Abstract
Free full text

Genomic characterisation of respiratory syncytial virus: a novel system for whole genome sequencing and full-length G and F gene sequences
Associated Data
Abstract
To advance our understanding of respiratory syncytial virus (RSV) impact through genomic surveillance, we describe two PCR-based sequencing systems, (i) RSVAB-WGS for generic whole-genome sequencing and (ii) RSVAB-GF, which targets major viral antigens, G and F, and is used as a complement for challenging cases with low viral load. These methods monitor RSV genetic diversity to inform molecular epidemiology, vaccine effectiveness and treatment strategies, contributing also to the standardisation of surveillance in a new era of vaccines.
At present, several RSV vaccine candidates are approved, pending regulatory body approval or in the final stages of clinical trials [1]. In addition to immunisations, the use of the monoclonal antibodies, palivizumab [2] and nirsevimab [3], is recommended to prevent RSV infection in infants and available in several countries [4]. The need to develop methods for monitoring the impact of monoclonal antibodies and vaccine effectiveness is becoming more urgent for the national and supranational surveillance systems.
In this study, we propose two methods for characterising respiratory syncytial virus (RSV) subtypes A and B: RSVAB-WGS, a novel amplicon-based system for RSV whole genome sequencing (WGS) and RSVAB-GF, a method focused on obtaining the specific sequences of the main antigens, glycoprotein (G) and fusion protein (F).
Design and evaluation of RSVAB-WGS targeted PCR
For the design, we aligned 922 selected sequences obtained from the GenBank and GISAID databases. We chose a total of 12 primers for the RSVAB-WGS method and two additional primers for RSVAB-GF method (Table 1). A mix of these primers collectively cover both RSV subtypes, generating PCR fragments of an average size of 1.5–2.5 kb in the WGS and 3.0–3.5 kb in the GF protocol. Both amplification protocols are openly available on the platform protocols.io [5].
Table 1
| Primer | Sequence (5’–3’) |
|---|---|
| Mix 1 RSVAB WGS | |
| RSVCombinitial | ACGCGAAAAAATGCGTACWACA |
| RSVWGS4R | CATGWTGWYTTATTTGCCCC |
| RSVWGS2F | CACTWACAATATGGGTGCC |
| RSVWGS1R | TCCATKGTTATTTGCCCC |
| RSVWGS3.2F | ACATGGAAAGAYATYAGCC |
| RSVWGS2R | CRTTYCTTAARGTRGGCC |
| RSVWGS3.2R | TTGCATCTGTAGCAGGAATGG |
| RSVCombending | ACGAGAAAAAAAGTGTCAAAAACTAA |
| Mix 2 RSVAB WGS | |
| RSVCombinitial | ACGCGAAAAAATGCGTACWACA |
| RSVWGS1R | TCCATKGTTATTTGCCCC |
| RSVWGS2F | CACTWACAATATGGGTGCC |
| RSVWGS8R | TCMAWYTCWGCAGCTCC |
| RSVWGS5R | CAAACATTTAATCTRCTAAGGC |
| RSVWGS6F | TTATAYAGATATCAYATGGGTGG |
| RSVWGS6R | CCCTCTCCCCAATCTTTTTC |
| RSVCombending | ACGAGAAAAAAAGTGTCAAAAACTAA |
| Mix RSVAB-GF | |
| OG1-21 | GGGGCAAATGCAACCATGTCC |
| RSVGF-R | TTCGYGACATATTTGCCCC |
The primers were organised by their position in the genome and the amplification mix in which they were employed. Primers RSVCombinitial, RSVWGS1R, RSVWGS2F and RSVCombending were used in both mixes.
The PCR parameters and the final sensitivity of the chosen protocol were evaluated using dilutions of well-characterised reference viruses, RSV A Long and RSV B CH-18537, as control samples (Table 2). Our method exhibited a sensitivity of 20 copies/mL for RSV A and 200 copies/mL for RSV B (Table 2).
