Abstract
Free full text

Exploring the Functional Basis of Epigenetic Aging in Relation to Body Fat Phenotypes in the Norfolk Island Cohort
Abstract
DNA methylation is an epigenetic factor that is modifiable and can change over a lifespan. While many studies have identified methylation sites (CpGs) related to aging, the relationship of these to gene function and age-related disease phenotypes remains unclear. This research explores this question by testing for the conjoint association of age-related CpGs with gene expression and the relation of these to body fat phenotypes. The study included blood-based gene transcripts and intragenic CpG methylation data from Illumina 450 K arrays in 74 healthy adults from the Norfolk Island population. First, a series of regression analyses were performed to detect associations between gene transcript level and intragenic CpGs and their conjoint relationship with age. Second, we explored how these age-related expression CpGs (eCpGs) correlated with obesity-related phenotypes, including body fat percentage, body mass index, and waist-to-hip ratio. We identified 35 age-related eCpGs associated with age. Of these, ten eCpGs were associated with at least one body fat phenotype. Collagen Type XI Alpha 2 Chain (COL11A2), Complement C1s (C1s), and four and a half LIM domains 2 (FHL2) genes were among the most significant genes with multiple eCpGs associated with both age and multiple body fat phenotypes. The COL11A2 gene contributes to the correct assembly of the extracellular matrix in maintaining the healthy structural arrangement of various components, with the C1s gene part of complement systems functioning in inflammation. Moreover, FHL2 expression was upregulated under hypermethylation in both blood and adipose tissue with aging. These results suggest new targets for future studies and require further validation to confirm the specific function of these genes on body fat regulation.
1. Introduction
Biological aging is a complex process that involves changes in many molecular systems, including cellular senescence, genomic instability, telomere attrition, epigenetic alteration, and loss of proteostasis [1]. In humans, these changes lead to an increase in the risk of many complex diseases such as cancer, cardiovascular disease, and Alzheimer’s disease [2,3,4]. As aging is a complex process with many biological systems changing over time, understanding the fundamental molecular mechanisms involved may lead to a better understanding of how to enhance the quality of life into old age.
The rate of biological aging varies substantially among humans, and the variation is due to the complex interplay of genetic profiles, environmental exposures, and epigenetic modifications. DNA methylation is a particular epigenetic modification involving the addition of a methyl group to cytosine-phosphate-guanine (CpG) sites. The addition or loss of methylation is to a large extent developmentally regulated, but can also be influenced by environmental exposures [5]. Moreover, methylation profiles at specific loci have been shown to change over time as a result of epigenetic drift [6]. Changes in DNA methylation levels can modulate gene expression levels due to the disruption of transcription factor binding or the recruitment of repressors [7,8]. While it is widely accepted that there is a negative correlation between methylation at the transcription start site and gene expression [9,10], studies have also shown a positive correlation between methylation in the gene body and an influence on expression [11]. DNA methylation could also control gene expression from a long distance away from promoters through its effect on distal regulatory elements such as enhancers. Enhancers can control gene expression by the formation of chromatin loops [12]. The methylation patterns on these regions were generally unmethylated [13]. Studies showed that the methylation patterns on enhancers could be used to classify different cells such as normal versus tumour cells [14].
Many studies have been performed to identify associations between DNA methylation and chronological aging. The Epigenome-wide Association Studies (EWAS) Atlas summarizes all EWASs conducted on various phenotypes [15]. Among them, the EWAS Atlas contains a list of more than 20,000 methylation CpGs annotated to 10,837 genes associated with aging in DNA collected from blood samples [15]. Age-related CpGs can also be considered a biomarker of aging from which biological age can be estimated (i.e., epigenetic clocks). By calculating epigenetic age acceleration, these clocks have been shown to accurately predict human lifespan and a large range of morbidities [3,4].
The mechanism by which these age-related CpGs affect the expression of the linked genes is not fully understood. To address this issue, Peters et al. conducted a meta-analysis of human peripheral blood in 14,983 individuals of European ancestry and identified 1497 genes differentially expressed with chronological age [16]. Of these, 1248 genes showed a potential enrichment of methylation sites (1 to 154 CpGs per gene) associated with chronological age and gene expression. These genes were enriched in various pathway clusters, including DNA replication, elongation, and mismatch repair; fatty acid metabolism; peroxisome activity; RNA metabolism; ribosome biogenesis; and purine metabolism. The study showed the link between the association of CpG methylations and gene expression (or eCpGs) with the change in many aging pathways.
Changes in body composition in terms of lipid metabolism and storage are essential factors in age-related diseases [17,18]. Excess body fat (especially visceral) is associated with decreased life expectancy [19]. Trim et al. showed similarities in the immunological profiles of aging and excess body fat [20], suggesting overlapping pathways between immunosenescence and obesity. Later, in 2019, a review paper discussed the correlation of aging and obesity in terms of nine critical hallmarks of the aging process [21], which also indicated a strong link between changes in body composition and aging. A recent meta-analysis using methylation and transcription data showed a younger epigenetic age with higher fitness level [22]. Therefore, understanding the pathways linking body fat composition and aging might provide better insight into the mechanisms that regulate the maintenance of good health.
Norfolk Island (NI) has been part of a long-term health study over the past 20 years, with a well-documented population history and known ancestry [23]. The island was founded by ten Bounty Mutineers and six Tahitian wives [24]. Since then, immigration has been limited due to its geographic location and restrictions. Due to the small number of founders and the isolated environment, the island provides less variation in environmental factors and, therefore, reduced genomic diversity [24]. Since aging involves complex changes affected by genetics together with the impact of the environment, the use of an isolated population could help to minimise the variation within the population.
This exploratory study aims to identify interactions between genes affected by age-related DNA methylation and its impact on gene expression and obesity-related phenotypes using the isolated NI population cohort.
2. Materials and Methods
The summary of the cohort and workflow is displayed in Figure 1.
