Abstract
Background
Abiotic stresses in plants include all the environmental conditions that significantly reduce yields, like drought and heat. One of the most significant effects they exert at the cellular level is the accumulation of reactive oxygen species, which cause extensive damage. Plants possess two mechanisms to counter these molecules, i.e. detoxifying enzymes and non-enzymatic antioxidants, which include many classes of specialized metabolites. Sunflower, the fourth global oilseed, is considered moderately drought resistant. Abiotic stress tolerance in this crop has been studied using many approaches, but the control of specialized metabolites in this context remains poorly understood. Here, we performed the first genome-wide association study using abiotic stress-related specialized metabolites as molecular phenotypes in sunflower. After analyzing leaf specialized metabolites of 450 hybrids using liquid chromatography-mass spectrometry, we selected a subset of these compounds based on their association with previously known abiotic stress-related quantitative trait loci. Eventually, we characterized these molecules and their associated genes.Results
We putatively annotated 30 compounds which co-localized with abiotic stress-related quantitative trait loci and which were associated to seven most likely candidate genes. A large proportion of these compounds were potential antioxidants, which was in agreement with the role of specialized metabolites in abiotic stresses. The seven associated most likely candidate genes, instead, mainly belonged to cytochromes P450 and glycosyltransferases, two large superfamilies which catalyze greatly diverse reactions and create a wide variety of chemical modifications. This was consistent with the high plasticity of specialized metabolism in plants.Conclusions
This is the first characterization of the genetic control of abiotic stress-related specialized metabolites in sunflower. By providing hints concerning the importance of antioxidant molecules in this biological context, and by highlighting some of the potential molecular mechanisms underlying their biosynthesis, it could pave the way for novel applications in breeding. Although further analyses will be required to better understand this topic, studying how antioxidants contribute to the tolerance to abiotic stresses in sunflower appears as a promising area of research.Free full text

Genetic control of abiotic stress-related specialized metabolites in sunflower
Abstract
Background
Abiotic stresses in plants include all the environmental conditions that significantly reduce yields, like drought and heat. One of the most significant effects they exert at the cellular level is the accumulation of reactive oxygen species, which cause extensive damage. Plants possess two mechanisms to counter these molecules, i.e. detoxifying enzymes and non-enzymatic antioxidants, which include many classes of specialized metabolites. Sunflower, the fourth global oilseed, is considered moderately drought resistant. Abiotic stress tolerance in this crop has been studied using many approaches, but the control of specialized metabolites in this context remains poorly understood. Here, we performed the first genome-wide association study using abiotic stress-related specialized metabolites as molecular phenotypes in sunflower. After analyzing leaf specialized metabolites of 450 hybrids using liquid chromatography-mass spectrometry, we selected a subset of these compounds based on their association with previously known abiotic stress-related quantitative trait loci. Eventually, we characterized these molecules and their associated genes.
Results
We putatively annotated 30 compounds which co-localized with abiotic stress-related quantitative trait loci and which were associated to seven most likely candidate genes. A large proportion of these compounds were potential antioxidants, which was in agreement with the role of specialized metabolites in abiotic stresses. The seven associated most likely candidate genes, instead, mainly belonged to cytochromes P450 and glycosyltransferases, two large superfamilies which catalyze greatly diverse reactions and create a wide variety of chemical modifications. This was consistent with the high plasticity of specialized metabolism in plants.
Conclusions
This is the first characterization of the genetic control of abiotic stress-related specialized metabolites in sunflower. By providing hints concerning the importance of antioxidant molecules in this biological context, and by highlighting some of the potential molecular mechanisms underlying their biosynthesis, it could pave the way for novel applications in breeding. Although further analyses will be required to better understand this topic, studying how antioxidants contribute to the tolerance to abiotic stresses in sunflower appears as a promising area of research.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10104-9.
Background
Abiotic stresses in plants can be defined as all the environmental conditions that decrease growth and yield below optimum levels [1]. They include, among others, drought, salinity, low and high temperatures, nutrient deficiencies, and ultraviolet radiation [2]. The impact of most of these stresses is becoming more severe because of climate change [1]. Abiotic stresses exert their effects in many complex and diverse ways, and plants have evolved a vast array of mechanisms to cope with them. Some of these mechanisms include the accumulation of wax and cutin on leaf surfaces, the desaturation of membrane lipids, and the accumulation of compatible solutes [2].
At the molecular level, one of the most significant effects of abiotic stresses is the accumulation of reactive oxygen species (ROS) [3], which arises from an imbalance between ROS production and scavenging [4, 5]. ROS are strong oxidizers and cause extensive damage to many biological molecules, like for instance proteins, lipids, and DNA [3, 5].
Plants use two mechanisms to counterbalance oxidative stress. The first is represented by detoxifying enzymes, such as superoxide dismutase, catalase, ascorbate peroxidase, and glutathione reductase [4]. The second corresponds to non-enzymatic antioxidants, i.e. ascorbic acid, reduced glutathione, α-tocopherol, and several classes of secondary or specialized metabolites such as carotenoids, flavonoids, and phenolic acids, whose ROS-scavenging activity has been demonstrated across many plant species [5, 6].