Table 2
| Strain | ATCC nomenclature | Cq value | Number of copies/mL | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Dilution of control virus | 10-1 | 10-2 | 10-3 | 10-4 | 10-1 | 10-2 | 10-3 | 10-4 | |
| RSV A Long | VR-26 | 15.12 | 18.88 | 22.09 | 25.57 | 2 × 104 | 2 × 103 | 2 × 102 | 2 × 101 |
| RSV B CH-18537 | VR-1580 | 16.21 | 19.5 | 22.8 | 26.2 | 2 × 104 | 2 × 103 | 2 × 102 | NA |
ATCC: American type culture collection; Cq: quantification cycle; NA: not applicable.
The standardisation process included quantifying the dilutions of each control virus based on the Cq value obtained in the RT-PCR used for viral detection and subtyping. Subsequently, we calculated the number of copies of viral nucleic acids in each dilution of the control viruses.
We constructed libraries following the instructions provided by the Illumina DNA library preparation kit (Illumina, United States) for 300-cycle cartridges in Illumina MiSeq and NextSeq sequencers. Sequences were analysed for viral genome reconstruction using the viralrecon pipeline v2.6.0. (https://github.com/nf-core/viralrecon) [6], implemented in Nextflow (https://www.nextflow.io). Phylogenetic analysis was conducted using FasTreeMP software [7] with a generalised time reversible (GTR) model and a bootstrap test of 1,000 iterations.
For validation, we analysed a total of 142 nasopharyngeal exudates (NPE) and nasopharyngeal aspirates (NPA) collected from RSV-positive patients during the epidemic seasons 2018/19, 2019/20 and 2021/22. To assess the RSV viral load and facilitate subsequent sequencing, we performed a real-time PCR to detect RSV and used the resulting quantification cycle (Cq) value as a preliminary indicator [1,8]. Despite the usefulness of setting a cut-off in the Cq values used to ensure the complete genome acquisition, in this work, a deliberate decision was made not to set it. The aim was to determine the limit of detection in the method by sequencing both our lowest Cq value sample from the season (14.33) and the highest (29.4).
Complete genome sequencing of respiratory syncytial virus A and B
For surveillance purposes, the RSVAB-WGS method exhibited practical versatility and proved to be applicable to specimens of both RSV subtypes across a spectrum of viral loads. Validation involved analysing 34 positive clinical specimens, 16 RSV A and 18 RSV B, previously typed [8]. These samples showed Cq values ranging from 15 to 20 (10 RSV A, eight RSV B), 21 to 26 (six samples of either subtype) and exceeding 27 (four RSV B). For samples with Cq values ≤
≤ 25, RSVAB-WGS achieved more than 90% genome coverage. For Cq values 26–27, coverage ranged from 60% to 90%, and for Cq values exceeding 27, coverage dropped to 50%. We append in Supplementary Table S1 the detailed sequencing parameters for RSV clinical specimens used in RSVAB WGS method including a description of subtypes, Cq values, sequencing/assembly metrics and GISAID accession numbers.
25, RSVAB-WGS achieved more than 90% genome coverage. For Cq values 26–27, coverage ranged from 60% to 90%, and for Cq values exceeding 27, coverage dropped to 50%. We append in Supplementary Table S1 the detailed sequencing parameters for RSV clinical specimens used in RSVAB WGS method including a description of subtypes, Cq values, sequencing/assembly metrics and GISAID accession numbers.
Recognising the importance of accurate RSV genomic characterisation, we developed the RSVAB-GF method to complement the genomic coverage for both major antigenic proteins in cases where WGS encountered coverage difficulties, such as in samples with low viral load or when the resources for sequencing were constrained. This method offers a simpler and more cost-effective approach to obtaining sequences of both antigens. Validation involved analysing 108 clinical specimens (74 RSV A, 34 RSV B) with Cq values ranging from 14.3 to 29.4 and in all cases, both genes were completely covered. We provide in Supplementary Table S2 a description of the samples used in the targeted RSV AB-GF method including sequence identification, subtype, Cq value and GISAID accession numbers.
