Abstract
Background
Gastric cancer (GC) stands as a prominent cause of cancer-related mortality and ranks second among the most frequently diagnosed malignancies in individuals with common variable immunodeficiency (CVID).Objective
We sought to conduct a comprehensive, large-scale genetic analysis to explore the CVID-associated germline variant landscape within gastric adenocarcinoma samples and to seek to delineate the transcriptomic similarities between GC and CVID.Methods
We investigated the presence of CVID-associated germline variants in 1591 GC samples and assessed their impact on tumor mutational load. The progression of GC was evaluated in patients with and without these variants. Transcriptomic similarities were explored by matching differentially expressed genes in GC to healthy gastric tissue with a CVID transcriptomic signature.Results
CVID-associated germline variants were found in 60% of GC samples. Our analysis revealed a significant association between the presence of CVID-related genetic variants and higher tumor mutational load in GC (P < .0001); high GC mutational load seems to be linked to immunotherapy response and worse prognosis. Transcriptomic similarities unveiled key genes and pathways implicated in innate immune responses and tumorigenesis. We identified upregulated genes related to oncogene drivers, inflammation, tumor suppression, DNA repair, and downregulated immunomodulatory genes shared between GC and CVID.Conclusions
Our findings contribute to a deeper understanding of potential molecular modulators of GC and shed light on the intricate interplay between immunodeficiency and cancer. This study underscores the clinical relevance of CVID-related variants in influencing GC progression and opens avenues for further exploration into novel therapeutic approaches.Free full text

Exploring gastric cancer genetics: A turning point in common variable immunodeficiency
Associated Data
Abstract
Background
Gastric cancer (GC) stands as a prominent cause of cancer-related mortality and ranks second among the most frequently diagnosed malignancies in individuals with common variable immunodeficiency (CVID).
Objective
We sought to conduct a comprehensive, large-scale genetic analysis to explore the CVID-associated germline variant landscape within gastric adenocarcinoma samples and to seek to delineate the transcriptomic similarities between GC and CVID.
Methods
We investigated the presence of CVID-associated germline variants in 1591 GC samples and assessed their impact on tumor mutational load. The progression of GC was evaluated in patients with and without these variants. Transcriptomic similarities were explored by matching differentially expressed genes in GC to healthy gastric tissue with a CVID transcriptomic signature.
Results
CVID-associated germline variants were found in 60% of GC samples. Our analysis revealed a significant association between the presence of CVID-related genetic variants and higher tumor mutational load in GC (P < .0001); high GC mutational load seems to be linked to immunotherapy response and worse prognosis. Transcriptomic similarities unveiled key genes and pathways implicated in innate immune responses and tumorigenesis. We identified upregulated genes related to oncogene drivers, inflammation, tumor suppression, DNA repair, and downregulated immunomodulatory genes shared between GC and CVID.
Conclusions
Our findings contribute to a deeper understanding of potential molecular modulators of GC and shed light on the intricate interplay between immunodeficiency and cancer. This study underscores the clinical relevance of CVID-related variants in influencing GC progression and opens avenues for further exploration into novel therapeutic approaches.
Cancer is widely recognized as a paradigmatic environmental condition that arises in genetically susceptible individuals. Chronic infection and the subsequent inflammation constitute significant contributing factors to tumorigenesis and the progression of established malignant lesions.1 Well-known cancer predisposing factors include inborn errors of immunity (IEI), which render the patients more susceptible to chronic viral and bacterial infections, immune dysregulation, defective immune surveillance, and DNA repair functions, among other factors.2 This is particularly relevant for gastric cancer (GC), one of the most common causes of cancer death worldwide, in which Helicobacter pylori is a major driver of tumorigenesis.3, 4, 5, 6 Intriguingly, although H pylori infects the gastric epithelial lining of more than half of the global population, cancer is an exceedingly rare complication, possibly indicating a contribution of the genetic background to precancerous and cancerous lesions.7 This has been exemplified by the lack of MyD88 signaling in a mouse model.8,9 Tumor-stroma interactions associated with immune alterations, such as Stat3 overexpression during H pylori natural infection, promote autocrine and paracrine mechanisms that favor accelerated tumor growth, angiogenesis, and immune evasion.10 Hence, exploring the crossover between IEI and GC may aid in unraveling the human genetic architecture and tumorigenesis mechanisms of IEI.
Common variable immunodeficiency (CVID) is the most frequent clinically manifested IEI, characterized by impaired antibody responses and a broad range of manifestations, including recurrent or severe infections, autoimmune diseases, B-cell lymphoproliferation, granulomas, and cancer.11 CVID was first described in the 1950s following the enthusiasm generated by the first patients of agammaglobulinemia.12,13 GC is the second most common cancer in CVID, with incidence rates ranging from 2% to 5.5% across various series and with a mortality rate approximately 10-fold higher than in the general population.14, 15, 16, 17, 18
With the availability of vast genetic databases and public gene repositories, it is now possible to analyze how somatic mutations, along with germline mosaicism, can contribute to a range of nonmalignant conditions, including CVID phenotypes. Conversely, certain congenital defects, such as CVID, may contribute to tissue-specific somatic mutations that converge into the development of cancer.19
In this study, we conducted an analysis of somatic mutations of extensive GC gene databases to seek possible crossovers with described CVID-like germline variants. In addition, we sought differentially expressed genes (DEGs) shared by both conditions. Our aim was to gain insight into their possible causative role in both diseases and to identify signaling pathways that could lead to novel approaches for the prevention and treatment of cancer.