Terpenes are another class of specialized metabolites with antioxidant properties. Although better known as constituents of essential oils, allelopathic agents, and attractants or repellants in plant–herbivore interactions [7], there is increasing evidence of their implication in ROS scavenging [8–10]. Taken together, it can be stated that most specialized metabolites induced by abiotic stresses show antioxidative activity [11], which makes these molecules key players for plant adaptation to more stressful environments and for breeding tolerant varieties.
Sunflower (Helianthus annuus L.) is the fourth most important oilseed worldwide. It can maintain stable yields across many conditions and, largely thanks to its well-developed tap roots, it is adapted to low water-input regimes in warm to semi-arid zones [12]. Although this crop is usually considered moderately drought tolerant, the challenges posed by climate change will require major efforts in terms of breeding and crop management.
To cope more efficiently with hydric stress, tolerant varieties will have to be developed [13], and tolerance to heat will have to be jointly prioritized, because high temperatures dramatically affect pollination, fertilization, and seed set [13]. Another way to avoid drought is early sowing. This strategy allows to anticipate the timing of flowering, thus avoiding the summer periods in which evaporative demand is higher [13, 14]. However, this practice presents side-effects, because the crop is more exposed to cold stress at germination [15]. From this perspective, developing hybrids with improved tolerance to cold will be another relevant goal.
The molecular mechanisms underlying tolerance to abiotic stresses in sunflower have been studied using different approaches over the last years. Non-targeted metabolomics and proteomics have been used to profile a set of inbred lines and hybrid genotypes [16, 17] and to find biomarkers for drought tolerance [18], while transcriptome and metabolome have been integrated to identify transcription factors regulated under the same condition [19].
Transcriptome profiling has been used to describe the impact of drought using co-expression networks [20], differential analysis [21], differential analysis coupled to association genetics [22] and gene-phenotype networks [23]. It has also been chosen to characterize low-nutrient stress and three water-related stresses [24]. Eventually, tolerance to salt stress and its link with vigor have been studied through association genetics [25]. Nevertheless, to date no information is available concerning the genetic control of specialized metabolome in sunflower under abiotic stress.
In this work, we present the results of the first GWAS performed in sunflower using specialized metabolites related to abiotic stresses as molecular phenotypes. Our approach consisted of three main steps. First, we analyzed the semi-polar fraction of leaf extracts of a panel of sunflower hybrids using untargeted liquid chromatography-mass spectrometry (LC-MS), which allowed us to focus our analysis on specialized metabolites. Second, we selected a subset of these compounds based on their genetic association with some previously known quantitative trait loci (QTLs) related to yield and abiotic stress tolerance. Third, we characterized in silico these compounds and the genes associated with them.
It has also to be noticed that, in addition to our own work, GWAS using molecular phenotypes in sunflower has been used so far only in another case, i.e. to disentangle the genetic basis of oil fatty acid content [26]. Our results can therefore be considered original by a methodological point of view.
Results
Association mapping
To study the genetic control of specialized metabolites in sunflower and how this relates to abiotic stress tolerance, we obtained the metabolomic profiles of the leaves of 450 hybrids originating from crosses among 36 restorer and 36 cmsPET1 sterile lines grown in agronomical conditions. After the partial removal of redundancy due to isotopes and adducts, the final metabolome dataset consisted of 2557 LC-MS features (Table S01) characterized by a retention time (RT) and a mass over charge ratio (m/z) (Table S02). A total of 21 features already had an annotation based on previous works (Table S02) [16, 18, 27].
A PCA performed using these data showed that the first two principal components accounted for 11% and 5% of total variability, which was consistent with the results obtained in similar contexts [28]. A clustering of most of the hybrids according to their male parental line was observed (Fig. 1). This was especially evident for SF295, SF324, SF330, SF342 and SF281 male lines, but could be observed in other cases as well. On the contrary, no clustering was observed according to female parental lines.
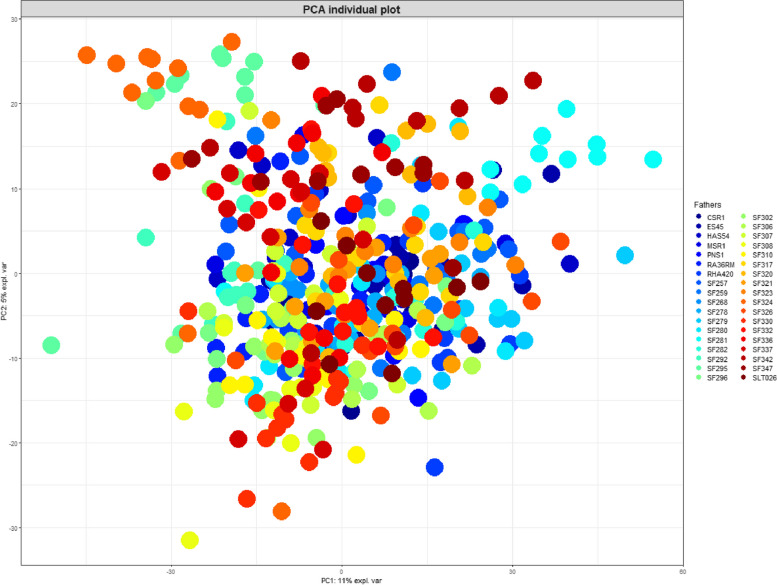
Individual plot of the first two components of the PCA based on the normalized intensities of 2557 LC-MS features measured in the leaves of 450 sunflower hybrids. The different hybrids are colored according to their male parental lines, which are indicated in the box on the right
The 2557 LC-MS features were then used as an input for the first step of the association analysis, which consisted in performing GWAS using reference SNPs. A visual overview of this analysis step, as well as all the other ones included in our workflow, is found in Fig. 2. Similarly to what observed in other plant species [29], 955 LC-MS features (i.e. 37.3% of the total number used) were associated to at least one SNP. This corresponded to 2560 associations (Table S03). On average, an LC-MS feature was therefore associated to 2.7 SNPs, with 472 features associated to only one SNP and 483 features associated to two to 19 SNPs.