Genetic analysis of the genomes obtained with RSVAB-WGS and RSVAB-GF
To compare taxonomic classifications obtained from full genome sequences and partial G and F sequences, we used 14 sequences (seven RSV A, seven RSV B) collected during the 2021/22 season (Table 3). We used the whole genome to assign RSV clades because either the complete sequence or the F gene sequence was available to us in addition to the G gene sequence. These clade definitions were based on the proposed nomenclatures by Goya et al. [9] and Ramaekers et al. [10] using Nextstrain platform. Analysis of the 14 RSV A sequences with both methods demonstrated complete agreement in clade assignation. For the seven RSV A sequences, identified subclades included A.D.1, A.D.3 and A.D.5.2. Taxonomic assignments from seasons 2018/19 and 2019/20 showed the presence of subclades A.D.1, A.D.3 (both also circulating in the 2021/22 season) and subclade A.D.2.3 (Table 3). In the RSV B subtype, clade assignment revealed total agreement in taxonomic classification. The seven sequences from the 2021/22 season were identified as subclade B.D.5.2.1.1 with both methods.
Table 3
| RSV sequence | Clade (RSVAB-WGS) | Clade (RSVAB-GF) |
|---|---|---|
| hRSV/A/Spain/MD-224273/2022 | A.D.1 | A.D.1 |
| hRSV/A/Spain/MD-224532/2022 | A.D.5.2 | A.D.5.2 |
| hRSV/A/Spain/MD-224512/2022 | A.D.5.2 | A.D.5.2 |
| hRSV/A/Spain/MD-224870/2022 | A.D.3.1 | A.D.3.1 |
| hRSV/A/Spain/MD-224747/2022 | A.D.3 | A.D.3 |
| hRSV/A/Spain/MD-224741/2022 | A.D.5.2 | A.D.5.2 |
| hRSV/A/Spain/MD-224199/2022 | A.D.3 | A.D.3 |
| hRSV/B/Spain/MD-220011/2021 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-220471/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-220289/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-224976/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-223807/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-224877/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
| hRSV/B/Spain/MD-224510/2022 | B.D.5.2.1.1 | B.D.5.2.1.1 |
F: fusion protein; G: glycoprotein; RSV: respiratory syncytial virus; WGS: whole genome sequencing.
For the phylogenetic analysis (Figure), we selected sequences from the main antigens G and F due to the high number of sequences with good quality and coverage in the GISAID database in comparison with those of the complete genome. We constructed a separate tree for each RSV subtype. In both subtypes, sequence clustering was primarily determined by mutations spanning consecutive seasons rather than by year or geographical location. The phylogenetic analysis of 2,314 RSV A sequences, including 74 sequences from this study, and 2,875 RSV B sequences, with the 34 G and F sequences obtained from this work, corroborated previous taxonomic results (Figure) [11].
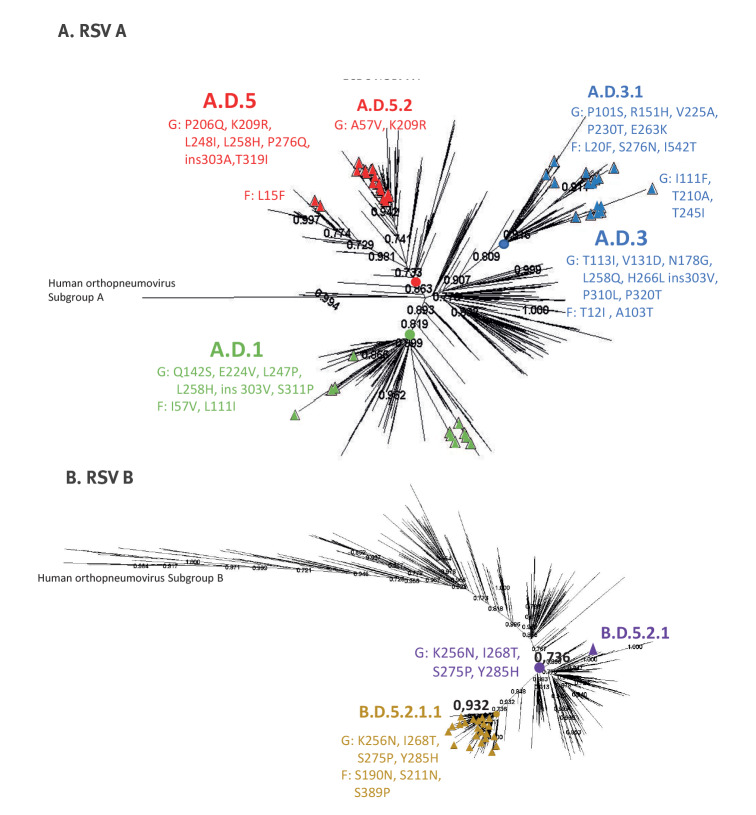
Phylogenetic tree of respiratory syncytial virus G and F sequences published in GISAID (n =
= 2,314 RSV A; n
2,314 RSV A; n =
= 2,875 RSV B)
2,875 RSV B)
F: fusion protein; G: glycoprotein; RSV: respiratory syncytial virus.