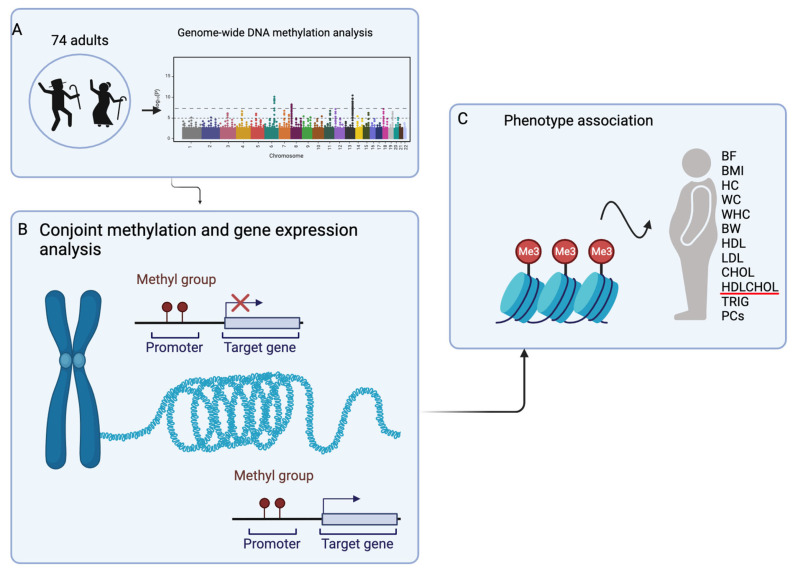
The summary of the workflow. (A) Genome-wide DNA methylation analysis with age. (B) The conjoint analysis of identified methylation and gene expression pairs. (C) The association of identified methylation CpGs in B to phenotypes. BF: body fat, P percentage, BMI: body mass index, HC: hip circumference, WC: waist circumference, WHR: waist-to-hip ratio, BW: body weight, HDL: high-density lipoprotein, LDL: low-density lipoprotein, CHOL: cholesterol, HDLCHOL: HDL to CHOL ratio, TRIGs: triglycerides, and PCs: principal components. Red line = example of blood lipid phenotype.
2.1. Sample and Phenotypic Characteristics
Through the Norfolk Island Health Study, participants volunteered to participate and completed the consent forms approved initially by the Griffith University Human Research Ethics Committee (HREC) and subsequently by the QUT HREC (no. 1600000464). Quantitative phenotypic data were collected at the same time as blood donation, including multiple obesity-related phenotypes. Body fat phenotypes included body fat percentage (BF, %, measured by infra-red refractance), body weight (BW, kg), body mass index (BMI, calculated as body weight divided by height squared in m2), waist circumference (cm), hip circumference (cm) and waist-to-hip ratio. Moreover, low- and high-density lipoprotein (LDL and HDL), total cholesterol (CHOL), triglyceride (TG), and the ratio of HDL to CHOL (HDLCHOL) were included in the analysis. The HDL, LDL, CHOL, and TG levels were measured in millimoles per litre (mmol/L). The details of these phenotypes can be found in a previously published study [25].
2.2. Principal Component Analysis
Missing values presented in the phenotypic variables were imputed by applying a parametric bootstrapping method in the missMDA package (version 1.14) [26]. Then, Principal Component Analysis (PCA) was performed using FactoMineR package (version 1.41) [27] to capture the correlation between body fat and lipid variables separately. This resulted in adding four principal components for body fat and lipid phenotypes, named BF.PC1, BF.PC2, LP.PC1, and LP.PC2, with the eigenvalue greater than 1. Each described measurement and PCs were treated as input phenotypes in the latter analysis.
2.3. Genomic Data
The extraction and purification of blood samples and DNA methylation analysis via Illumina methylation 450 K arrays have previously been described in Benton MC et al. [28]. DNA methylation raw data were obtained, and the methylation matrix quality control was performed in R through the ChAMP package (release 3.17) via the “champ.filter” function without removing CpGs annotated to contain SNPs [29]. Then, 433,525 CpGs remained for further analysis, and CpGs were annotated using the CpG feature profiles provided in the ChAMP package.
Gene expressions for corresponding samples were obtained from Illumina HumanHT-12 v.4 Expression BeadChip Kit arrays as previously described in Benton MC et al. [30]. There were 23,323 transcripts included after the log2 transformation and quantile normalisation. Genes were annotated using the illuminaHumanv4.db package (version 1.26.0).
2.4. Statistical Analysis
2.4.1. Identification of Methylation Sites Associated with Age
A simple linear regression was performed with age as the outcome variable and CpG methylation as the independent variable to identify methylation sites associated with age. Significant CpGs were filtered based on(rle the genome-wide threshold of 2.4 × 10−7 [31].
2.4.2. Conjoint Analysis of Methylation and Gene Expression Associated with Age
For genes with CpGs significantly associated with age, pairs of gene expression and methylation CpGs were used in the conjoint analysis. A pair of expression and methylation was selected when the methylation CpG mapped to the same gene as a gene expression CpG (i.e., was in cis). Therefore, each CpG for gene expression may be paired with multiple methylation CpGs. Using these, we tested for the interaction between methylation on gene expression to indicate age-related CpGs that act via gene expression. For this model, age was included as the outcome, while methylation, transcript, and its interaction term (methylation × transcript) were the independent variables. Significant pairs of methylation and transcript were selected if the p-value of CpGs, transcript, and interaction term were all less than 0.05. If there was more than one pair for the same gene, the one with the largest R2 was selected as the representative (index CpG).
2.4.3. Association of Functional Age-Related CpGs on Body Fat Phenotypes
A series of linear regressions was performed where significant CpGs from the above conjoint analysis were considered independent variables and phenotypes as the dependent variables. This analysis adjusted the phenotype according to age, sex, and kinship. Kinship was the score of relatedness between individuals ranging from 0 to 1. The kinship score was included due to the potential confounder of family relationships between members in the cohort [32]. Sex was included as a covariate, given the significant difference in most phenotypes by sex, as seen in Table 1. The association was considered significant if the p-value was less than 0.05.
Table 1
Summary of body fat and lipid phenotypes in NI cohort.
| Female (N = 24) | Male (N = 50) | Total (N = 74) | p | |
|---|---|---|---|---|
| Age | 51 (12.8) | 52.3 (12.1) | 51.9 (12.2) | 0.6 |
| Body Fat Percentage (%) | 34.1 (5.3) | 25.4 (7.8) | 28.2 (8.1) | <0.001 * |
| Body Mass Index (kg/m2) | 23.3 (2.3) | 27.9 (4.3) | 26.4 (4.3) | <0.001 * |
| Hip Circumference (cm) | 98.2 (6.1) | 105.9 (8.1) | 103.4 (8.3) | <0.001 * |
| Waist Circumference (cm) | 81.3 (9.4) | 99.1 (12.8) | 93.3 (14.4) | <0.001 * |
| Waist-to-Hip Ratio | 0.8 (0.06) | 0.9 (0.1) | 0.9 (0.1) | <0.001 * |
| Body Weight (kg) | 63.3 (7.67) | 89.36 (14.6) | 81 (17.7) | <0.001 * |
| Total Cholesterol (mmol/L) | 5.6 (0.85) | 5.71 (1.01) | 5.7 (0.96) | 0.586 |
| HDL-to-CHOL ratio (HDLCHOL) | 3.4 (0.74) | 4.53 (1.57) | 4.15 (1.46) | <0.001 * |
| Low Density Lipoprotein (mmol/L) | 3.4 (0.77) | 3.68 (0.98) | 3.6 (0.9) | 0.24 |
| High Density Lipoprotein (mmol/L) | 1.7 (0.33) | 1.37 (0.4) | 1.48 (0.41) | <0.001 * |
| Triglycerides (mmol/L) | 1.01 (0.35) | 1.68 (1.7) | 1.47 (1.45) | 0.063 |
Data were summarized as mean (standard deviation). * statistically significant.
