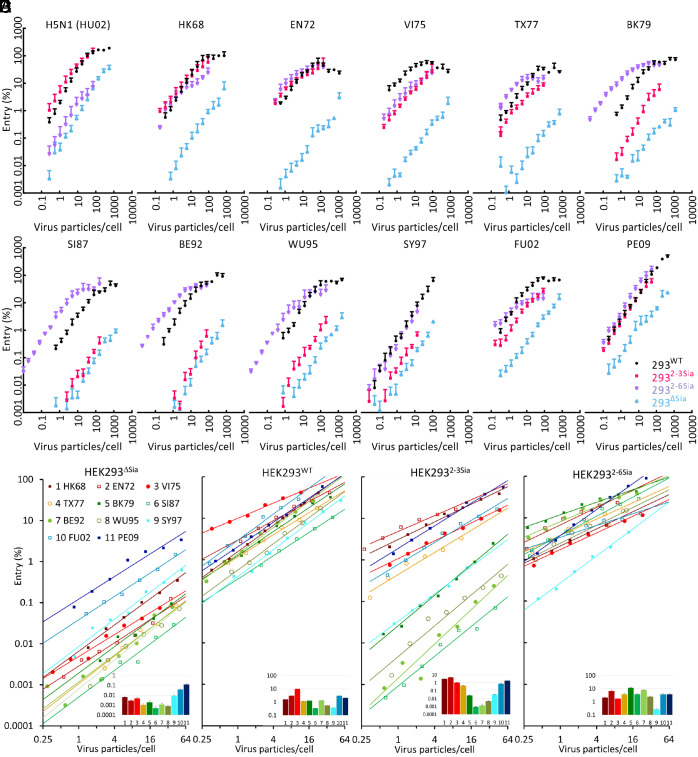
Determination of virus entry efficiency into HEK293WT, HEK293ΔSia, HEK2932-6Sia and HEK2932-3Sia cells. (A) HEK293WT, HEK293ΔSia, HEK2932-6Sia, and HEK2932-3Sia cells were infected with a dose range between 0.01 and 10.000 particles/cell (x axis) in 96-well plates (25.000 cells per plate). Single round infection was measured 16 h p.i. and plotted on the y axis after normalization to HK68 maximum infection level into HEK293WT cells. Data points represent the average of 9 measurements (three replicates, each performed in triplicate). Error bars show SD. (B–E) Data points from exponential parts of the curves (within a range of ~0.25 to ~50 particles per cell) were fitted to y = axb. In this equation, a is the entry efficiency at a dose of 1 particle/cell and b displays the slope of the linear curve when plotted double logarithmic (exponential increase). Statistics of curve fitting are shown in SI Appendix, Table S5. Curves for all H3N2 strains were plotted together per cell line. Inserts show infection levels (y axis) of the different strains at an infection dose of 1 particle/cell.
For infectious doses lower than 100 particles the entry curves fitted well to a function y = axb (SI Appendix, Table S5), where y is infection level, x is particle number, and a value of b= 1 implies that infection doubles when particle number is doubled. Infection of HEK293WT cells displayed a 30-fold infection efficiency range for the different H3N2 strains. Entry of HEK293WT cells, displaying low levels of 2-6Sia and high levels of 2-3Sia (SI Appendix, Fig. S2), did not correlate with the affinity for either 3SLN (−0.33) or 6SLN (−0.09). Entry efficiency of HEK2932-3Sia cells (Fig. 3D) varied largely between strains (~5,300-fold at 1 particle/cell) and is most efficient for the early H3N2 isolates that display similar affinity for 3SLN and 6SLN (HK68, EN72, VI75 and PE09). However, a correlation of absolute entry levels with absolute affinity for 3SLN was lacking (−0.31). Entry efficiency of HEK2932-6Sia cells (Fig. 3E) displayed much less variation between strains (only sevenfold when PE09 is left out) but also here a correlation with absolute affinity for 6SLN was lacking (−0.23).