The numbers represent percentage bootstrap support. The sequences obtained in this work were represented by triangles. Clusters, where these sequences were located, are identified by three colours (red, blue and green) according to the defining mutations. The colours yellow and purple mark the nodes with the defining mutations of the group.
Discussion
Several methods have already been designed for sequencing RSV isolates or samples, employing diverse approaches [12-16]. However, the primary challenges in successful RSV sequencing often involve obtaining complete genomes from clinical samples with low viral loads [17] and high diversity of RNA genomes [18]. Previous methods often required prior subtyping and quantification due to differences in primer design based on subtypes [13,19], making the process expensive and labour-intensive. PCR amplicon sequencing, using amplicons generated through specific primers [20,21] or sequence-independent single primer amplification (SISPA) [13], exhibits notable advantages, particularly in clinical specimens with low viral loads [13]. The effectiveness of double-stranded cDNA generation before targeted amplification has proven effective for other respiratory viruses like severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [22,23], and it was also validated in this work for RSV. The use of cDNA in our RSVAB- WGS method provided an augmented amount of viral nucleic acids using only a few microlitres of the clinical sample extract, optimising sample usage. Amplicon size is critical, as shorter amplicons can introduce biases in genome reconstruction and prevent the successful assembly of a full genome sequence for unknown viruses. The method described in this work produces amplicons compatible with both long and short-read sequencers, offering a versatile solution for laboratories, irrespective of the type of sequencer available.
To demonstrate the utility of our RSVAB-WGS and RSVAB-GF methods in molecular epidemiology studies of RSV, we conducted phylogenetic and genomic analyses using samples collected during the 2018/19, 2019/20 and 2021/22 epidemic RSV seasons. The phylogenetic analysis revealed that changes in the G protein gene had a larger impact on grouping than the timing of virus collection or geographical location. While WGS for monitoring viral transmission chains can offer great accuracy, studies indicate that the relationships identified with the G region are similar to the patterns observed with full-genome sequences [16]. Furthermore, a previous world-wide analysis of the RSV clades [11] demonstrated that entirely sequenced complete G gene sequences yielded a phylogenetic tree topology comparable to that obtained using whole genomes. This underscores the utility of the RSVAB-GF method in providing evolutionary information through a simplified system that could be used when the viral load is low and also when the resources for sequencing are not enough to support a WGS system.
In addition, we currently find it highly pertinent to characterise the F protein gene variability with an accurate and robust complementary method such as the RSVAB-GF PCR. This is especially crucial since the F protein is the main target for most vaccines, which are either already approved [1] or soon expected to be. Moreover, there are several monoclonal antibodies for RSV prevention [3], such as nirsevimab or palivizumab, all of which also target the F protein.
Despite the advantages presented by these methods, we also have to address limitations in the WGS method, such as a decrease in coverage starting from Cq = 25 and the need to improve the complete genome sequencing in RSV B. Currently, the method is being improved to solve these issues. Although this work was confined to the sequences received in our laboratory, we believe this does not restrict the capability of these methods to sequence any type of RSV because the primer design was performed using sequences from various locations and different seasons.
The World Health Organization has started a global effort to homogenise RSV surveillance, based on the Global Influenza Surveillance and Response System [11]. The surveillance system needs to be supported by reliable and practical methods.
Conclusions
While implementing WGS of RSV remains technically demanding and expensive, many laboratories express the need for guidance. Our universal amplicon-based system RSVAB-WGS is an accurate and cost-effective means to obtain genomic data. Coupled with the RSVAB-GF method, it enhances capacity and performance for high-quality sequence acquisition from both genes in a single run, even when processing hundreds of samples. These methods to characterise the complete RSV genome and the variability of G and F protein genes can serve as a useful tool for RSV surveillance, including monitoring of preventive measures to limit the spread of the virus.