Due to the small sample size, the method used in conjoint analysis and functional age-related CpGs did not account for multiple testing to capture the preliminary result. All statistical analyses were performed in R (version 4.1.1) [33] and RStudio (version 1.4.1717) [34].
3. Results
3.1. Cohort Characteristics
The descriptive statistics of our cohort are summarised in Table 1. The cohort consisted of 24 females and 50 males. The mean age was 51.9 years, ranging from 31 to 83 years. The phenotypes included six anthropomorphic measurements for body fat compositions, summarised in Table 1. All of these exhibited significant differences between male and female subjects. Of five measured lipid phenotypes, only high-density lipoprotein (HDL) and HDL to cholesterol ratio (HDLCHOL) showed significant differences between males and females.
3.2. Epigenome-Wide Association Study of Age in NI Cohort
After the initial quality control of methylation data from the 74 blood samples, 433,525 CpGs remained for analysis. The association testing of CpG methylation with age was performed by applying a per-CpG linear regression. This identified 226 CpGs across 183 genes with p-values less than the epigenome-wide threshold (Figure 2). Of these 226 CpGs, more than half were located in gene regulatory regions (Figure 3). The top 10 CpGs ranked by R2 are summarized in Table 2. All CpGs, ranked by significance level and chromosome position, are shown in Supplementary Table S1.
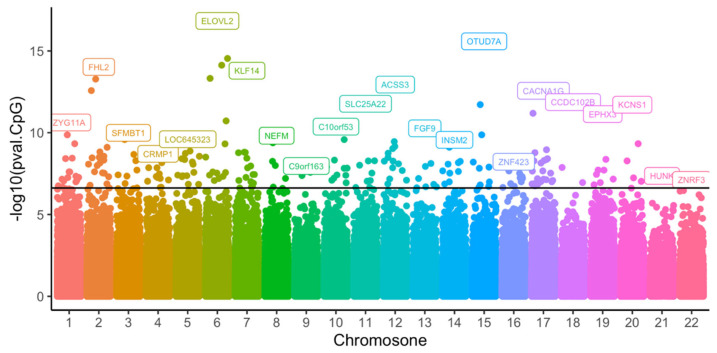
Epigenome-wide association of age in Norfolk Island cohort identified 226 CpGs annotated to 183 genes. Gene names shown in boxes indicate the most significant genes in each chromosome. The dark line indicates the significant threshold (p < 2.4 × 10−7).
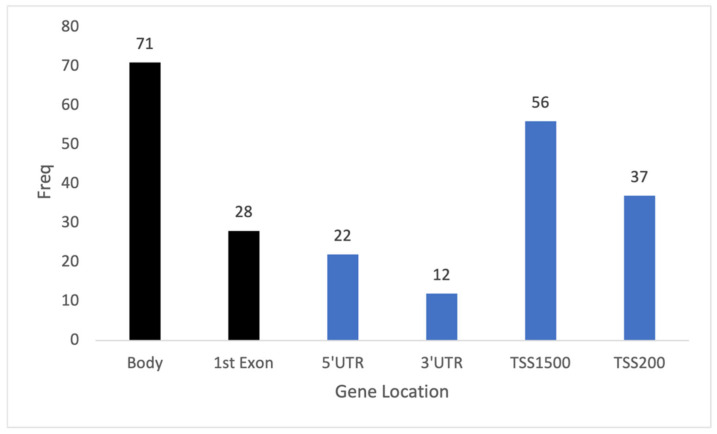
The distribution of 226 significant methylation sites within genes associated with age. The x-axis showed the list of gene locations while the y-axis showed the frequency of significant genes identified based on their positions. Black bars are non-regulatory regions. Blue bars are regulatory regions.
Table 2
The summary statistics of the top 10 significant CpGs associated with age (p < 2.4 × 10−7).
| CpG | Beta | p | R2 | CHR | MAPINFO | Gene | Feature |
|---|---|---|---|---|---|---|---|
| cg16867657 | 0.8 | 1.4 × 10−17 | 0.64 | 6 | 11044877 | ELOVL2 | TSS1500 |
| cg04875128 | 0.78 | 2.6 × 10−16 | 0.61 | 15 | 31775895 | OTUD7A | Body |
| cg22736354 | 0.76 | 2.9 × 10−15 | 0.58 | 6 | 18122719 | NHLRC1 | 1stExon |
| cg22454769 | 0.75 | 1.1 × 10−14 | 0.57 | 2 | 106015767 | FHL2 | TSS200 |
| cg24079702 | 0.75 | 9.5 × 10−15 | 0.57 | 2 | 106015771 | FHL2 | TSS200 |
| cg24724428 | 0.76 | 7.4 × 10−15 | 0.57 | 6 | 11044888 | ELOVL2 | TSS1500 |
| cg14361627 | 0.75 | 1.6 × 10−14 | 0.56 | 7 | 130419116 | KLF14 | TSS1500 |
| cg23606718 | 0.74 | 5.3 × 10−14 | 0.55 | 2 | 131513927 | FAM123C | 5′UTR |
| cg21572722 | 0.74 | 4.7 × 10−14 | 0.55 | 6 | 11044894 | ELOVL2 | TSS1500 |
| cg11649376 | −0.73 | 1.1 × 10−13 | 0.54 | 12 | 81473234 | ACSS3 | Body |
All of these 10 genes were associated with age in previous studies. Beta: standardised regression coefficients.
The EWAS atlas was built to summarise findings from all EWAS studies and includes many studies aimed at age as the dependent variable. Filtering for age-associated CpGs in blood samples returned a list of 21,082 CpGs. Compared to the public gene list found in the EWAS atlas, there was an overlap of 164 CpGs annotated to 136 genes in the NI cohort.
3.3. Identification of Age-Related Expression CpGs
The aim of this study was to explore the functional mechanisms of age-related CpGs by linking these with gene expression levels within the same study subjects. We tested for age-related expression CpGs (eCpGs) by performing statistical interaction modelling. This modelling structure was used to identify CpGs that are associated with age in a manner dependent on gene expression (i.e., an interaction effect). Intersecting the measured gene sets derived from the methylation and expression arrays showed that 77 of the 183 age-related genes identified had data for both DNA methylation and gene expression. These CpGs and transcripts were included in the statistical models. This analysis resulted in the identification of 35 genes that showed evidence of an interaction effect between methylation on gene expression (i.e., eCpGs) in relation to aging (Table 3). Supplementary Table S2 displays the complete summaries of regression results, highlighting the highest R2 within each gene, as shown in the index column.