The slope of the curves in Fig. 3 B–E, showing infectious dose-dependent increase in infection, slightly varied between strains and cell lines. In particular, entry of 2-6Sia-selective strains into HEK2932-3Sia cells displayed steeper slopes than observed for its entry into HEK2932-6Sia cells. Thus, virus entry displays maximal receptor selectivity at a low-infectious dose (entry into HEK2932-6Sia cells divided by entry into HEK2932-3Sia cells; Fig. 4A). Based on three biological replicates, this effect was significant for strains BK79, SI87, BE92, WU95, and FU02 (Fig. 4 B and C).

Analysis of IAV entry selectivity. Three replicates of entry into HEK2932-6Sia or HEK2932-3Sia cells, each performed in triplicate, were fitted to the expression y = axb in order to determine the significance of a difference in slopes (b from the equation) observed in Fig. 3. (A) Entry selectivity was calculated from the fitted curves and plotted for three particle/cell infection doses, showing that selectivity decreases at increased dose for TX77 to FU02. (B) Example of fitted curves for BK79. (C) A plot of b (from y = axb) for all H3N2 strains comparing entry into HEK2932-6Sia and HEK2932-3Sia cells. All data of the analysis are listed in SI Appendix, Table S6. Error bars are SD; *P < 0.05; **P < 0.01. (D) Entry efficiency into HEK293ΔSia cells, HEK2932-6Sia cells or HEK2932-3Sia cells relative to HEK293WT cells. (E) Entry selectivity (entry into HEK2932-6Sia cells/entry into HEK2932-3Sia cells; left y axis) and affinity selectivity (KD 3SLN/6SLN; right y axis) plotted against year of virus strain isolation. Correlation between entry and affinity selectivity is 0.93 (P = 3.51E-05). (F) Entry efficiency into HEK293ΔSia cells (left yaxis) and KD for 6SLN (right y axis) plotted against year of virus strain isolation. Correlation between entry efficiency and KD is 0.92 (P = 5.35E-05).
Entry into HEK2932-3Sia, HEK2932-6Sia, and HEK293ΔSia cells was normalized to entry into HEK293WT cells in order to compare the effects of linkage-type-specific sialylation on entry selectivity (Fig. 4D). Clearly, strains TX77 to WU95 infected HEK2932-6Sia cells 4- to 11-fold better than HEK293WT cells (Fig. 4D, red bars), whereas they infected HEK2932-3Sia cells 50- to 1,000-fold less well than HEK293WT cells (Fig. 4D, green bars). Entry selectivity (HEK293Sia2-6 entry/HEK293Sia2-3 entry) and binding specificity (KD 3SLN/KD 6SLN, Table 1) are plotted in Fig. 4E and showed a strong correlation of 0.93 (P = 3.51E-05). Interestingly, IAV entry into Sia-deficient HEK293ΔSia cells (Fig. 3B) displayed a ~250-fold range of entry efficiencies between H3N2 strains, which correlated well (0.92; P = 5.35E-05) with the KD for 6SLN (Fig. 4F). Thus, in other words, a loss of affinity for 2-6Sia matched with improved entry via the Sia-independent pathway that is supposedly utilized in HEK293ΔSia cells. Notably, this entry route is much less efficient than Sia-dependent entry and the infection dose versus entry curves display, in contrast to Sia-dependent entry, similar slopes between strains (b = 1.03 ± 0.08; Fig. 3B).
In conclusion, infection of Sia linkage-type-specific cell lines has provided a strong correlation between affinity selectivity for 2-3Sia and 2-6Sia and entry selectivity. This correlation is stronger than the correlation between affinity and virus binding to artificial surfaces on which glycans are immobilized in fixed ways. Selectivity of infection for 2-6Sia-expressing cell lines over 2-3Sia-expressing cell lines decreased with increased infection dose. Sia-independent entry correlated positively with a decrease in affinity for 6SLN. Remarkably, the direction in which these properties evolve is changeable over time.