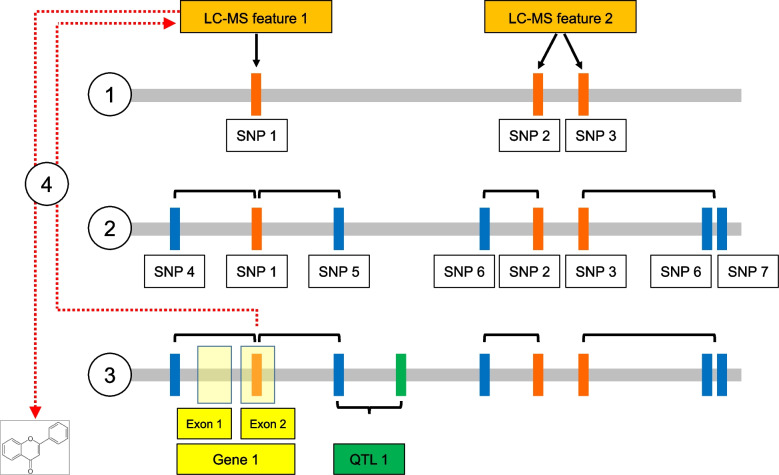
Graphical abstract illustrating the main steps of our analysis workflow. (1) First step of GWAS: detection of the associations among LC-MS features (orange boxes) and reference SNPs (red vertical bars); (2) Second step of GWAS: reference SNPs are linked to co-inherited SNPs sets (blue vertical bars); (3) Co-localization among co-inherited SNPs and SNPs belonging to abiotic stress-related QTLs (green vertical bar). Co-inherited SNPs falling in a 50 kb interval downstream or upstream of an SNP belonging to a QTL are considered co-localizing with the same QTL. All the SNPs in complete linkage disequilibrium (LD) with co-localizing SNPs and mapping to exons (yellow transparent boxes) are then used to identify putative candidate genes; (4) LC-MS features associated to reference SNPs in complete LD with co-localizing SNPs are selected and tentatively annotated
kb interval downstream or upstream of an SNP belonging to a QTL are considered co-localizing with the same QTL. All the SNPs in complete linkage disequilibrium (LD) with co-localizing SNPs and mapping to exons (yellow transparent boxes) are then used to identify putative candidate genes; (4) LC-MS features associated to reference SNPs in complete LD with co-localizing SNPs are selected and tentatively annotated
Among the 955 significantly associated LC-MS features, 798 (i.e. 83.6%) presented genomic heritability (h2g) values higher than 0.50, the average being 0.66 (Fig. S01), which was in line with what reported in the literature [29, 30]. The 2560 detected associations corresponded to 1716 unique reference SNPs, with 378 SNPs associated to two to 43 features. This suggested that many LC-MS features were under pleiotropic control, as already described in other species [31, 32], or that some biochemical information redundancy was still present in our data set after LC-MS data filtering. Five features already possessed an annotation based on previous works (Table S03).
The second step of association analysis consisted in linking reference SNPs to their corresponding sets of co-inherited SNPs, which included all the SNPs in complete linkage disequilibrium (LD) with them (see Fig. Fig.22 and the Methods section). After this step, 62,134 associations were found. The number of associations per LC-MS feature ranged from one to 3666, the average being of 65.1 (Table S04). Overall, these associations corresponded to 27,246 unique SNPs.
To gather a first functional understanding of the genetic control of sunflower specialized metabolome, we then investigated all the possible associations among LC-MS features and genes in an unsupervised way, i.e. without any further biological information. A gene was considered associated to a feature if at least one SNP of a co-inherited set mapped to one of its exons (Fig. (Fig.2).2). Our analysis highlighted that 1768 SNPs belonging to co-inherited sets out of 27,246 (i.e. 6.5%) were found in exons.
Exonic SNPs corresponded to 533 genes, with an average of 3.3 associations per gene (Table S05). These genes appeared to be involved in several pathways, with a slight over-representation of those related to glutathione, lipids and specialized metabolite biosynthesis, like for instance flavonoids. Anyway, it must be considered that the levels of enrichment, especially in terms of numbers of genes associated to each ontology, were rather low (Table S06).