Methods
Overall study design
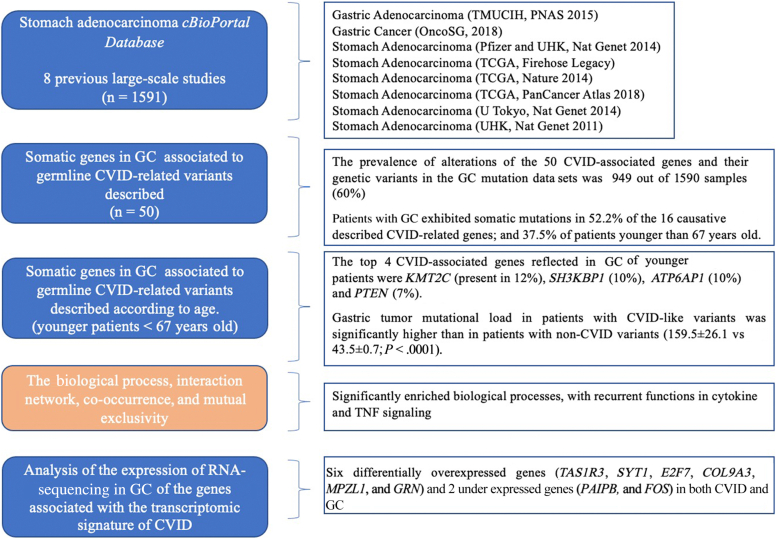
This is a noninterventional study based on the analysis of real-world data publicly available in the cBioPortal database and using web-based analytical tools. We explored the presence of the CVID-like signature in the genomic and transcriptomic profile of GC to identify common pathophysiological mechanisms between both conditions (Fig 1). All the analyses were performed in the Cancer Immunomonitoring and Immuno-Mediated Pathologies Support Unit of the Clinical Immunology Department, in close collaboration with the Oncology Department, at the Hospital Clínico San Carlos in Madrid, Spain.
Data collection and processing
Analysis of CVID-related pathogenic variants in mutational landscape of GC
cBioPortal database, gene ontology, and functional analysis
An integrative analysis of GC (gastric adenocarcinoma) was performed using the cBioPortal database (https://www.cbioportal.org; accessed in September 2022)—a publicly available database for tumor genomics20 including 8 previous large-scale studies (n = 1591) to explore the presence and distribution of CVID-related gene variants among somatic alterations in gastric tumor (see Table E1 in this article’s Online Repository at www.jaci-global.org). A total of 50 of the most frequent CVID-associated genes described as causative and CVID-associated pathogenic variants were selected for analysis,21,22 among which 20 causative CVID-like genes stood out: CR2, LRBA, NFKB1, NFKB2, IL21, TNFRSF13B, TNFRSF13C, CD81, IKZF1, PRKCD, MS4A1, NOD2, MSH5, TNFRSF13B, HLA, CTLA4, PLCG2, PIK3CD, PIK3R1, and CD19 according to the Online Mendelian Inheritance in Man (OMIM) database and recent publications.21,23
The analogy between CVID-related pathogenic variants found within the altered genes in GC was then verified through bibliographic sources as well as consulted using the ClinVar-NCBI tool (https://www.ncbi.nlm.nih.gov/clinvar/). Genes from the KMT2C family, for example, play an essential role in immune functions and have been depicted as possibly harmful with a high frequency of heterozygotes compared with controls, despite not being considered as CVID-related pathogenic variants.24, 25, 26, 27 These genes also were considered in the analysis. Variant allele frequency data were analyzed when available.
Given the 2 age peaks at 6 to 10 years and 21 to 35 years for CVID onset,11,28,29 and that GC in these series would be expected as the first clinical manifestation, special attention was given to those genes with an equal or greater prevalence of alterations in young patients (age of diagnosis below median age) compared with those above median age.
The biological functions of these genes were obtained from the 2018 Molecular Function Gene Ontology Terms using the publicly available EnrichR online platform (https://maayanlab.cloud/Enrichr/; accessed October 4, 2022) (see Tables E2 and E3 in this article’s Online Repository at www.jaci-global.org). To predict protein-protein interaction (PPI) networks, we used the Search Tool for the Retrieval of Interacting Genes (http://www.string-db.org).30 The database defaults were deployed to map the interaction network and biological distance. A local clustering coefficient closer to 1 indicates that the network is more likely to form functional clusters. Statistical significance was determined by the PPI enrichment P value. Proteins with more interactions than the average are considered hubs. Mutation co-occurrence, mutual exclusivity, and mutations per patient analyses were performed using the cBioPortal online platform (see Tables E4 and E5 in this article’s Online Repository at www.jaci-global.org). This tool calculates the odds ratio for each pair of query genes, indicating the likelihood that the alterations for the 2 genes are co-occurrent or mutually excluded in the selected cases, using the Fisher exact test (statistical significance, P < .05).