Ethical statement
Nasopharyngeal exudates and nasopharyngeal aspirates were obtained using standard clinical procedures. The study was conducted in accordance with the Declaration of Helsinki. The Institutional Review Board and the Research Ethic Committees of participant’s hospitals approved the study: CEIM A1045, PI-2255, PI-3546 and PI-4676.
Data availability statement
GISAID accession number of sequences obtained by RSVAB-WGS method: EPI_ISL_18463183,EPI_ISL_18463185,EPI_ISL_18463167,EPI_ISL_18463162,EPI_ISL_18463188,EPI_ISL_18463190,EPI_ISL_18463170,EPI_ISL_18463181,EPI_ISL_18463177,EPI_ISL_18463184,EPI_ISL_18463165,EPI_ISL_18463174,EPI_ISL_18463178,EPI_ISL_18463186,EPI_ISL_18463171,EPI_ISL_18463173,EPI_ISL_18463187,EPI_ISL_18463189,EPI_ISL_18463191,EPI_ISL_18463172,EPI_ISL_18463192,EPI_ISL_18463179,EPI_ISL_18463168,EPI_ISL_18463166,EPI_ISL_18463169,EPI_ISL_18463180,EPI_ISL_18463175,EPI_ISL_18463176,EPI_ISL_18463182,EPI_ISL_18463164 and EPI_ISL_18463163
GISAID accession number of sequences obtained by RSVAB-GF method:
EPI_ISL_18463183,EPI_ISL_18469715,EPI_ISL_18469716,EPI_ISL_18469676,EPI_ISL_18469707,EPI_ISL_18469655,EPI_ISL_18469662,EPI_ISL_18469664,EPI_ISL_18469667,EPI_ISL_18463162,EPI_ISL_18469696,EPI_ISL_18463167,EPI_ISL_18469711,EPI_ISL_18469708,EPI_ISL_18469656,EPI_ISL_18469709,EPI_ISL_18469700,EPI_ISL_18469661,EPI_ISL_18469713,EPI_ISL_18469706,EPI_ISL_18469668,EPI_ISL_18463170,EPI_ISL_18469675,EPI_ISL_18469669,EPI_ISL_18469701,EPI_ISL_18463181,EPI_ISL_18469691,EPI_ISL_18469657,EPI_ISL_18469704,EPI_ISL_18469666,EPI_ISL_18469717,EPI_ISL_18469697,EPI_ISL_18469687,EPI_ISL_18469714,EPI_ISL_18469688,EPI_ISL_18469698,EPI_ISL_18469679,EPI_ISL_18469695,EPI_ISL_18469705,EPI_ISL_18469681,EPI_ISL_18469670,EPI_ISL_18469710,EPI_ISL_18469699,EPI_ISL_18469702,EPI_ISL_18469680,EPI_ISL_18469665,EPI_ISL_18469685,EPI_ISL_18469720,EPI_ISL_18469712,EPI_ISL_18469703,EPI_ISL_18469686,EPI_ISL_18469658,EPI_ISL_18469673,EPI_ISL_18469659,EPI_ISL_18469718,EPI_ISL_18469690,EPI_ISL_18469663,EPI_ISL_18463184,EPI_ISL_18469660,EPI_ISL_18469693,EPI_ISL_18469683,EPI_ISL_18469654,EPI_ISL_18469692,EPI_ISL_18469677,EPI_ISL_18469671,EPI_ISL_18469719
EPI_ISL_18469678,EPI_ISL_18469684,EPI_ISL_18469682,EPI_ISL_18469694,EPI_ISL_18469689,EPI_ISL_18469674,EPI_ISL_18463186,EPI_ISL_18469672,EPI_ISL_18470178,EPI_ISL_18470179,EPI_ISL_18470180,EPI_ISL_18463172,EPI_ISL_18470181,EPI_ISL_18470182,EPI_ISL_18470183,EPI_ISL_18470184,EPI_ISL_18470185,EPI_ISL_18474127,EPI_ISL_18470186,EPI_ISL_18470187,EPI_ISL_18470188,EPI_ISL_18470189
EPI_ISL_18470190,EPI_ISL_18470191,EPI_ISL_18470192,EPI_ISL_18463168,EPI_ISL_18470193,EPI_ISL_18470194,EPI_ISL_18470195,EPI_ISL_18470196,EPI_ISL_18470197,EPI_ISL_18470198,EPI_ISL_18463180
EPI_ISL_18463176,EPI_ISL_18470199,EPI_ISL_18463182,EPI_ISL_18463164,EPI_ISL_18470200,EPI_ISL_18463163,EPI_ISL_18470201,EPI_ISL_18470202 and EPI_ISL_18470203
Funding statement
This research was partially funded by Instituto de Salud Carlos III through the Project PI18/00167, PI21CIII/00019 (MPY 439-21) and PI21/00377, and partially by UNESPA donation-“COVIDSEQUNESPA” (grant number MPY 226/22). Also, funding was received from grant MSD MISP: IISP 60255.