Table 3
Interactive effect of methylation and gene expression on relation to age.
| CpG | Transcript | CpG | Transcript | CpG × Transcript | R2 | CHR | Position | Gene | Feature | R a | Known b | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | p | Beta | p | Beta | p | |||||||||
| cg18651026 | ILMN_1748166 | 5.35 | 4.30 × 10−2 | 3.16 | 2.90 × 10−2 | −6.87 | 2.50 × 10−2 | 0.4 | 6 | 33140660 | COL11A2 | Body | 0.04 | 1 |
| cg00743094 | ILMN_1663538 | −3.09 | 4.30 × 10−2 | −1.21 | 7.30 × 10−3 | 3.9 | 1.70 × 10−2 | 0.39 | 13 | 100547968 | CLYBL | 3′UTR | 0.09 | 0 |
| cg11872672 | ILMN_1728844 | −7.36 | 2.60 × 10−2 | −5.7 | 3.70 × 10−2 | 7.75 | 3.90 × 10−2 | 0.36 | 7 | 157514730 | PTPRN2 | Body | −0.23 | 0 |
| cg13096208 | ILMN_1855910 | 9.05 | 4.60 × 10−3 | 1.49 | 3.10 × 10−3 | −8.63 | 6.40 × 10−3 | 0.24 | 18 | 55019843 | ST8SIA3 | 1stExon | −0.11 | 0 |
| cg05638739 | ILMN_1690465 | −10.44 | 4.40× 10−5 | −12.94 | 5.40× 10−5 | 18 | 5.30× 10−5 | 0.22 | 16 | 89440324 | ANKRD11 | 5′UTR | 0.19 | 0 |
| cg23095192 | ILMN_1820767 | −8.89 | 1.90 × 10−2 | −3.94 | 3.20 × 10−2 | 8.89 | 2.20 × 10−2 | 0.2 | 2 | 145271307 | ZEB2 | Body | −0.19 | 0 |
| cg20332195 | ILMN_1682449 | −10.72 | 9.20 × 10−3 | −2.15 | 9.00 × 10−3 | 11.19 | 7.30 × 10−3 | 0.2 | 4 | 10459929 | ZNF518B | TSS1500 | −0.04 | 0 |
| cg06799422 | ILMN_2372379 | 7.08 | 1.70 × 10−2 | 1.54 | 1.70 × 10−2 | −7 | 2.30 × 10−2 | 0.19 | 15 | 41952235 | MGA | TSS1500 | 0.07 | 1 |
| cg22088743 | ILMN_2133675 | 9.56 | 4.70 × 10−3 | 16.9 | 3.50 × 10−3 | −18.97 | 3.70 × 10−3 | 0.19 | 17 | 78183317 | SGSH | 3′UTR | −0.06 | 0 |
| cg10666081 | ILMN_2355831 | −6.92 | 8.40 × 10−3 | −12.79 | 1.20 × 10−2 | 14.17 | 1.10 × 10−2 | 0.17 | 2 | 105985002 | FHL2 | Body | −0.05 | 0 |
| cg17761990 | ILMN_2391750 | 8.32 | 1.40 × 10−3 | 14.69 | 1.00 × 10−3 | −17.13 | 1.10 × 10−3 | 0.17 | 3 | 53042940 | SFMBT1 | 5′UTR | 0.03 | 0 |
| cg07198402 | ILMN_1749667 | 8.69 | 2.40 × 10−2 | 10.48 | 2.00 × 10−2 | −14.3 | 2.00 × 10−2 | 0.17 | 1 | 228395145 | OBSCN | TSS1500 | 0.07 | 0 |
| cg14082919 | ILMN_2352295 | 11.16 | 8.20 × 10−3 | 14.14 | 7.30 × 10−3 | −16.69 | 6.80 × 10−3 | 0.17 | 11 | 129814820 | PRDM10 | Body | −0.17 | 0 |
| cg18443378 | ILMN_1746552 | −5.65 | 4.20 × 10−2 | −1.52 | 3.10 × 10−2 | 6.09 | 3.20 × 10−2 | 0.16 | 4 | 176986950 | WDR17 | TSS200 | −0.04 | 0 |
| cg04662983 | ILMN_1708508 | −9.33 | 6.80 × 10−3 | −1.88 | 3.20 × 10−3 | 9.43 | 5.80 × 10−3 | 0.16 | 17 | 56834321 | PPM1E | Body | −0.14 | 0 |
| cg20671534 | ILMN_1658576 | −6.04 | 6.50 × 10−3 | −2.56 | 1.20 × 10−2 | 6.65 | 8.80 × 10−3 | 0.15 | 2 | 220174629 | PTPRN | TSS1500 | 0.12 | 0 |
| cg13053396 | ILMN_1781626 | −10.19 | 4.50 × 10−3 | −8.53 | 5.10 × 10−3 | 12.23 | 5.40 × 10−3 | 0.15 | 12 | 7168545 | C1S | 5′UTR | −0.13 | 0 |
| cg16765387 | ILMN_1719975 | −5.3 | 7.10 × 10−4 | −3.14 | 8.30 × 10−4 | 5.9 | 7.20 × 10−4 | 0.15 | 12 | 54411245 | HOXC4 | 5′UTR | −0.09 | 0 |
| cg19929852 | ILMN_1774948 | 9.08 | 1.00 × 10−2 | 13.15 | 8.60 × 10−3 | −15.21 | 9.60 × 10−3 | 0.14 | 4 | 15068643 | CPEB2 | 3′UTR | −0.08 | 0 |
| cg02764478 | ILMN_1696279 | 6.66 | 3.70 × 10−2 | 1.85 | 4.90 × 10−2 | −6.46 | 4.50 × 10−2 | 0.14 | 6 | 100904316 | SIM1 | Body | −0.1 | 1 |
| cg19262958 | ILMN_1687958 | 4.49 | 4.80 × 10−2 | 8.21 | 4.20 × 10−2 | −9.57 | 3.80 × 10−2 | 0.14 | 11 | 792861 | SLC25A22 | Body | −0.02 | 0 |
| cg26365090 | ILMN_2082209 | −10.07 | 1.20 × 10−2 | −0.55 | 2.70 × 10−3 | 10.11 | 1.30 × 10−2 | 0.13 | 20 | 42574362 | TOX2 | 5′UTR | 0.21 | 0 |
| cg13539203 | ILMN_1768483 | 9.55 | 7.80 × 10−3 | 10.02 | 7.80 × 10−3 | −14.08 | 7.10 × 10−3 | 0.12 | 2 | 26950545 | KCNK3 | Body | 0.01 | 0 |
| cg08858926 | ILMN_1808587 | −7.5 | 8.90 × 10−3 | −3.74 | 1.20 × 10−2 | 7.97 | 1.00 × 10−2 | 0.12 | 16 | 72918832 | ZFHX3 | Body | −0.09 | 0 |
| cg22743761 | ILMN_1739366 | −5.7 | 3.20 × 10−2 | −2.22 | 2.70 × 10−2 | 6.85 | 2.60 × 10−2 | 0.11 | 2 | 162273648 | TBR1 | Body | 0.26 | 0 |