Analysis of the expression of RNA-sequencing in GC of the genes associated with the transcriptomic signature of CVID
Public data and preprocessing
The CVID-associated transcriptomic signature and pathway analysis data were based on the associated data previously described by van Schouwenburg et al.31
Differential expression analysis and gene-set enrichment analysis between GC and normal gastric tissue
Raw paired-end reads of the RNA-sequencing samples of GC in The Cancer Genome Atlas (TCGA) were downloaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/) using the R/Bioconductor package “TCGAbiolinks.”32 RNA-sequencing expression data for normal gastric tissue were downloaded from the database of the Genotype-Tissue Expression project (https://gtexportal.org/home/).30 The unification of cancer and normal RNA-sequencing data was carried out following these steps: alignment, quality control, quantification, and checking for batch effect correlation, as described by Wang et al.33 The genome annotation was obtained from the Genome-wide annotation for Human using the R/Bioconductor package “org.Hs.eg.db.”34 The bioinformatic analysis of differential gene expression (DGE) and gene-set enrichment was performed using the R/Bioconductor package “DESeq2.”35
CVID transcriptomic signature in the GC gene-set enrichment
DEGs were analyzed in GC compared with normal gastric tissue and then compared with CVID-associated transcriptomic signature. We used a threshold of an adjusted P value less than .005 and log2FoldChange greater than 0.58 (FoldChange = 1.5) or log2FoldChange less than −0.58 (FoldChange = −1.5) to define upregulated and downregulated genes, respectively. Within these DEGs, we selected those sharing a similar transcriptional pattern between GC and CVID.31 To visualize DGE between individuals in both categories, we created volcano plots, heat maps, and boxplots. The volcano plots were represented using the R/Bioconductor package “EnhancedVolcano.”34 Each heat map featured the log10 transformation of the fold changes and was created using the heatmap.2 function in the R package “gplots.” For hierarchical clustering of expression data, we used the R function heatmap.2 using default parameters. Data analysis was performed using R (version 4.1.2).
Results
Clinical, demographic, and histopathologic characteristics
The 8 public GC data sets combined accounted for a total of 1590 GC samples of 853 patients. The distribution according to sex was as follows: 60.2% men, 34.4% women, and 5.4% not specified. The median age at diagnosis was 67 years (range, 30-90 years, n = 702; younger/older than 67 years, n = 187 [not applicable]).
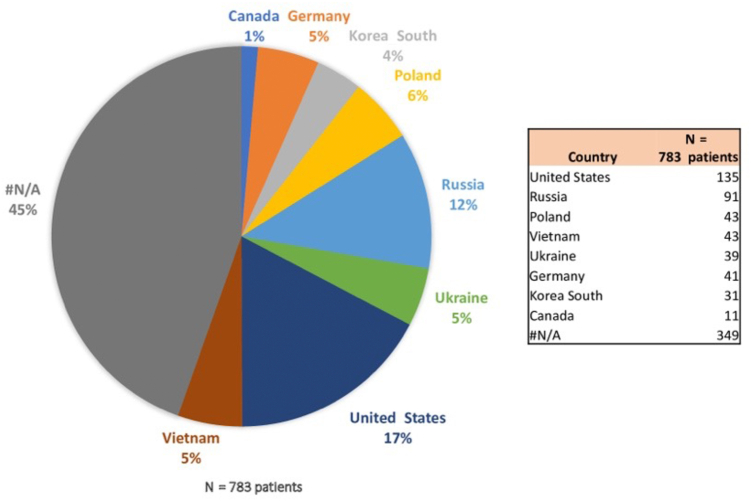
In terms of demographic information, in the cohort of 853 patients, comprehensive data were attainable for 783 individuals, with most of the subjects predominantly hailing from the United States, Russia, Poland, Vietnam, Ukraine, and Germany (Fig 2). A total of 76 patients possessed documented records of H pylori infection, 231 individuals exhibited negative findings for this infection, and 476 patients lacked this specific infection status within the registry. Furthermore, within this cohort, 71 patients exhibited the presence of EBV, 416 individuals tested negative for EBV, and such information was unrecorded in the case of 296 patients. Eighty percent of the samples were derived from the primary tumor (gastric adenocarcinoma), but the remaining 20% were not specified within the studies.