Discussion
IAV particles, displaying multivalent binding, are, assisted by receptor-destroying enzymes, prototypic examples of molecular walkers (31). The extreme binding avidity (KD in pM to fM range) seemingly contrasts with motility being essential for infectivity. However, dynamic HA–receptor interactions with a half-life time of 0.8 s (Fig. 1F) enable the exchange of Sia moieties between HA and NA that drives motility and determines viral fitness. Multivalency augments receptor specificity as small differences in affinity for different receptors support a receptor density-dependent phenomenon designated super selectivity (32). Whereas this may direct the initial binding location of a virus on epithelial tissues, subsequent virus motility is determined by the kinetic parameters (KD=koff/kon) of individual interactions involving a diversity of receptors displayed on the target cell. Structural analysis has provided detailed understanding of specificity of IAV–receptor interactions (17) but does not reveal details on kinetic parameters (15). Here, systematic analysis of the kinetics unveils striking features of the evolution of receptor specificity of seasonal H3N2 strains. Dual-specific affinity of 1968 pandemic H3N2 was maintained for 8 y followed by a gradual increase in human-type 2-6Sia specificity that declined again after 1995. Remarkably, this was solely due to changes in the association rate (Fig. 1E). The implications of these findings are discussed below.
Early gradual evolution toward human-type receptor specificity with a return to dual-specificity after 1995, implies that dual specificity is compatible with efficient spread of seasonal H3N2. Avian-type 2-3Sia receptors are abundantly displayed on O-linked glycans of mucins, making secreted mucins and the mucus layers decoy receptor-loaded barriers and clearance mechanisms for infection by 2-3Sia-binding viruses (33, 34). However, dual-specific binding of SY97, FU02, and PE09 is of very low affinity. Notably, while these bound 2-6Sia-(LN)3 efficiently, binding of 2-6Sia-LAMP1 required elongation of antennae on N-linked glycans by cotransfection with B3GNT2 and B4GALT1 (Fig. 2D). Likely, 2-6Sia-(LN)3 chains attached to two juxtaposed biotin-binding sites (2 nm apart) on streptavidin (SA) resemble the elongated biantennary N-linked glycans that were shown to be preferentially bound by later H3N2 isolates (7, 9, 27). Molecular dynamics simulations suggested that elongated bidentate glycans provide enhanced avidity by interacting simultaneously with two protomers of an HA trimer (27). Such a binding mode may compensate for the low affinity for 2-6Sia while the low affinity for 2-3Sia prevents binding to short, mostly single-chain, mucin-type O-linked glycans. Still, the high Sia density presented by mucin domains may result in high avidity binding to such domains and needs to be investigated in more detail.
Binding affinity only moderately correlates with virus-binding rate to receptor-coated surfaces (Fig. 2B). Individual deviations from this trend show that additional factors affect binding rate. The rate of the initial, intermolecular interaction between a virus-embedded HA and a receptor-coated surface is determined by virus concentration and the KD. However, formation of subsequent, intramolecular interactions leading to multivalent binding will meet spatial constraints. Sia moieties rigidly bind to the RBS in a conserved orientation, regardless of linkage-type (17). This restricts formation of additional interactions by accessibility, motility, and identity of nearby Sia moieties, as well as by the lateral and rotational motility of HAs in the viral envelope, which may differ between strains. Collectively these factors, which also include features of the glycan chain (e.g., length) as discussed above (Fig. 2D), will determine an apparent KD2 for a second intramolecular interaction, followed by a KD3 and so on (18).
Still, these apparent KDs will also be affected by the absolute KD determined here, and the absolute KD was also shown to correlate positively (0.84; P = 0.0012) with the receptor density at which virus-binding rate is half-maximal (SI Appendix, Table S4). In comparison to artificial receptor surfaces used in most virus binding studies, Sia density and structural diversity on the cell surface is high (35). This will raise the chance of finding suitably oriented receptors for intramolecular interaction. Heteromultivalent binding effects, by which low-affinity receptors decrease the threshold density at which receptors of higher affinity support binding (18), are supported by receptors of various affinities (SI Appendix, Table S4) indicating that a large spectrum of receptors can be used for virus to cell surface binding. In contrast to the mediocre [2-6Sia-(LN)3]-to-moderate (2-6Sia-LAMP1) correlation of KD with virus binding to receptor-coated surfaces (Fig. 2 B and C), affinity correlates strongly with entry selectivity (Fig. 4E). Likely, cell surface–embedded glycans pose less spatial constraints on formation of intramolecular multivalent interactions due to their motility and flexibility in comparison to the immotile glycans on a fixed, density-determining grid, of artificial surfaces. Consequentially, binding rate to cells may be mostly determined by the first, KD-dependent, monovalent interaction.