As observed in other species [29, 30], metabolite-associated SNPs and genes were not randomly distributed across the genome, but appeared to be especially concentrated in some specific ‘hot spots’ (Fig. S02). To explore this pattern of distribution, we first used a sliding window approach and subsequently, by applying a threshold based on the proportion and on the absolute number of metabolite-associated genes in each window, we defined six hot spots on chromosomes 5, 6, 7, 9, 12 and 16. These regions contained 88 metabolite-related genes and spanned 48 Mb, which corresponds to a sixth of all metabolite-related genes on slightly less than 1.5% of the sunflower genome (Table S07). In some instances, these genes were arranged in small families, like in the case of the hot spot on chromosome 5, which contained six putative quinate O-hydroxycinnamoyltransferase, and in the case of the hot spot on chromosome 6, where seven putative glutathione transferases were detected (Table S07). Anyway, despite the presence of these specific patterns, we could not find any evidence of functional metabolic clusters as defined by Nützmann and coworkers [33].
Mb, which corresponds to a sixth of all metabolite-related genes on slightly less than 1.5% of the sunflower genome (Table S07). In some instances, these genes were arranged in small families, like in the case of the hot spot on chromosome 5, which contained six putative quinate O-hydroxycinnamoyltransferase, and in the case of the hot spot on chromosome 6, where seven putative glutathione transferases were detected (Table S07). Anyway, despite the presence of these specific patterns, we could not find any evidence of functional metabolic clusters as defined by Nützmann and coworkers [33].
Co-localization of SNPs associated to LC-MS features with QTLs related to abiotic stresses
As illustrated in Fig. Fig.2,2, to study the genetic control of specialized metabolites linked to abiotic stresses we tested the co-localization of co-inherited SNPs associated to LC-MS features with previously identified QTL regions for drought, cold, and nutrient stress tolerance and for productivity and development-related traits [15, 34]. These last two groups of traits were added because they were considered as indirectly related to abiotic stress tolerance.
We detected a total of 638 SNPs that co-localized with 20 QTLs, among which seven were related to drought stress tolerance, five to cold stress tolerance, four to nutrient stress tolerance and four to productivity and development-related traits, with the QTLs for abiotic stress tolerances showing some overlap among them (Table S08). We then took these 638 co-localizing SNPs and searched for all the other SNPs which were in complete LD with them, and which were 10,793. On the one hand, these co-inherited SNPs were associated to 137 LC-MS features (Table S09), of which 16 were related to drought stress tolerance, 92 to cold stress tolerance, four to nutrient stress tolerance and 17 to productivity and development-related traits. A further eight LC-MS features were associated to two QTLs at the same time (Table S09). On the other hand, the same co-inherited SNPs were also associated to 155 putative candidate genes (Table S09).
Annotation of LC-MS features of interest and identification of the most likely candidate genes
Because the LC-MS protocol used in this work did not involve data dependent MS/MS (i.e. tandem mass spectrometry; see the Methods section), the only way to perform the annotation of the 137 previously identified LC-MS features of interest was by relying on an in silico workflow. This procedure allowed to tentatively annotate 30 features, among which one was related to drought stress tolerance, 21 to cold stress tolerance, one to nutrient stress tolerance and four to productivity and development-related traits. A further three features were associated to two QTLs at the same time. Most of the annotated molecules (Table 1) belonged to the biochemical classes of terpenes (30%), flavonoids (17%), polyacetylenes (17%) and cinnamic acids (10%).
Table 1
Putative annotation of the 30 LC-MS features measured in the leaves of sunflower hybrids and co-localizing with QTLs of interest. All the metabolites were assigned an MSI level 3 (see Methods)