| cg17949162 | ILMN_1796855 | 13.11 | 1.60 × 10−2 | 1.51 | 9.10 × 10−3 | −13.17 | 1.70 × 10−2 | 0.11 | 10 | 121355153 | TIAL1 | Body | 0.03 | 0 |
| cg04256697 | ILMN_1684440 | 5.15 | 1.30 × 10−2 | 3.05 | 8.50 × 10−3 | −6.22 | 1.20 × 10−2 | 0.11 | 12 | 120688557 | PXN | Body | 0.1 | 0 |
| cg10912240 | ILMN_2149946 | −5.43 | 3.20 × 10−2 | −1.76 | 2.40 × 10−2 | 5.99 | 2.60 × 10−2 | 0.11 | 14 | 29235907 | FOXG1 | TSS1500 | 0.06 | 1 |
| cg00866814 | ILMN_1668194 | −5.78 | 7.00 × 10−3 | −2.98 | 9.10 × 10−3 | 6.46 | 8.40 × 10−3 | 0.11 | 19 | 49017364 | LMTK3 | TSS1500 | 0.03 | 0 |
| cg09628707 | ILMN_1761903 | 7.53 | 8.70 × 10−3 | 3.09 | 9.00 × 10−3 | −7.86 | 9.70 × 10−3 | 0.11 | 20 | 43729410 | KCNS1 | 5′UTR | −0.05 | 0 |
| cg17370322 | ILMN_1788457 | 7.85 | 1.40 × 10−2 | 0.88 | 7.80 × 10−3 | −7.95 | 1.50 × 10−2 | 0.1 | 13 | 95953482 | ABCC4 | Body | 0.16 | 0 |
| cg03578473 | ILMN_1669425 | −9.72 | 1.30 × 10−2 | −1.15 | 1.30 × 10−2 | 9.9 | 1.20 × 10−2 | 0.09 | 2 | 182546504 | NEUROD1 | TSS1500 | 0.05 | 0 |
| cg22509041 | ILMN_2119692 | 5.42 | 4.10 × 10−2 | 1.18 | 2.40 × 10−2 | −5.6 | 4.30 × 10−2 | 0.09 | 12 | 53574524 | CSAD | TSS200 | 0.15 | 0 |
| cg11718501 | ILMN_1666310 | 5.18 | 4.20 × 10−2 | 1.13 | 2.30 × 10−2 | −5.09 | 4.10 × 10−2 | 0.08 | 11 | 122850972 | BSX | Body | −0.21 | 0 |
| cg08198377 | ILMN_1691181 | −3.6 | 2.70 × 10−2 | −1.84 | 3.20 × 10−2 | 3.62 | 2.80 × 10−2 | 0.07 | 14 | 51707975 | TMX1 | Body | −0.24 | 0 |
a: Correlation value between methylation and gene expression. b: Known age-related CpG (no = 0; yes = 1). According to largest effect size (R2), the Collagen Type XI Alpha 2 Chain (COL11A2) gene located on the gene body of chromosome 6 contained a functional eCpG associated with age. The following four most significant genes were Citramalyl-CoA Lyase (CLYBL), Protein Tyrosine Phosphatase Receptor Type N2 (PTPRN2), ST8 Alpha-N-Acetyl-Neuraminide Alpha-2,8-Sialyltransferase 3 (ST8SIA3), and Ankyrin Repeat Domain Containing 11 (ANKRD11), located on chromosomes 13, 7, 18, and 16, respectively. Except for ST8SIA3, which had a sole CpG associated with age, the other four genes had more than 5 CpGs associated with age through the interaction with gene expression, forming functional differentially methylated regions (DMRs) related to age.
To further investigate whether the 35 genes identified were enriched in any specific biological processes, they were submitted to the ToppGene webpage [35]. Results of this analysis indicted two significant Gene Ontology hits involving molecular function on DNA-binding transcription factor activity, namely GO:0000981 (pbonferroni = 0.032, 10 genes) and GO:0003700 (pbonferroni = 0.044, 10 genes).
3.4. Age-Related eCpGs and Body Fat Phenotypes
The identified 35 age-related eCpGs were followed up to explore associations in relation to obesity-related phenotypes. To do this, a series of linear regressions was performed and adjusted by sex, age, and kinship. Table 4 summarizes the significant CpGs associated with phenotypes using the CpGs lists highlighted in Table 3. CpGs with a p-value less than 0.05 were considered significant. As a result, ten genes contained CpGs for which methylation levels were associated with one or more phenotypes, with R2 ranging from 0.2 to 0.53. Specifically, there were four genes with methylation located on the gene body and six genes with methylated CpGs found in surrounding regulatory regions. The top four genes with the highest R2 were Cytoplasmic Polyadenylation Element Binding Protein 2 (CPEB2), TIA1 cytotoxic granule associated RNA binding protein like 1 (TIAL1), Complement C1s (C1S), and ANKRD11. Among these, cg05638739, located on 5′UTR of the ANKRD11, was associated with nine out of fifteen tested phenotypes. Most of the associations between cg05638739 and phenotypes were positive, except for the negative associations with HDL.
Table 4
Age-related eCpGs associated with body fat phenotypes.