Analysis of CVID-like variants in the genomic landscape of GC
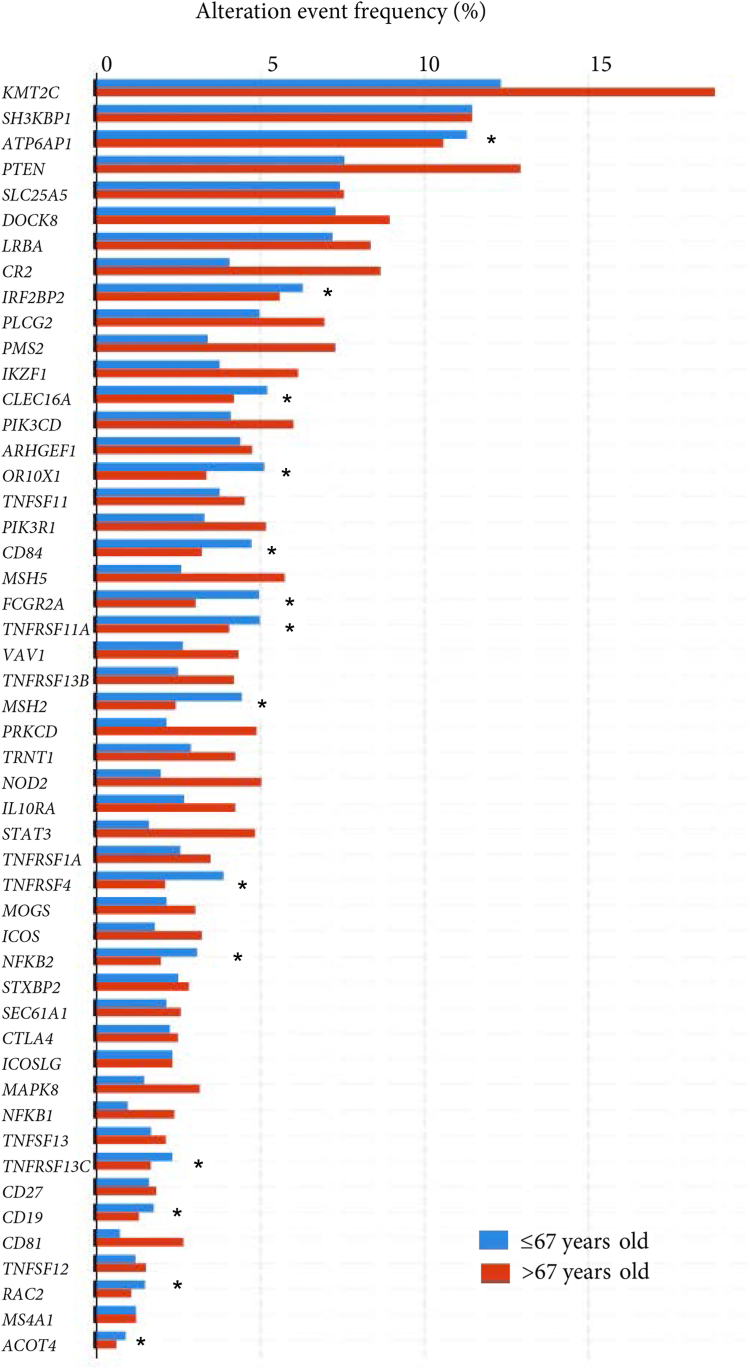
The prevalence of the 50 CVID-associated gene alterations was investigated in the GC data sets. A total of 949 of 1590 samples (60%) harbored CVID-associated genes being mainly distributed among the studies associated with TCGA (750 of 1213 [61.83%]). The most frequent CVID-related mutations were as follows: KMT2C (15% of patients with GC), SH3KBP1 (12%), ATP6AP1 (11%), PTEN (10%), SLC25A5 and DOCK8 (8%), LRBA (7%), and CR2 (6%) (as provided in Table I). The functions of the main CVID-like genes described in all GC studies are provided in Table E6 (in the Online Repository available at www.jaci-global.org).
Table I
Prevalence of the main CVID-associated genetic alterations observed in the GC data set
| Gene symbol | No. of altered samples (all patients) | Percentage of altered samples (all patients) | No. of altered samples (< 67 y old) | Percentage of altered samples (< 67 y old) |
|---|---|---|---|---|
| KMT2C | 234 | 15 | 76 | 12 |
| SH3KBP1 | 183 | 12 | 63 | 10 |
| ATP6AP1 | 169 | 11 | 63 | 10 |
| PTEN | 153 | 10 | 46 | 7 |
| SLC25A5 | 130 | 8 | 44 | 7 |
| DOCK8 | 127 | 8 | 41 | 6 |
| LRBA | 116 | 7 | 43 | 7 |
| CR2 | 97 | 6 | 26 | 4 |
| IRF2BP2 | 87 | 5 | 38 | 6 |
| PLCG2 | 81 | 5 | 26 | 4 |
| PMS2 | 75 | 5 | 18 | 2.9 |
| IKZF1 | 75 | 5 | 22 | 3 |
| CLEC16A | 73 | 5 | 30 | 5 |
| PIK3CD | 69 | 4 | 17 | 2.7 |
| ARHGEF1 | 69 | 4 | 24 | 4 |
| OR10X1 | 67 | 4 | 27 | 4 |
| TNFSF11 | 64 | 4 | 22 | 3 |
| PIK3R1 | 64 | 4 | 19 | 3 |
| CD84 | 63 | 4 | 29 | 5 |
| MSH5 | 60 | 4 | 13 | 2.1 |
| FCGR2A | 57 | 4 | 26 | 4 |
| TNFRSF11A | 57 | 4 | 21 | 3 |
| MSH2 | 49 | 3 | 25 | 4 |
| TNFRSF4 | 40 | 2.5 | 19 | 3 |
Only gene variants observed in at least 0.5% of cases are displayed. The prevalence in the subgroup of patients younger than 67 y is shown.