Acknowledgements
We thank Noelia Reyes, Silvia Moreno, M Jose Casas and Mar Molinero from the National Reference Laboratory for their exceptional technical support. We thank Pilar Jimenez, Mercedes Jimenez and Angel Zaballos from the Genomics unit at the Instituto de Salud Carlos III who collaborated in obtaining genomic sequences. We gratefully acknowledge all data contributors, authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing them via the GISAID initiative and GenBank database (www.ncbi.nlm.nih.gov/genbank/), from which we retrieved the sequences used in this research.
Supplementary Data
Notes
Disclaimer
The authors are responsible for the views expressed in this article and they do not necessarily represent the views, decisions or policies of the institutions with which they are affiliated.
Notes
Conflict of interest: None declared.
Authors’ contributions: MIC and ICa: conceptualisation, methodology, validation, data curation (lead); writing – original draft (lead); formal analysis (lead); visualisations (lead); writing – review and editing (equal). SCS, SM and SV: methodology, validation and review and editing (equal). SVM: phylogenetic analysis and visualisation; writing – review and editing; CC, MLG and JGC: sampling providing, review and editing (equal); FP, AC, ICu and VM: data curation, analysis, visualisation and review and editing (equal).
References
Articles from Eurosurveillance are provided here courtesy of European Centre for Disease Prevention and Control
Full text links
Read article at publisher's site: https://doi.org/10.2807/1560-7917.es.2023.28.49.2300637
Read article for free, from open access legal sources, via Unpaywall:
https://www.eurosurveillance.org/deliver/fulltext/eurosurveillance/28/49/eurosurv-28-49-2.pdf?itemId=%2Fcontent%2F10.2807%2F1560-7917.ES.2023.28.49.2300637&mimeType=pdf&containerItemId=content/eurosurveillance
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/157287041
Article citations
Respiratory syncytial virus infections - recent developments providing promising new tools for disease prevention.
Euro Surveill, 28(49), 01 Dec 2023
Cited by: 4 articles | PMID: 38062943 | PMCID: PMC10831406
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Robust and sensitive amplicon-based whole-genome sequencing assay of respiratory syncytial virus subtype A and B.
Microbiol Spectr, 12(4):e0306723, 27 Feb 2024
Cited by: 1 article | PMID: 38411056 | PMCID: PMC10986592
A Recombinant Respiratory Syncytial Virus Vaccine Candidate Attenuated by a Low-Fusion F Protein Is Immunogenic and Protective against Challenge in Cotton Rats.
J Virol, 90(16):7508-7518, 27 Jul 2016
Cited by: 30 articles | PMID: 27279612 | PMCID: PMC4984630
Packaging and Prefusion Stabilization Separately and Additively Increase the Quantity and Quality of Respiratory Syncytial Virus (RSV)-Neutralizing Antibodies Induced by an RSV Fusion Protein Expressed by a Parainfluenza Virus Vector.
J Virol, 90(21):10022-10038, 14 Oct 2016
Cited by: 25 articles | PMID: 27581977 | PMCID: PMC5068507
Molecular epidemiology of respiratory syncytial virus.
Rev Med Virol, 28(2), 29 Jan 2018
Cited by: 40 articles | PMID: 29377415
Review