| Phenotype | CpG | CHR | MAPINFO | Gene | Feature | Beta | p | R2 |
|---|---|---|---|---|---|---|---|---|
| BF | cg10666081 | 2 | 105985002 | FHL2 | Body | 0.25 | 3.5 × 10−2 | 0.31 |
| HC | cg20671534 | 2 | 220174629 | PTPRN | TSS1500 | 0.25 | 2.4 × 10−2 | 0.29 |
| BF | cg19929852 | 4 | 15068643 | CPEB2 | 3′UTR | −0.3 | 3.6 × 10−3 | 0.35 |
| BMI | −0.27 | 9.9 × 10−3 | 0.35 | |||||
| BW | −0.21 | 1.5 × 10−2 | 0.53 | |||||
| HC | −0.24 | 2.6 × 10−2 | 0.29 | |||||
| BF.PC1 | −0.23 | 1.8 × 10−2 | 0.41 | |||||
| BF.PC2 | −0.26 | 2.7 × 10−3 | 0.56 | |||||
| WC | cg18651026 | 6 | 33140660 | COL11A2 | Body | 0.28 | 1.9 × 10−2 | 0.44 |
| WHR | 0.28 | 1.3 × 10−2 | 0.50 | |||||
| BF.PC1 | 0.26 | 3.5 × 10−2 | 0.40 | |||||
| BF | cg17949162 | 10 | 121355153 | TIAL1 | Body | 0.41 | 6.1 × 10−4 | 0.38 |
| BMI | 0.45 | 1.0 × 10−4 | 0.43 | |||||
| BW | 0.29 | 3.9 × 10−3 | 0.55 | |||||
| HC | 0.32 | 9.2 × 10−3 | 0.30 | |||||
| WC | 0.39 | 2.7 × 10−4 | 0.50 | |||||
| WHR | 0.35 | 6.8 × 10−4 | 0.54 | |||||
| BF.PC1 | 0.41 | 2.1 × 10−4 | 0.47 | |||||
| BF.PC2 | 0.28 | 5.1 × 10−3 | 0.55 | |||||
| BF | cg11718501 | 11 | 122850972 | BSX | Body | −0.29 | 2.3 × 10−2 | 0.32 |
| WHR | −0.23 | 4.2 × 10−2 | 0.48 | |||||
| BF.PC2 | −0.21 | 4.9 × 10−2 | 0.53 | |||||
| BMI | cg13053396 | 12 | 7168545 | C1S | 5′UTR | 0.24 | 2.1 × 10−2 | 0.34 |
| BW | 0.24 | 6.3 × 10−3 | 0.54 | |||||
| HC | 0.25 | 2.2 × 10−2 | 0.29 | |||||
| WC | 0.22 | 2.2 × 10−2 | 0.44 | |||||
| BF.PC1 | 0.25 | 1.3 × 10−2 | 0.41 | |||||
| BF | cg05638739 | 16 | 89440324 | ANKRD11 | 5′UTR | 0.27 | 1.3 × 10−2 | 0.33 |
| BMI | 0.34 | 1.7 × 10−3 | 0.38 | |||||
| BW | 0.21 | 2.4 × 10−2 | 0.52 | |||||
| HC | 0.25 | 3.0 × 10−2 | 0.28 | |||||
| WC | 0.24 | 1.8 × 10−2 | 0.45 | |||||
| HDL | −0.23 | 4.9 × 10−2 | 0.20 | |||||
| CHOLHDL | 0.24 | 4.6 × 10−2 | 0.22 | |||||
| BF.PC1 | 0.27 | 7.7 × 10−3 | 0.42 | |||||
| BF.PC2 | 0.21 | 2.2 × 10−2 | 0.53 | |||||
| BMI | cg26365090 | 20 | 42574362 | TOX2 | 5′UTR | −0.21 | 4.1 × 10−2 | 0.33 |
| BW | cg09628707 | 20 | 43729410 | KCNS1 | 5′UTR | 0.18 | 3.9 × 10−2 | 0.52 |
3.5. Exploring Causal Links of eCpGs with Adiposity
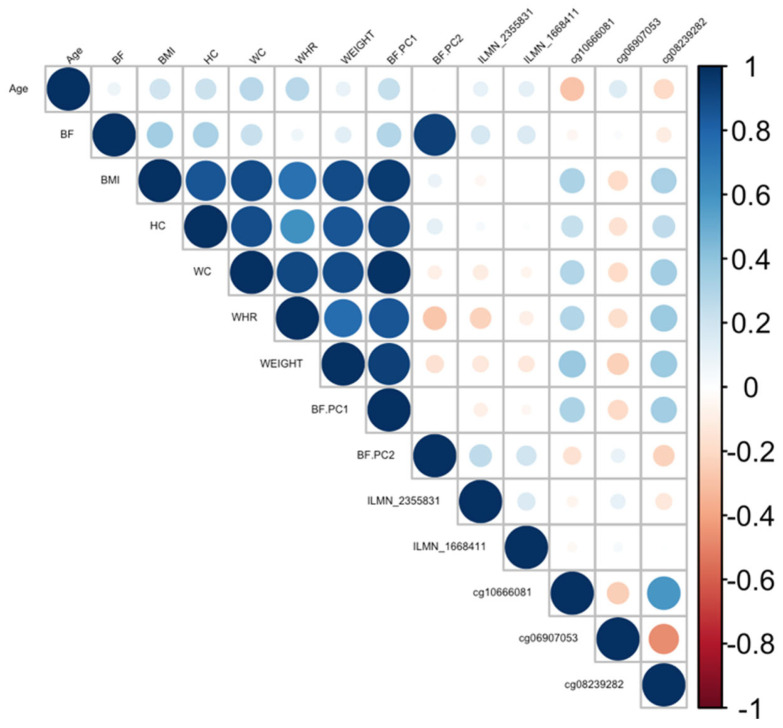
Ranking genes by R2 may only be partially useful, as some genes will be indirectly associated with body fat phenotypes due to age association and not functional causality. To best interpret the functional (causal) importance of the top 10 obesity-related genes, a critical and systematic method was required. Notably, our examined transcripts were derived from blood cells, not adipose tissue. Thus, we ideally required a link to adipose tissue for the genes to be functionally causal. To this end, a literature-based assessment of the functional role of age-related CpGs in adipose tissue and obesity was performed using the terms “gene AND obesity” and “gene AND adipose” for all 10 genes. Priority was then assigned to genes based on whether any published studies showed evidence of functional involvement (Supplementary Table S3). This assessment showed that only three out of the 10 genes provided good functional evidence for involvement in adiposity. Ranked by max R2, these were C1S, COL11A2, Four and FHL2. Specifically, these genes are epigenetically and transcriptionally variable in adipose tissue among subjects with obesity and/or metabolic syndrome [36,37,38,39]. To visually assess the DNA methylation patterns in relation to gene expression, body fat, and age, a correlation plot between all CpGs, transcripts, body fat phenotypes, and age was generated for each gene (Figure 4, Figure 5 and Figure 6).
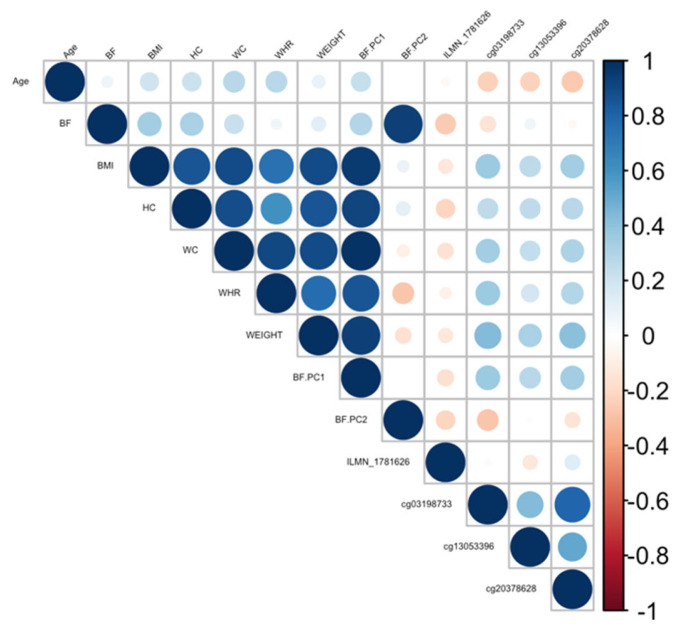
Correlation plot between all CpGs, transcripts, body fat phenotypes, and age generated for the C1S gene. BF: body fat percentage, BMI: body mass index, HC: hip circumference, WC: waist circumference, WHR: waist-to-hip ratio, Weight: body weight, BF.PC1: principal component 1, BF.PC2: principal component 2.