Globally, 52.2% (830 of 1590) of patients with GC exhibited somatic mutations in the 16 causative CVID-related genes described, of which 37.5% belonged to the group of younger patients (<67 years) (Table I). The most common types of variants (data available in 4 of the 8 studies: OncoSG, 2018; TCGA, Nature 2014; TCGA, PanCancer Atlas 2018; and TCGA, Firehose Legacy) were 1031 missense mutations, 860 amplifications, 421 truncating mutations (putative drivers), 414 deep deletions, and 29 inframe mutations (putative passenger). MSH2 genes were considered as driver mutations, present in 3% of all adenocarcinomas and 4% in younger patients.
The PPI map displaying the significant functional network integrated by the selected genes involved in patients with GC (described in Table I) is shown in Fig 3. The spectrum of CVID-related genes differed according to age in the different studies and the proportion and number of altered samples (see Table E7 in this article’s Online Repository available at www.jaci-global.org). To further investigate the functional characterization of the set of CVID-associated genes in younger patients, we searched the EnrichR database and the gene set was run through the Reactome pathway analysis tool. Significantly enriched biological processes related with cytokine and TNF signaling (significant ways adjusted by q value are provided in Table E2).
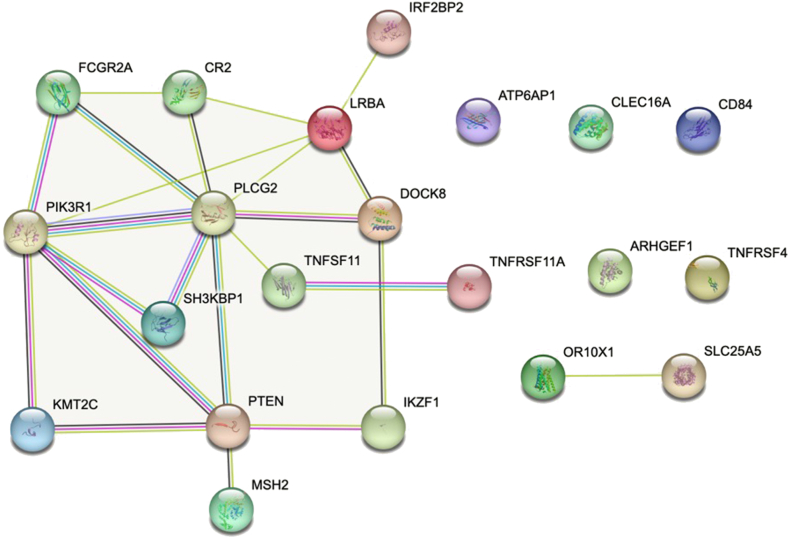
PPI map displaying the functional network integrated by the CVID-like selected genes involved in patients with GC. Known interactions are displayed in blue and pink lines. Predictive interactions include the following: green line, gene neighborhood; red line, gene fusions; blue line, gene co-occurrence; yellow line, text mining; black line, coexpression; and gray line, protein homology. Connections are filtered by the highest confidence (0.7). PPI enrichment P value less than 2.6 × 10−8.
DGEs in CVID-like genes
Taking the intersection of the DGEs in the 8 data sets and CVID-like genes, we identified 15 genes with an equal or greater prevalence of gene alterations in younger patients with GC compared with older patients (>67 years) (Fig 4; Table E3). This enrichment may suggest that these variants were involved in the early stages of GC formation. The CVID-associated gene alterations significantly more prevalent in younger patients included IRF2BP2, CLEC16A, OR10X1, CD84, FCGR2A, MSH2, TNFRSF11A, TNRFSF4, SLC25A5, CD27, NFKB2, TNFRSF13C, TNFSF12, CD19, and ACOT4. The genes MSH2, TNFRSF4, FCGR2A, OR10X1, and CD84 reached statistical significance, but were not significant after correction (q value). Regarding the genes with the highest prevalence in the older group, CR2, KMT2C, PTEN, ARHGEF1, STAT3, PMS2, NOD2, MSH5, PRKCD, and CD81 stood out, all of them being significant even after statistical correction.
CVID-like variants associate with mutational load in GC
The mutational load observed in GC samples was significantly higher in those exhibiting a CVID-like profile (“CVID-positive group,” ≥2 CVID-like variants, n = 949; median, 4; mean, 12 [range, 2-22]) and also in those with non-CVID variants (“CVID-negative group,” n = 641) (159.5 ± 26.1 vs 43.5 ± 0.7; P < .0001), suggesting a role in tumor immune escape and tumor progression. However, the CVID-positive group had a significantly higher female/male ratio than the CVID-negative group, especially in the younger group (first quartile, <45 years, 73.5% [n = 25]/26.5% [n = 9] vs 18% [n = 7]/82% [n = 32]; P = 5.688 × 10−6). Regarding the co-occurrence in the CVID-positive group, the possible synergy between KMT2C and PLCG2, ARHGEF1, PRKCD, and PIK3R1 as well as between CD84 and FCGR2A, OR10X1, IRF2BP2, and CR2 is striking. In mutual exclusivity, PTEN stands out with genes such as ATP6AP1, SLC25A5, IKZF1, OR10X1, TNFRSF4, and MOGS and KMT2C with genes OR10X1 and FCGR2A, all of them statistically significant (Table II). The co-occurrence and mutual exclusivity of altered CVID-like genes in GC samples are provided in Tables E4 and E5.