Our finding of a strong correlation between affinity and entry into HEK2932-6Sia or HEK2932-3Sia cells carrying a receptor repertoire similar to the soluble glycans used for affinity measurements, contrasts with reported poor correlations between entry and binding avidity. Selectivity was maximal at infectious doses much lower than commonly used (Fig. 4C). The lack of selectivity at high-infectious dose is readily explainable as curves for entry into 2-6Sia or 2-3Sia cells converge to a maximally sustainable entry level (Fig. 3A). Still, at low-infection doses entry curves for 2-3Sia and 2-6Sia cells are convergent too (Fig. 4A), implying that the chance of infecting either 2-3Sia or 2-6Sia cells does not equally increase by doubling infection dose (Fig. 4B). Several factors can affect this apparently unequal chance distribution. For instance, Sia moieties are presented as complex receptor repertoires of variable density, on diverse subterminal glycan structures that may differ as ST3Gal4 and ST6Gal1 likely have dissimilar fine acceptor substrate specificity. Second, 2-3Sia and 2-6Sia linkage types result in a different conformation of the underlaying glycan chain, imposing different rules on formation of (hetero)multivalent interactions (15). Third, entry of multiple particles was deemed necessary (36) for starting an infection (note the low infectious titer to particle ratio, SI Appendix, Fig. S2) despite the highly selective (37) but imperfect (38) packaging of the segmented IAV genome. Importantly, high selectivity at low infectious dose implies that initial infection via respiratory droplets or aerosols, carrying few particles, requires cells exposing sufficient high-affinity receptors. Once infection is established, thousands of particles will spread from an infected cell to neighboring cells, suggesting that at this phase of infection selectivity could be lower.
Previous studies have demonstrated the ability of IAVs to enter virtually any immortalized cell line, without a strong correlation with receptor-binding avidity and specificity (39, 40). This was extended here to our genetically depleted sialyltransferase-deficient HEK293ΔSia cells. Infection efficiency of HEK293ΔSia cells was up to 250-fold different between the H3N2 strains that were tested here (Fig. 3B) and, remarkably, displayed a strong negative correlation with affinity for 6SLN (Fig. 4F). Possibly, binding to non-Sia receptors is an evolutionary compensation for a lowered affinity for Sia receptors potentially involving heteromultivalent binding. Non-Sia receptors have been identified in recent years (41, 42) but their significance for in vivo infection is still unclear.
The evolution of H3N2 viruses from dual-Sia specificity to high 2-6Sia- and back to dual-Sia specificity shows that neither high selectivity, nor high affinity, for human-type 2-6Sia receptors is necessary for efficient virus spreading. Receptor-binding plasticity is likely bolstered by a recently described heteromultivalent binding mode (18), showing that avian-type receptors of relatively low affinity contribute to the binding and entry of 2-6Sia-specific human IAVs. This enables optimal utilization of the receptor diversity on epithelial cells and may expand the options for antigenic change. This occurs at seven amino acid positions around the RBS (43) and may, as a collateral effect, alter binding properties. More than optimizing human-type receptor binding, evasion of the immune response is therefore the likely driving force of the evolution of H3N2 binding properties. Antigenic cluster-transition substitutions are accompanied by multiple accessory substitutions (43), making prediction of mutations that change binding properties tedious. Yet, strains TX77 to WU95 uniquely carry a glutamic acid at position 158 (cluster-transition substitution G158E from VI75 to TX77 followed by E158K from WU95 to SY97) that corresponds to their enhanced 2-6Sia binding selectivity. Accessory substitution K135T from WU95 to SY97 introduces an N-linked glycosylation site located in the RBS at position 133 that could cause the low affinity of SY97 and later strains.