| LC-MS feature | [M + + H]+ H]+m/z | RT (min) | isotopic pattern ion | putative chemical formula for [M + + H]+ H]+ | putative metabolite name | metabolite ID | metabolite class |
|---|---|---|---|---|---|---|---|
| M201T581 | 201.1637 | 9.69 | M0 | C15H21 | 4,5,9,10-dehydroisolongifolene isomer A | CID: 588771 | sesquiterpenes |
| M203T379 | 203.1066 | 6.32 | M0 | C13H15O2 | demethoxyencecalin | CID: 177040 | chromenes |
| M287T548 | 287.0550 | 9.14 | M0 | C15H11O6 | cyanidin | CID: 128861 | flavonoids |
| M289T534 | 289.0706 | 8.89 | M0 | C15H13O6 | pentahydroxychalcone | CID: 129636553 | flavonoids |
| M307T374 | 307.1288 | 6.23 | M0 | C15H19O5N2 | brachystemidine A | CID: 10892118 | pyrroles |
| M349T519 | 349.0917 | 8.64 | M0 | C17H17O8 | hexahydroxydimethylflavanone | – | flavonoids |
| M131T321 | 131.0490 | 5.36 | M0 | C9H7O | 2-nonene-4,6,8-triyn-1-ol | CID: 57449462 | polyacetylenes |
| M294T138 | 294.1547 | 2.31 | M0 | C12H24O7N | deoxyfructosyl-leucine | CID: 131752244 | amino acid derivatives |
| M145T650 | 145.1011 | 10.83 | M0 | C11H13 | 1,9-undecadiene-5,7-diyne | CID: 15736690 | polyacetylenes |
| M151T299 | 151.0754 | 4.99 | M0 | C9H11O2 | coumaryl-alcohol isomer A | CID: 5280535 | cinnamic acids |
| M151T342 | 151.0754 | 5.71 | M0 | C9H11O2 | coumaryl-alcohol isomer B | CID: 5280535 | cinnamic acids |
| M165T301 | 165.0545 | 5.02 | M0 | C9H9O3 | coumaric acid | CID: 637542 | cinnamic acids |
| M173T648 | 173.1324 | 10.80 | M0 | C13H17 | 1,4-tridecadiene-7,9-diyne | CID: 101410972 | polyacetylenes |
| M175T609 | 175.0753 | 10.14 | M0 | C11H11O2 | (2E,4E)-5-phenylpenta-2,4-dienoic acid | CID: 1549512 | styrenes |
| M201T687 | 201.1637 | 11.45 | M0 | C15H21 | 4,5,9,10-dehydroisolongifolene isomer B | CID: 588771 | sesquiterpenes |
| M202T649 | 202.1671 | 10.81 | M1 | C15H21 | 4,5,9,10-dehydroisolongifolene isomer C | CID: 588771 | sesquiterpenes |
| M217T336 | 217.1585 | 5.60 | M0 | C15H21O | (3S,9Z)-pentadeca-9,14-dien-4,6-diyn-3-ol | CID: 163193273 | polyacetylenes |
| M231T299 | 231.1378 | 4.99 | M0 | C15H19O2 | dehydrocostus lactone isomer A | CID: 73174 | sesquiterpenes |
| M231T362 | 231.1378 | 6.03 | M0 | C15H19O2 | dehydrocostus lactone isomer B | CID: 73174 | sesquiterpenes |
| M231T604 | 231.1378 | 10.06 | M0 | C15H19O2 | dehydrocostus lactone isomer C | CID: 73174 | sesquiterpenes |
| M243T272 | 243.0881 | 4.54 | M0 | C12H11O2N4 | lumichrome | CID: 5326566 | alloxazines |
| M247T334 | 247.1327 | 5.57 | M0 | C15H19O3 | annuolide A | CID: 44583825 | sesquiterpenes |
| M249T273 | 249.1484 | 4.55 | M0 | C15H21O3 | annuolide E | CID: 131752335 | sesquiterpenes |
| M266T572 | 266.1467 | 9.53 | M1 | C15H21O4 | heliannuol F | CID: 10730325 | sesquiterpenes |
| M273T341 | 273.1485 | 5.69 | M0 | C17H21O3 | 8-acetoxy-1,9,14-pentadecatriene-4,6-diyn-3-ol | CID: 14037439 | polyacetylenes |
| M273T635_2 | 273.2213 | 10.59 | M0 | C19H29O | androst-5-en-4-one | CID: 22213540 | steroids |
| M299T342 | 299.1278 | 5.70 | M0 | C18H19O4 | 3-phenyl-1-(2,3,4-trimethoxyphenyl)prop-2-en-1-one | CID: 67199147 | flavonoids |
| M346T682 | 346.1001 | 11.37 | M1 | C18H17O7 | tambulin | CID: 5281700 | flavonoids |
| M372T338 | 372.1288 | 5.64 | M0 | C16H22O9N | 4-(2-amino-3-hydroxyphenyl)-4-oxobutanoic acid glucoside | CID: 10948689 | phenolic compounds |
| M515T343 | 515.2122 | 5.72 | M0 | C24H35O12 | eugenol acetylrhamnosylglucoside | – | phenolic compounds |
Subsequently, the results obtained from GWAS showed that 13 out of the 30 aforementioned metabolites were associated to at least one gene, corresponding to a total of 80 ‘initial’ genes (Table S10). Two metabolites, namely 1,4-tridecadiene-7,9-diyne (a polyacetylene) and 4,5,9,10-dehydroisolongifolene (a terpene), were associated to the same genes, potentially suggesting pleiotropy.
After the process of functional characterization of all of these associations, we focused on a final set of four metabolites that could be related to seven most likely candidate genes (Table 2). Three associations involved metabolites and enzyme-encoding genes, namely (i) the flavonoid pentahydroxychalcone and a member of the P450 cytochrome family; (ii) the sesquiterpene heliannuol F and a uridine diphosphate (UDP) glucosyltransferase (UGT); (iii) the flavonoid hexahydroxydimethylflavanone and three UGTs. The sesquiterpene 4,5,9,10-dehydroisolongifolene, instead, was associated to two transcription factors (TFs) of the AP2/ERF family (Table (Table22).