Table II
Main genes with co-occurrence and mutual exclusivity of CVID-positive group in GC samples
| Gene A | Gene B | Both | P value | q value | Tendency |
|---|---|---|---|---|---|
| SH3KBP1 | ATP6AP1 | 115 | <.001 | <0.001 | Co-occurrence |
| ATP6AP1 | SLC25A5 | 109 | <.001 | <0.001 | Co-occurrence |
| SH3KBP1 | SLC25A5 | 107 | <.001 | <0.001 | Co-occurrence |
| CD84 | FCGR2A | 36 | <.001 | <0.001 | Co-occurrence |
| CR2 | SH3KBP1 | 34 | <.001 | 0.002 | Co-occurrence |
| OR10X1 | FCGR2A | 32 | <.001 | <0.001 | Co-occurrence |
| OR10X1 | CD84 | 32 | <.001 | <0.001 | Co-occurrence |
| KMT2C | PLCG2 | 32 | .001 | 0.014 | Co-occurrence |
| DOCK8 | LRBA | 30 | <.001 | <0.001 | Co-occurrence |
| CR2 | IRF2BP2 | 27 | <.001 | <0.001 | Co-occurrence |
| KMT2C | ARHGEF1 | 27 | .002 | 0.025 | Co-occurrence |
| KMT2C | PRKCD | 26 | <.001 | <0.001 | Co-occurrence |
| KMT2C | PIK3R1 | 25 | .001 | 0.018 | Co-occurrence |
| IRF2BP2 | CD84 | 24 | <.001 | <0.001 | Co-occurrence |
| CR2 | CD84 | 24 | <.001 | <0.001 | Co-occurrence |
| PTEN | ATP6AP1 | 153 | .006 | 0.047 | Mutual exclusivity |
| PTEN | SLC25A5 | 123 | <.001 | <0.001 | Mutual exclusivity |
| PTEN | IKZF1 | 68 | .006 | 0.044 | Mutual exclusivity |
| PTEN | OR10X1 | 63 | .004 | 0.033 | Mutual exclusivity |
| KMT2C | OR10X1 | 60 | <.001 | 0.012 | Mutual exclusivity |
| KMT2C | FCGR2A | 51 | .005 | 0.043 | Mutual exclusivity |
| PTEN | TNFRSF4 | 40 | <.001 | 0.012 | Mutual exclusivity |
| PTEN | MOGS | 35 | .002 | 0.023 | Mutual exclusivity |
Analysis of a CVID-associated transcriptomic signature in GC
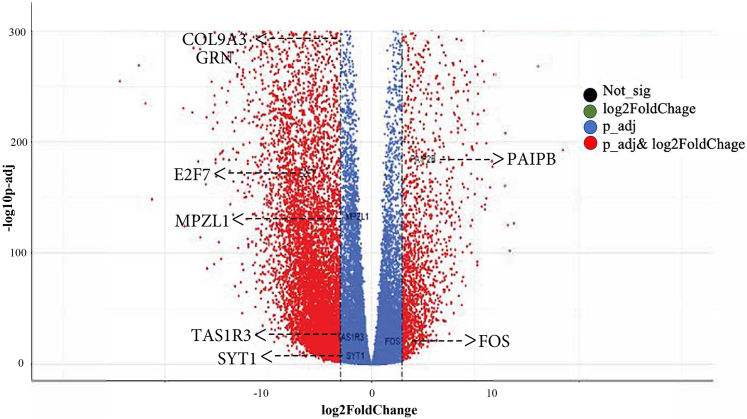
First, we filtered out the expression pattern of healthy gastric tissue. Expression analysis was performed on 766 samples, including 407 GC samples from TCGA and 359 normal gastric tissue samples from GTEx. Individual expression data of each gene were normalized. We detected 20,198 DEGs between both conditions, which included most of the annotated human reference genes. We found 14,717 upregulated genes and 5,481 downregulated genes in GC in comparison with healthy tissue (adjusted P < .005; log2FoldChange > 0.58 [FoldChange = 1.5]).
We then cross-filtered these 20,198 genes differentially expressed in GC with the 112 DEGs in CVID transcriptomic signature31 and matched those genes that showed similar upregulation and downregulation, identifying 6 genes with increased expression (TAS1R3, SYT1, E2F7, COL9A3, MPZL1, and GRN) and 2 genes with decreased expression (PAIPB and FOS) in both CVID and GC (Fig 5).
Discussion
Our results reveal that GC samples have a high prevalence of somatic variants in CVID-associated genes. The alteration of the somatic genes could imply an alteration of host immunity similar to that of CVID but within a local tumor microenvironment, thereby predisposing to and/or directly influencing the development of GC. Three potential scenarios may explain our results: (1) pure de novo somatic CVID-associated variants in the gastric tumor burden; (2) genome mosaicism at the postzygotic stage of CVID-associated variants affecting diverse patients’ tissues including the immune system and gastric tissue; and (3) CVID-associated germline variants in undiagnosed patients with CVID included into the pool of patients with GC. All 3 may be present in different proportions to be deciphered when paired somatic-germline studies become available. All 3 might be driving immune escape and inflammation advantageously for the tumor. Indeed, the mutational load differed between patients with GC harboring CVID-like variants and those who did not, which could be linked to better immunotherapy response because of a higher neoantigen load and tumor immunogenicity36,37 and to worse prognosis.38, 39, 40 These data highlight the biological and clinical relevance of CVID-like variants in GC with direct effect within the tumor and tumor niche affecting prognosis. Alternatively, our findings may also provide unique insight into the mechanisms of GC in CVID and could explain, in part, the worse prognosis of patients with CVID affected with GC.