HA and NA are generally assumed to require balanced receptor binding and cleavage activity toward a specific receptor repertoire for optimal virus motility and entry (10). Binding-affinity changes are expected to change this balance and compensatory, but as yet unidentified, changes in NA may be required. Also, antigenic evolution of NA may cause accompanying changes in the HA/NA balance. Here, NA activity changes expectedly involve a shift toward higher cleavage activity for 2-6Sia of which the analysis is technically challenging. Hydrolysis of the commonly used NA substrate 2′-(4-Methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA) does not distinguish linkage-type-specific activity and, in addition, is measured in solution, whereas NA activity on a receptor surface is largely determined by the binding properties of its HA partner in the virus particle.
In conclusion, the use of a straightforward method for determination of HA-Sia receptor affinities has enabled the systematic study of the relation between binding affinity and receptor-binding specificity and entry selectivity. Opposing the canonical view, preferential binding to human-type 2-6Sia receptors evolved slowly for human H3N2 virus strains and was not maintained over time. Systematic analysis of kinetic-binding parameters will provide a basis for a detailed understanding of the enigmatic HA/NA balance and of the requirements for heteromultivalent binding and its potential role in zoonotic transmission.
Materials and Methods
Virus Strains.
The human H3N2 recombinant influenza virus strains used in this paper were generated and described previously and represent consecutive antigenic clusters (43). They are 2:6 reassortants containing the 6 internal gene segments of strain A/Puerto Rico/8/1934(H1N1) and the HA and NA gene segments of: A/Bilthoven/16190/68 (HK68); A/Bilthoven/21793/72 (EN72); A/Bilthoven/1761/76 (VI75); A/Bilthoven/2271/76 (TX77); A/Netherlands/233/82-193N (BK79); A/Netherlands/620/89 (SI87); A/Netherlands/179/93 (BE92); A/Netherlands/178/95-145K (WU95); A/Netherlands/427/98 (SY97); A/Netherlands/213/03 (FU02). A/Perth/16/2009 (PE09) is a H3N2 vaccine strain from 2009. A/duck/Hunan/795/2002(H5N1) (HU02) was described in ref. 18. Virus strains were propagated by inoculating seed stocks into MDCK-II as described (44) and stored at −80 °C. Enterovirus strain RlucCVB3 was obtained by transfection of infectious clones pRluc-53CB3/T7 (45) and used at MOI 0.1 in infection and luciferase reporter assays (18).
Nanoparticle Tracking Analysis (NTA).
Virus particle numbers were quantified by NTA (NanoSight NS3000, Malvern Panalytical) as described (5). Particle numbers were determined as the average of five measurements. Virus particle numbers were used to normalize virus binding rate in binding assays and infection dose of virus strains for infection assays.
Expression and Purification of Biotinylated Glycoprotein Receptors.
Recombinant glycoprotein LAMP1, of which the transmembrane domain was replaced by a BAP-tag, yielding a secretable and C-terminally biotinylated N-glycoprotein (18), was expressed in a HEK293-derived knockout cell line HEK293ΔSia that lacks all Siaα2-3Gal and Siaα2-6Gal glycotopes as eight sialyltransferases (ΔST3GAL1/2/3/4/5/6 and ST6GAL1/2) were knocked out (4). To install specific sialylation patterns on HEK293ΔSia cells and the secreted and C-terminally biotinylated recombinant glycoprotein LAMP1, expression vectors encoding sialyltransferases (pCDNA-ST3Gal4 or pCDNA-ST6Gal1) (Genscript, Piscataway, NJ, USA, NM_1254757.2; NM_173216.2) and an expression vector (pCD5) encoding a BirA biotin ligase were cotransfected with an expression vector coding for the extracellular domain of lysosomal membrane protein I (LAMP 1) fused to a C-terminal biotinylation site. Transfection with ST3Gal4 yielded 2-3Sia-LAMP1 and with ST6Gal1 yielded 2-6Sia-LAMP1. Details on HEK293ΔSia cell culture, transfection procedures, and recombinant protein purification by Ni-NTA binding chromatography are described in refs. 18 and 46.
Real-Time Virus-Binding and Lectin-Binding Assays by BLI.