Table 2
List of the four tentatively annotated metabolites that were associated to seven most likely candidate genes. Column seven reports the results of the tBlastN analysis when they appear to be helpful in clarifying the already available gene descriptions
| LC-MS feature | putative metabolite name | metabolite class | v2.1 gene | v2.1 gene description | tBLASTn result | co-inherited v1.0 SNP | type of mutation | base mutation | amino acid change | reference v1.0 SNP |
|---|---|---|---|---|---|---|---|---|---|---|
| M201T581 | 4,5,9,10-dehydroisolongifolene | sesquiterpenes | HanXRQr2_Chr07g0314841 | putative transcription factor AP2-EREBP family | AP2/ERF transcription factor | HanXRQChr07_98876273 | synonymous | HanXRQChr07_97029247 | ||
| HanXRQr2_Chr07g0314871 | putative transcription factor AP2-EREBP family | AP2/ERF transcription factor | HanXRQChr07_98748814 | missense | T > > G G | ASP>GLU | HanXRQChr07_97029247 | |||
| M266T572 | heliannuol F | sesquiterpenes | HanXRQr2_Chr09g0363371 | putative UDP-glucuronosyl/UDP-glucosyltransferase | Glycosyltransferase 74G1, likely family 1 | HanXRQChr09_10756950 | missense | G > > A A | ALA>THR | HanXRQChr09_10756950 |
| HanXRQChr09_10757103 | missense | G > > T T | ALA>SER | HanXRQChr09_10756950 | ||||||
| M289T534 | pentahydroxychalcone | flavonoids | HanXRQr2_Chr03g0130651 | putative cytochrome P450 | Cytochrome P450 | HanXRQChr03_139760168 | synonymous | HanXRQChr03_139760112 | ||
| M349T519 | hexahydroxydimethylflavanone | flavonoids | HanXRQr2_Chr11g0515571 | putative UDP-glucuronosyl/UDP-glucosyltransferase | Glycosyltransferase 73E1, likely family 1 | HanXRQChr11_157852754 | missense | G > > A A | ASP>ASN | HanXRQChr11_157849826 |
| HanXRQChr11_157852773 | missense | C > > T T | SER > > LEU LEU | HanXRQChr11_157849826 | ||||||
| HanXRQr2_Chr11g0515581 | hypothetical protein | Glycosyltransferase 73E1, likely family 1 | HanXRQChr11_157868396 | missense | A > > G G | LYS > > ARG ARG | HanXRQChr11_157849826 | |||
| HanXRQChr11_157868891 | missense | C > > T T | ALA>VAL | HanXRQChr11_157849826 | ||||||
| HanXRQChr11_157868836 | missense | G > > T T | ASP>TYR | HanXRQChr11_157849826 | ||||||
| HanXRQr2_Chr11g0515591 | Putative trans-zeatin O-beta-D-glucosyltransferase | Glycosyltransferase 73E1, likely family 1 | HanXRQChr11_157874063 | synonymous | HanXRQChr11_157849826 | |||||
| HanXRQChr11_157873968 | missense | C > > T T | TYR > > HIS HIS | HanXRQChr11_157849826 | ||||||
| HanXRQChr11_157874469 | missense | A > > G G | ARG > > GLY GLY | HanXRQChr11_157849826 |
Overall, these genes were linked to 13 SNPs belonging to co-inherited sets, 10 of which caused missense mutations (Table (Table2).2). Eventually, the genotypic boxplots corresponding to the four characterized metabolites showed in all of the cases a good correlation among the different allelic states and the phenotypic values of the LC-MS features (Fig. 3).
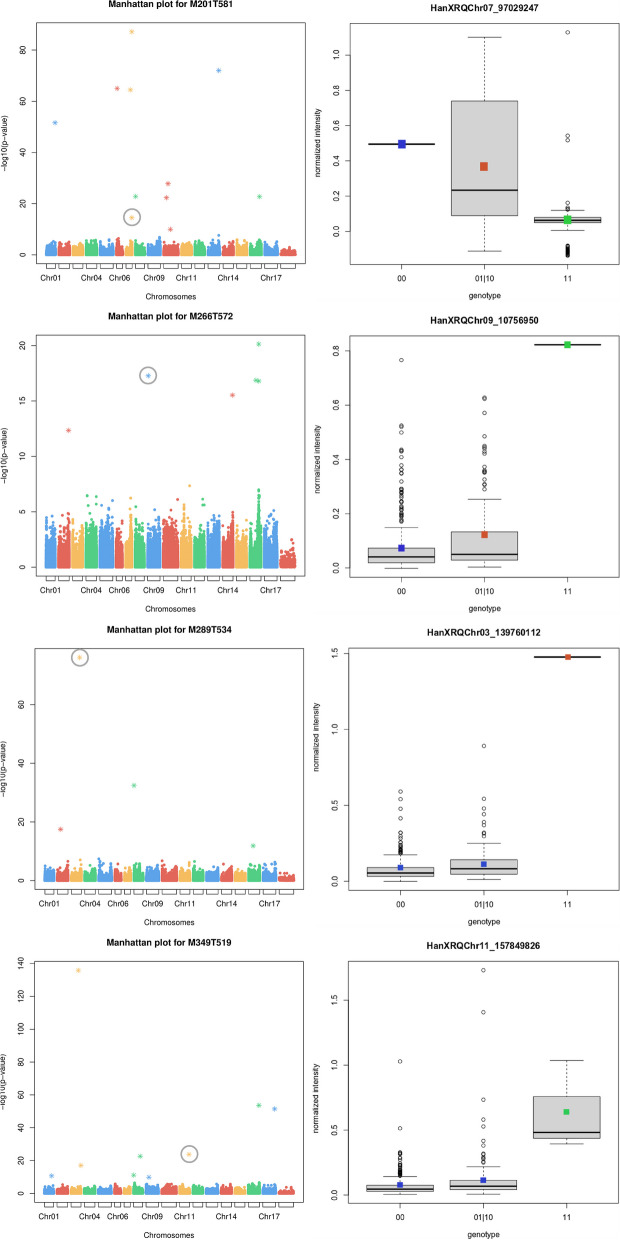
Manhattan plots and genotypic boxplots of the functionally characterized metabolites, here indicated using the codes of LC-MS features. For each metabolite, the corresponding Manhattan plot (left) and genotypic boxplot (right) are shown. Manhattan plots show the reference SNPs obtained from GWAS on the X-axis and the corresponding p-values on the Y-axis. SNPs filtered according to the eBIC criterion are shown as asterisks (Table S03). The reference SNP associated to a most likely candidate gene is highlighted by a grey circle. Genotypic boxplots show the normalized intensity values of the corresponding LC-MS feature (Y-axis) grouped according to the three possible allelic states (i.e. 00, 01|10, and 11). The classes identified with the Tukey’s test are indicated using colored squares. It is to note that the SNPs used to produce the genotypic boxplots are, by definition, reference SNPs, and therefore they are not the same ones found associated to the corresponding most likely candidate gene, which belong instead to co-inherited sets. The correspondences among the reference SNPs used for genotypic plots and the co-inherited used to identify most likely candidate genes are given in Table Table22