Despite the genetic heterogeneity of GC and CVID, we observed putative shared alterations in regulators of key processes such as histone methylation, mainly by lysine methyltransferases (KMTs), B-cell activation (SH3KBP1), and proton secretion within the microenvironment (ATP6AP1). KMTs (lysine methyltransferases) have a distinctive impact among the cluster of diverse histone methylation modifiers that play a vital role in GC. The SH3 domain–containing kinase binding protein 1 (SH3KBP1) gene encodes an adapter protein involved in the regulation of diverse signal transduction pathways and is required in the control of cell shape and migration. Patients with SH3KBP1 mutations suffer from severe bacterial infections because of defective antibody production,41 which has also been described as a prognostic factor of pancreatic cancer infiltration by immune cells in tumor microenvironment.42 SH3KBP1 has been found to form a complex with MUC-1 and to play a role in the signaling pathways of gastrointestinal epithelial cells during health and disease.43 Cancer occurrence and progression may be facilitated by aberrant expression of ATPase H+ transporting accessory protein 1 (ATP6AP1) through its ability to increase H+ secretion, resulting in tumor cell survival in hypoxia and an acidic tumor microenvironment.44 The ATP6AP1 protein is altered in numerous tumors, including head and neck carcinoma, lung cancer, and leukemia.45
Zheng et al46 described the prevalence of gastrointestinal cancer in IEI integrating 149 patients from 76 studies, in which CVID was the most frequent immunodeficiency linked to this type of malignancy. Of note, 12% of patients had molecular genetic diagnoses, the 3 most frequently implicated genes being ATM, CARMIL2, and CTLA4; these genes are more involved with DNA repair and cancer development than with the own pathogenesis of CVID.
In line with the genetic and immunologic theory of infection proposed by Jean-Laurent Casanova, according to which severe infections can be due to IEI,47,48 we can extend the reasoning into that cancer might also be due to more complex forms of inheritance of IEI together with triggering infections, because cancer is indeed a major clinical phenotype of IEI. Likewise, to unravel underlying mechanisms by which chronic inflammation promotes many human cancers, it should be noted that chronic infections are associated with an inflammatory state that is increasingly being recognized to induce immune exhaustion and promote tumor growth and progression.49 Cancer occurs because of cumulative oncogenetic variants, raising the question of how such a random process can lead to the described enrichment of CVID-related genes. In parallel to that described for severe infections, it could be possible to infer general mechanisms of protective immunity to a particular cancer, which may pave the way to new avenues of immunotherapy. This approach should be complemented by experimental functional models in specific cell subsets or molecular pathways in host protection against GC, as well as bioinformatic pipelines to identify the potential functional interactions of microbial ligand-human receptor pairs in ex vivo samples. To understand whether and how increased mutation loads have a functional effect and predispose to CVID and cancer or the other way around, single-cell analysis or clones grown out of a single cell may be of great relevance.50
In our study, we also conducted an analysis of DGE with the aim of identifying the transcriptomic signature of CVID in GC samples. An enrichment was observed in pathways involved in the innate immune response against pathogen-associated molecular patterns and type 2 immune responses to parasites and protozoa in the distal small intestine and to viruses (TAS1R3 and SYT1, respectively)51, 52, 53, 54; DNA repair (E2F7)55; cell proliferation, migration, and invasion in several types of cancers (MPZL1)56, 57, 58; and regulation of cell cycle, inflammation, and host-defense response (PGRN).59, 60, 61
Particularly, PGRN, which encodes the precursor protein progranulin, expressed in cells of several tissues, including stromal cells in the bone marrow and cells of the innate and acquired immune system, has been described as a promoter of antibody diversity in B cells and an immune-regulatory molecule of TNF signaling pathways.62,63 It has also been shown to stimulate tumorigenesis by regulating cells’ growth, division, and survival.64, 65, 66 PGRN expression is constitutively high in various solid (breast cancer, ovarian cancer, and cholangiocarcinoma)63,64 and hematological malignancies (diffuse large B-cell lymphoma–non-Hodgkin lymphoma),66,67 the latter being the most frequent malignancy associated with CVID.31 Interestingly, a significant overexpression of PGRN in gastric epithelial cells has been described after prolonged infection by H pylori, inducing a chronic inflammatory state that contributes to an immune dysregulation profile associated with proinflammatory cytokine production.63,68,69
Other interesting gene dysregulations bridging GC and CVID were the elevated expression of the deubiquitinase ubiquitin-specific protease 3 (USP3), an essential GC oncogene and a biomarker of worse prognosis, as well as the upregulation of COL9A3, known to promote epithelial-to-mesenchymal transition, GC progression, and metastasis.70 In contrast, FOS, a member of the leucine zipper protein family encoding genes, was found to be downregulated in patients with CVID. FOS can dimerize with proteins of the JUN family to form the transcription factor complex AP-1. The AP-1 transcription factor JunD activates LILRB1, which plays pivotal roles in immunologic balance.71 LILRB1 binds MHC-I molecules to regulate immune cell function through transduction signals recruiting protein tyrosine phosphatase molecules, leading to negative regulation of immune cell activation.71, 72, 73, 74 Reduced expression of LILRB1 on B cells has been observed in patients with SLE.75
All these genes are closely related to the development of tumors and may provide essential clues about the link between CVID and GC. Ideally, the expression analysis of GC samples with and without a concurrent CVID diagnosis would help understand the pathophysiological implications of these gene dysregulations and their potential impact in early disease and prognosis, but existing data on CVID in published cancer series and repositories remain unfortunately very limited.76
Our study has several limitations. First, it is based on a compilation of heterogeneous data, in which clinical and pathological annotation is not universally consistent or available. Although this limitation is inherent to large-scale mining of genomic databases, further efforts are highly needed to generate databases focused on immunodeficiencies. Second, we explored the representation of CVID-related germline mutations in data of somatic mutations. Although most of the tumor mutations reported in the source studies did not filter out germline mutations, we cannot exclude some bias in the different processing pipelines including mutation calling among the studies. Third, our results are hypothesis-generating and posit a novel avenue of research but will require both dedicated patient cohorts and preclinical models for validation.