Real-time virus binding was studied by BLI analysis using an Octet RED384 (Fortebio). All experiments were carried out at 30 °C in Dulbecco’s PBS with calcium and magnesium (PBS+/+) (Lonza) as standard assay buffer. BLI protocols are described stepwise in detail in ref. 46. In short, SA biosensors were loaded with biotinylated receptors. These were either biotinylated synthetic glycans [2-6Sia-(LN)3, Siaα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc; 2-6Sia-(LN)2, Siaα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1 or the Siaα2-3 versions 2-3Sia-(LN)3 or 2-3Sia-(LN)2 of these] or recombinant glycoprotein LAMP1 (2-6Sia-LAMP1 or 2-3Sia-LAMP 1). Receptors were loaded to saturating levels and real-time virus association was examined for 900 s in the presence of 10 µM oseltamivir carboxylate (OC; Roche) to block NA activity. Absolute initial virus-binding rates were calculated and plotted (nm/109 virus particles). LAMP1 sialylation levels were analyzed by lectin binding assays [(Maackia amurensis lectin I (MAL I, Vector Labs) binds to α2-3Sia-Galβ1-4GlcNAc; Sambucus nigra lectin (SNA, Vector Labs) binds preferentially to α2- 6Sia-Galβ1-4GlcNAc; and Erythrina cristagalli lectin (ECA, Vector Labs) binds to terminal LacNAc (Galβ1-4GlcNAc)].
KD Determination.
For analysis of KD by equilibrium binding and dissociation experiments by BLI, virus particles were associated to a level of ~3 nm to SA sensors loaded with 2-6Sia-(LN)3 or 2-36Sia-(LN)3 receptors in the presence of 10 µM OC. After washing in PBS+/+ plus 10 µM OC sensors were dipped in a concentration range (twofold dilutions series)s of soluble glycans Siaα2-6Galβ1-4GlcNAc (6SLN), Siaα2-3Galβ1-4GlcNAc (3SLN), Siaα2-6Galβ1-4Glc (6SLac), or Siaα2-3Galβ1-4Glc (3SLac). The concentration series ranged from 0.25 mM to 100 mM depending on the virus and glycan combination (maximally 25 mM for 6SLN and 3SLN). After association for 15 s, sensors were moved to PBS+/+ plus 10 µM OC for measuring dissociation. All steps were performed at 30 °C and each association/dissociation cycle was repeated 3 times in a row. Each virus/glycan combination was tested in biological replicates. Association and dissociation curves (5 points per sec) were analyzed using GraphPad Prism 8.4.0 choosing the “association than dissociation” algorithm from the binding kinetics package. Curves from several concentrations around the KD concentration were used from different replicates. Average values and SDs were calculated and reported in Table 1 and SI Appendix, Tables S1 and S2.
Infection and Luciferase Reporter Assays.
Single-round virus infection assays of wild-type HEK293WT cells, HEK293ΔSia cells (genetically depleted of ST6Gal 1 and 2 and ST3Gal1 to 6) HEK2932-6Sia cells (HEK293ΔSia transfected with ST6Gal1) and HEK2932-3Sia cells (HEK293ΔSia transfected with ST3Gal4) were performed using a luciferase reporter system employing the Gaussia luciferase-encoding vector pHH-Gluc as described (44). HEK293WT or HEK293ΔSia cells were seeded in a 96-well plate (25,000 cells/ well) to reach 90% confluency the following day at which cells were transfected with 50 ng pHH-Gluc. For HEK2932-6Sia or HEK2932-3Sia cells 0.8 ng of, respectively, ST6Gal1 or ST3Gal4 was cotransfected using Lipofectamine 2000 (InVitrogen) according to the manufacturer’s protocol. The cell culture medium was replaced with Opti-mem (Gibco) 6 h post transfection. After 48 h, the transfected cells were infected with a twofold dilutions series of virus particles as indicated in Fig. 3. Each condition was tested in triplicate, and three biological replicates were performed. At 17 h, p.i. samples from the supernatant were assayed for luciferase activity as described (44). For normalization, infection of Sia-independent enterovirus strain RlucCVB3, which was shown to give identical infection of the different cell types (18), was taken along in triplicate for every plate. An MOI of 0.1 for RlucCVB3 was used.