Discussion
Potential antioxidants represent a large proportion of sunflower leaf metabolome under abiotic stress
Oxidative damage is one of the most important modifications induced by abiotic stresses in plant cells. It is caused by the accumulation of ROS and has been reported, for instance, in the cases of drought, cold and salinity [5, 11]. Plants use many strategies to mitigate the impact of ROS, one of which is the biosynthesis of non-enzymatic antioxidants. These compounds include, among others, several important groups of semi-polar specialized metabolites like carotenoids, flavonoids, and phenolic acids [6]. Indeed, the majority of specialized metabolites produced by plants under abiotic stress show antioxidative activity in vitro, even if an in vivo experimental confirmation of their function is still lacking in many cases [11].
In this work, we studied the metabolome of 450 sunflower hybrids by performing LC-MS on the semi-polar fraction of leaf extracts, thus specifically targeting specialized metabolites. We then selected a subset of these compounds based on their genetic association to some previously known QTLs which were mainly related to abiotic stresses and, to a lesser extent, development-related traits. Eventually, we annotated this subsect of molecules. However, it must be considered that because tandem mass spectrometry had not been performed and because MS commercial standards were not available for the large majority of these metabolites, only putative annotations could be assigned.
Most of the tentatively annotated metabolites belonged to four biochemical classes, namely terpenes, flavonoids, polyacetylenes and cinnamic acids (Table (Table1).1). These findings were in line with our previous research on sunflower, and specifically with the works focusing on drought stress [16, 18]. However, the detection of polyacetylenes was unique to this study.
Flavonoids, represented by five molecules in our data, are among the best characterized ROS scavengers in plants [7, 35]. Their activity is known to provide tolerance towards many abiotic stresses, like drought [36], cold [37], and nutrient depletion [38].
Even if better known as constituents of essential oils or allelopathic agents [7], there is increasing evidence that terpenes are also involved in ROS scavenging, as described for instance in tea plant [10], sage, and rosemary [8, 9]. In other instances, the involvement of terpenes in abiotic stress tolerance has been demonstrated, even if the underlying antioxidant mechanism has not been proven yet [39].
The class of cinnamic acids, here intended as including all the derivatives of cinnamic acid, is found in most plant families, including Asteraceae [18, 27, 40]. Chlorogenic acid shows antioxidant properties, although this could be true also in the case of other cinnamic derivatives [41]. However, no ROS-scavenging activity has been demonstrated in the case of coumaric acid and coumaryl alcohol. This latter molecule, instead, is one of the precursors of lignin [42] whose accumulation, in turn, is increased for instance under drought and cold [43].
Polyacetylenes are found in a few botanical families, Apiaceae and Asteraceae being among the most relevant [44, 45]. They exhibit a wide range of antibacterial, antifungal, and insecticidal activities [45, 46].
The remaining annotated compounds belonged to several different classes. Phenolic acids, represented by 4-(2-amino-3-hydroxyphenyl)-4-oxobutanoic acid glucoside and eugenol acetylrhamnosylglucoside, are well characterized as antioxidants in plants [6, 47].
The mammal steroid androstenone is found in many plant species [48]. Mammal steroids in plants are known to be involved in processes such as root and shoot growth [49], but no information is specifically available for androstenone.
Demethoxyencecalin is a chromene. The compounds in this class have been described in plants such as mulberry [50] and Hypericum polyanthemum [51], but they are especially frequent in Asteraceae [52]. They are repellent towards herbivorous insects.