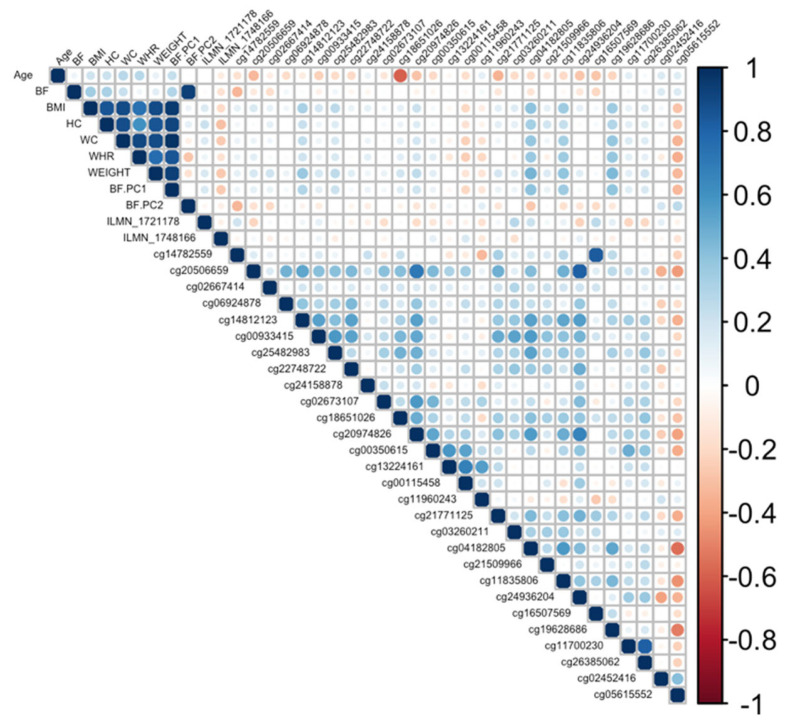
Correlation plot between all CpGs, transcripts, body fat phenotypes, and age generated for the COL11A2 gene. BF: body fat percentage, BMI: body mass index, HC: hip circumference, WC: waist circumference, WHR: waist-to-hip ratio, Weight: body weight, BF.PC1: principal component 1, BF.PC2: principal component 2.
Correlation plot between all CpGs, transcripts, body fat phenotypes and age generated for the FHL2 gene. BF: body fat percentage, BMI: body mass index, HC: hip circumference, WC: waist circumference, WHR: waist-to-hip ratio, Weight: body weight, BF.PC1: principal component 1, BF.PC2: principal component 2.
4. Discussion
Here, we investigated functional epigenetic sites associated with age- and obesity-related phenotypes in the NI cohort. We identified 226 significant CpG CpGs associated with age annotated to 183 genes; 164 of the CpG CpGs overlapping with the Aging EWAS Atlas at 77 of the 183 genes had both DNA methylation and gene expression profiles, from which 35 genes showed a mediation effect between methylation and gene expression.
Of 35 indexed CpGs highlighted in Table 3, only two CpGs were presented in the 226 significant CpGs to be associated with age, indicating that these CpGs directly affected gene expression on the same gene in which the CpGs were located. Interestingly, the remaining CpGs only showed an association with age on the same gene after the addition of gene expressions and the interaction of these factors. This might suggest that these CpGs act as mediators to age rather than having a direct effect. Moreover, many of the significant CpGs associated with age were eliminated after adding gene expressions and their interactions. The potential reason for this elimination could be due to the insignificance of transcription or the interaction term in the regression model. Another reason could be the small sample size, potentially decreasing statistical power.
In our study, COL11A2 was the most significant gene, with 28 CpGs associated with both age and gene expression. In particular, CpG cg18651026 was associated with three phenotypes: WC, WHR, and BF.PC1. The COL11A2 is located on chromosome 6, and encodes one of two alpha chains of type XI collagen, a fibrillary collagen found in multiple tissues across the body, such as the cartilage, tendons, trachea, etc. The disruption of type XI collagen loosens extracellular matrix (ECM) connections by disrupting the correct fibril assembly [40]. Mutations in the COL11A2 gene cause skeletal abnormalities [41]. Interestingly, COL11A2 has also been implicated as an insulin-resistant gene epigenetically regulated in visceral adipose tissue in a morbidly obese cohort [37]. Furthermore, in an investigation of SNPs associated with the methylation of COL11A2 genes, two identified rare alleles of these methylation sites showed an association with plasma fasting glucose levels in an obese cohort of metabolic syndrome [36]. However, methylation of the COL11A2 sites was not associated with body fat phenotypes. The direct relationship between the COL11A2 gene and WC remains unclear. The waist is comprised of many adipose cells, which typically increase with age. Therefore, the enlargement of these tissues requires a remodelling of the ECM [42]. Collagen is involved in correct fibril assembly and the correct formation of some of the other components in the ECM to ensure a healthy function of adipose tissues and insulin sensitivity. Further functional analysis of the COL11A2 gene would be beneficial to understand the impact of collagen on the ECM and other connective tissues and a potential relationship with body fat phenotypes.
Another gene found to be associated with five body fat phenotypes (BF, BMI, HC, WC, and BF.PC1) was the C1S gene. The C1S gene belongs to a C1 complex including C1q, C1r, and C1s subunit. This is part of the classical pathway, in which the activation of C1s triggers the formation of a membrane complex to promote inflammation and autoimmunity. The function of C1s was discussed in a previous review by Ye et al. [43]; however, a direct role for C1s in body fat phenotypes has not previously been described. It is known that C1q binding to antigen–antibody immune complexes will activate C1s and the C1 complex, and that C1q can activate the WNT signalling pathway that is known to play a role in obesity [44]. In addition, the upregulation of C1q was observed in adipose tissue in the ob/ob mice [45]. Furthermore, a study on rare monozygotic twin pairs showed the upregulation of the C1s gene in adipose tissue in heavier BMI co-twins and obese males, suggesting the increased inflammatory activity of complement pathway components [38]. Our study showed a negative association between DNA methylation and gene expression and a negative association between DNA methylation and body fat phenotypes. The methylation CpGs are situated on the promoter of the C1S gene and therefore potentially control the expression of C1S during the activation of the immune system.