Software and Statistical Analysis.
All entry and KD experiments were performed in triplicate with 2 or 3 biological replicates as indicated in the text. Second- or third-order polynomial curve fitting using GraphPad Prism 8.4.0 was performed for Figs. 1 and and2.2. Two or three biological and technical replicates of BLI binding assays were performed. Virus initial binding rates, corresponding to the slopes of the binding curves during the first 5 min, were corrected by virus particle numbers (46). Statistical significance for bar diagram plots of Fig. 4B was determined using the unpaired t test (GraphPad Prism 8.4.0). Curve fitting of the virus entry data by a power regression model to the function y = axb was performed by the data analysis package of Excel2016. Data were first natural log transformed enabling linear log-log regression (ln y = b ln x) yielding R square, significance (P) and CIs for a and b as listed in SI Appendix, Tables S5 and S6.
Acknowledgments
Support by a personal grant from the Chinese Scholarship Council to M.L. was obtained (201908350116). Support by the Lundbeck Foundation, Novo Nordisk Foundation, and the Danish National Research Foundation was obtained (DNRF107L: Y.N. and H.C.). We thank Dr. Ron Fouchier (Viroscience, Erasmus Medical Center, Rotterdam, the Netherlands) for critical reading of the manuscript and for providing virus strains. Biotinylated synthetic glycans were kindly provided by Dr. Geert-Jan Boons (Utrecht University, Utrecht, the Netherlands).
Author contributions
M.L., F.J.M.v.K., C.A.M.d.H., and E.d.V. designed research; M.L. and A.S.B. performed research; Y.N. and H.C. contributed new reagents/analytic tools; M.L., A.S.B., and E.d.V. analyzed data; Y.N. and H.C. reviewed and edited the manuscript; F.J.M.v.K. edited the paper; secured funding; C.A.M.d.H. reviewed and edited the manuscript; supervised the study; E.d.V. supervised the study; and M.L. and E.d.V. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. C.R.P. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/151719525
Article citations
Molecular Markers and Mechanisms of Influenza A Virus Cross-Species Transmission and New Host Adaptation.
Viruses, 16(6):883, 30 May 2024
Cited by: 0 articles | PMID: 38932174 | PMCID: PMC11209369
Review Free full text in Europe PMC
Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32(2):261-275.e4, 01 Feb 2024
Cited by: 1 article | PMID: 38307019
The pathogenesis of influenza in intact alveoli: virion endocytosis and its effects on the lung's air-blood barrier.
Front Immunol, 15:1328453, 26 Jan 2024
Cited by: 0 articles | PMID: 38343548 | PMCID: PMC10853445
Review Free full text in Europe PMC
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 6GalView structure
-
(1 citation)
PDBe - 3GalView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human-type sialic acid receptors contribute to avian influenza A virus binding and entry by hetero-multivalent interactions.
Nat Commun, 13(1):4054, 13 Jul 2022
Cited by: 25 articles | PMID: 35831293 | PMCID: PMC9279479
Adaptation of influenza viruses to human airway receptors.
J Biol Chem, 296:100017, 22 Nov 2020
Cited by: 51 articles | PMID: 33144323 | PMCID: PMC7948470
Review Free full text in Europe PMC
H3N2 Influenza Viruses with 12- or 16-Amino Acid Deletions in the Receptor-Binding Region of Their Hemagglutinin Protein.
mBio, 12(6):e0151221, 07 Dec 2021
Cited by: 2 articles | PMID: 34872354 | PMCID: PMC8649756
Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin.
Proc Natl Acad Sci U S A, 109(52):21474-21479, 10 Dec 2012
Cited by: 194 articles | PMID: 23236176 | PMCID: PMC3535595
Funding
Funders who supported this work.
CSC | Chinese Government Scholarship (1)
Grant ID: 201908350116
Danmarks Grundforskningsfond (1)
Grant ID: DNRF107L

 a
,
1
and
a
,
1
and 