We are aware that the data presented in this work strongly indicate the potential genetic association of both conditions. Still, it is necessary to continue the efforts to provide a definitive cause-effect linkage between them. Currently, we are working on several cause-effect studies to resolve this issue.
Our main project is focused on multi-omics integration, using multiomics approaches (genomics, transcriptomics, and proteomics of patients with CVID and GC) to comprehensively understand the molecular pathways linking CVID-related genetic variants to GC to identify the key regulatory networks. In the same way, we planned to explore the cellular and molecular interactions between the immune system, genetic variants, and GC through single-cell analysis and to explore how immune deficiencies associated with CVID may create an environment that could foster GC development.
Conclusions
We provide a first report on the common genomic and transcriptomic grounds of GC and CVID, describing a putative molecular connection between these 2 entities, in which immunodeficiency facilitates tumorigenesis. Our findings may have important clinical implications in different scenarios such as the primary and secondary prevention strategies of early-onset GC in patients with CVID or the investigation of immunotherapies once a GC diagnosis is established. Although challenging, studies involving the omic characterization of prospective cohorts of patients with CVID with gastric and other malignancies are required, including an ongoing institutional effort, to further investigate our findings and improve patient management and outcomes.
Data availability statement: All the data analyzed in the study are included in this article’s Online Repository at www.jaci-global.org. Further inquiries can be directed to the corresponding author.
Ethical considerations: Approval for the study was obtained from the Hospital Clínico San Carlos Institutional Research Ethics Committee (21/504-E). The INSIGHT project (Intersection of Shared Immunodeficiencies Grounded by High-throughput analytical Tools) continues to be active in the Cancer Immunomonitoring and Immuno-Mediated Pathologies Support Unit of the Research Institute San Carlos Health (IdISSC) in the Clinical Immunology Department, Institute of Laboratory Medicine at San Carlos University Hospital, Madrid, Spain. The authors vouch for the accuracy and completeness of the data in this report. This article does not contain any studies with human participants or animals performed by any of the authors.
Disclosure statement
K.G.-H. is supported by the European Social Fund through a Río Hortega Grant (grant no. CM20/00098) and a Juan Rodés Grant (grant no. JR22/00018) for health research projects by the Carlos III Health Institute. J.F.A. is supported by a Fundación Sociedad Española de Oncología Médica fellowship. N.L.R. receives funding from the National Institutes of Health (grant no. R21AI164100) and Takeda Pharmaceuticals.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
References
Articles from The Journal of Allergy and Clinical Immunology: Global are provided here courtesy of Elsevier
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/157751064
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genomic crossroads between non-Hodgkin's lymphoma and common variable immunodeficiency.
Front Immunol, 13:937872, 05 Aug 2022
Cited by: 7 articles | PMID: 35990641 | PMCID: PMC9390007
The Dysfunctional Immune System in Common Variable Immunodeficiency Increases the Susceptibility to Gastric Cancer.
Cells, 9(6):E1498, 19 Jun 2020
Cited by: 7 articles | PMID: 32575504 | PMCID: PMC7349552
Telomere biology disorders may manifest as common variable immunodeficiency (CVID).
Clin Immunol, 257:109837, 08 Nov 2023
Cited by: 1 article | PMID: 37944684
Understanding the genetic and epigenetic basis of common variable immunodeficiency disorder through omics approaches.
Biochim Biophys Acta, 1860(11 pt b):2656-2663, 15 Jun 2016
Cited by: 14 articles | PMID: 27316315
Review
Funding
Funders who supported this work.
European Social Fund Plus (2)
Grant ID: JR22/00018
Grant ID: CM20/00098
Instituto de Salud Carlos III
National Institutes of Health (1)
Grant ID: R21AI164100