In our study, the methylation of the FHL2 gene was positively associated with BF. Both methylation and the expression of the FHL2 gene in the blood and adipose tissue increase with age [39]. Interestingly, hyper-methylation of FHL2 increases its expression [39]. A previous study reported higher expressions of the FHL2 gene in the white adipose tissue when comparing obese and lean individuals [46]. They also showed in mice that a decrease in FHL2 could be beneficial in in preventing weight gain when exposed to a high-fat diet. In addition, the FHL2 gene affects the formation of adipocytes [47]. Altogether, these data support our findings, which suggest the association of FHL2 and BF.
As well as the significantly associated DMR on COL11A2, a large group of 116 CpGs annotated to the ANKRD11 gene on chromosome 16 were associated with age after adding mediation terms. The index CpG of this gene was cg05638739, which was associated with nine out of fifteen tested phenotypes. Notably, this CpG was positively associated with seven phenotypes in the body fat sub-group, except for WHR. It was also associated with HDL and HDLCHOL. Increased ANKRD11 expression elevated the expression of the tumour suppressor p53 to protect normal cells against cell cycle dysregulation. In addition to suppressing tumour growth, recent reviews suggest a vital role of p53 in adipogenesis [48] and glucose metabolism [49], both processes crucial to the development of obesity. In our study, the index CpG (cg05638739) was located on the 5′UTR of the ANKRD11 gene, which may result in enhanced p53 gene transcription, suggesting that ANKRD11 may play a role in controlling fat cells by activating p53 pathways.
The CPEB2 gene was associated with six phenotypes within the body fat subgroup, except for WHR and WC. CPEB2 is a protein-coding gene belonging to the CPEB family, which act as transcription regulators. While other genes within the CPEB family, including CPEB1, CPEB3, and CPEB4, have been associated with aging, a role for CPEB2 has not been previously reported. The interaction of CPEB2 with eukaryotic elongation factor 2 (eEF2) regulates the translation of hypoxia-inducible factor 1α (HIF1A) mRNA, which has been proven to increase in adipose tissue in obese mice [50,51]. In addition, CPEB2 expression is upregulated in renal cancer cell proliferation and migration via the inactivation of tumour suppressor p53 [52]. In obesity research, the knockout of the CPEB2 gene showed its ability to reduce thermogenesis in brown adipose tissue [53].
Interestingly, the TIAL1 gene was positively associated with all body fat subgroups. TIAL1 is TIA-1-related/like protein. Research has shown that TIAL1 acts as a cellular sensor; knocking out this protein promoted the expression of many mRNAs in cell proliferation and apoptosis [54,55,56,57]. However, our study is the first to show a positive association of the TIAL1 gene with multiple body fat phenotypes. How this gene is involved in the control of body fat cells is not clear; however, we suggest that TIAL1 expression may control cell proliferation and apoptosis to maintain a healthy balance of adipose cells.
Our study analysed genomic data from an isolated healthy cohort of 74 individuals. Due to the small sample size, the statistical method did not address multiple testing in model 2 and model 3, as it was an exploratory study. Furthermore, this study only considered genes that have been found to be associated with age in model 1 to model 2, resulting in 77 out of 183 genes with both gene expression and methylation data. An independent and larger general population would be required to validate these results. In addition, a larger sample size would help investigate additional covariates such as cell proportions and SNP genotypes on these associations.
In conclusion, there were 35 age-related expression CpGs. Among these eCpGs, ten genes were associated with at least one obesity-related phenotype. While the function of these genes’ impact on body fat metabolism is unclear, these genes were shown to participate in multiple related biological processes, including the arrangement of ECM and cell proliferation/cell cycles as well as the immune system. While promising targets for future studies, these new findings still need further validation and functional evaluation to clarify their role in aging and obesity.
Acknowledgments
We would love to express our appreciation to all the volunteers who participated in this study from Norfolk Island.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cimb45100497/s1, Table S1: The summary statistic of significant probes associated with age (p < 2.4 × 10−7) ordered by Chromosome and position; Table S2: Age-related eCpGs; Table S3: Literature-based assessment of the functional role of age-related eCpG genes in adipose tissue and obesity.
Funding Statement
The National Health and Medical Research Council of Australia, NHMRC, funded this research with project grant APP1058806. The Australia Government Research Training Program Scholarship provided funding for this work as a scholarship to T.V.C.
Author Contributions
Conceptualization, R.A.L. and T.V.C.; Methodology, T.V.C. and R.A.L.; Software, T.V.C.; Validation, T.V.C. and R.A.L.; Formal Analysis, T.V.C. and R.A.L.; Investigation, R.A.L.; Resources, R.A.L.; Data Curation, T.V.C. and R.A.L.; Writing—Original Draft Preparation, T.V.C.; Writing—Review and Editing, H.G.S., M.C.B., L.M.H., R.A.L. and L.R.G.; Visualization, T.V.C.; Supervision, R.A.L.; Project Administration, T.V.C.; Funding Acquisition, R.A.L. and L.R.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethics approval for this study was granted by the QUT human ethics committee (Project code: 3923 Approval date: 10 May 2021).
Informed Consent Statement
All subjects included in this study provide informed consent prior to participation.
Data Availability Statement
Data will be shared with researchers upon request and on a collaborative basis.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
Articles from Current Issues in Molecular Biology are provided here courtesy of Multidisciplinary Digital Publishing Institute (MDPI)
Full text links
Read article at publisher's site: https://doi.org/10.3390/cimb45100497
Read article for free, from open access legal sources, via Unpaywall:
https://www.mdpi.com/1467-3045/45/10/497/pdf?version=1695807126
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/156747012
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Quick GO (2)
- (1 citation) GO - GO0003700
- (1 citation) GO - GO0000981
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A genome-wide methylation study of body fat traits in the Norfolk Island isolate.
Nutr Metab Cardiovasc Dis, 31(5):1556-1563, 12 Feb 2021
Cited by: 5 articles | PMID: 33810959
An integrated -omics analysis of the epigenetic landscape of gene expression in human blood cells.
BMC Genomics, 19(1):476, 19 Jun 2018
Cited by: 32 articles | PMID: 29914364 | PMCID: PMC6006777
Smoking-related changes in DNA methylation and gene expression are associated with cardio-metabolic traits.
Clin Epigenetics, 12(1):157, 22 Oct 2020
Cited by: 26 articles | PMID: 33092652 | PMCID: PMC7579899
Visceral adiposity and inflammatory bowel disease.
Int J Colorectal Dis, 36(11):2305-2319, 09 Jun 2021
Cited by: 17 articles | PMID: 34104989
Review