(2E,4E)-5-phenylpenta-2,4-dienoic acid belongs to styrenes, which are naturally synthesized for instance by mulberry [50] and styrax [53]. Some styrenes show antifeedant activity against insects in pear [54], but no evidence is available in the case of (2E,4E)-5-phenylpenta-2,4-dienoic acid.
The last three molecules highlighted by our study are lumichrome (i.e. an alloxazine), brachystemidine A, (i.e. a pyrrole), and deoxyfructosyl-leucine (i.e. an amino acid derivative). Their role is not clear and therefore difficult to discuss in our biological context.
Altogether, it is possible to affirm that a relevant fraction of the metabolites that we have tentatively annotated, i.e. from seven (considering flavonoids and phenolic acids) to 16 (including also terpenes) out of 30, fit in biochemical classes with oxygen scavenging properties.
Although the number of characterized molecules in our work is relatively small, and even if their specific in vivo activity has not been proven yet, our findings can be considered in agreement with the high proportion of antioxidants observed by many authors in abiotic stress-related specialized metabolome [11, 31, 55, 56].
The most likely candidate genes mainly belong to two highly diverse superfamilies
Interestingly, the five enzyme-encoding most likely candidate genes found in our study belonged to only two families, namely cytochromes P450 (CYPs) and glycosyltransferases (GTs). Although very different in their functionalities, both of them are large and catalyze greatly diverse reactions that create a vast array of chemical modifications. This is consistent with the high level of plasticity of specialized metabolism pathways in plants [57].
The reactions catalyzed by CYPs play a basic role in defining the skeletal structure of many metabolites, such as flavonoids and terpenes [58, 59]. These reactions include hydroxylations, reductive activations, ring couplings, ring formations, ring expansions and oxidative aryl migrations [60, 61].
Today, a large number of CYPs involved in flavonoid biosynthesis are known in many plant families, including Asteraceae [62, 63]. It is therefore possible to hypothesize that the enzyme encoded by the HanXRQr2_Chr03g0130651 gene performs one of the molecular reactions that lead to the biosynthesis of the flavonoid pentahydroxychalcone.
Unlike cytochromes P450, all glycosyltransferases catalyze the same type of reaction, i.e. the transfer of a sugar moiety to an acceptor. Anyway, they act on a broad range of compounds such as lipids, proteins, nucleic acids and other molecules [64].
GTs are classified in many families according to the CAZy database (http://www.cazy.org/). Family 1 is defined by the presence of a specific domain [64, 65] and is usually referred to as UDP-glycosyltransferases (UGTs). Plant UGTs are especially involved in the glycosylation of specialized metabolites such as flavonoids and terpenes [66].
Glycosylation increases the activity and the availability of both these categories of compounds and hence the ROS-scavenging capacity of the plant, which confers tolerance to several abiotic stresses. In Arabidopsis, for instance, the enzymes encoded by UGT79B2 and UGT79B3 add a UDP-rhamnose to the flavonoids cyanidin and cyanidin 3-O-glucoside. An increased concentration of these two molecules provides tolerance to cold, salinity and drought [67]. In tea plant, instead, CsUGT78A14–1 and CsUGT78A14–2 are involved in the biosynthesis of the flavonoids kaempferol 3-O-glucoside and kaempferol diglucoside, which reduces oxidative damage and increases tolerance to cold stress [68]. Another similar example is provided by the action of sesquiterpene nerolidol in tea plant. Again, this metabolite is glycosylated by the protein encoded by CsUGT91Q2, which causes an enhanced level of tolerance against cold stress [10].
In light of this, it could be speculated that the three UGTs associated to the flavonoid hexahydroxydimethylflavanone, i.e. HanXRQr2_Chr11g0515571, HanXRQr2_Chr11g0515581 and HanXRQr2_Chr11g0515591, could be involved in the glycosylation of this molecule, thus contributing to reduce the impact of oxidative damage in sunflower. Likewise, the UGT HanXRQr2_Chr09g0363371 could be involved in ROS scavenging through the glycosylation of the terpene heliannuol F.
Besides enzyme-encoding genes, we also found that two transcription factors of the AP2/ERF family, i.e. HanXRQr2_Chr07g0314841 and HanXRQr2_Chr07g0314871 were associated to the terpene 4,5,9,10-dehydroisolongifolene. Because some transcription factors of this family are implicated in the biosynthesis of terpenes, as in the cases of orange [69] and Litsea cubeba [70], it is possible to imagine a potential link with the aforementioned metabolite.
As already stated, 4,5,9,10-dehydroisolongifolene was associated to the same co-inherited SNPs sets that were linked to the polyacetylene 1,4-tridecadiene-7,9-diyne. This could suggest a potential case of pleiotropic control of metabolite biosynthesis, which has already been described in other plants [31, 32]. Because the biochemical pathways of terpenes and polyacetylenes are completely different, the only genes that could explain this case of pleiotropy are indeed the previously indicated AP2/ERF transcription factors. Anyway, to date information about which TFs could be involved in the biosynthesis of polyacetylenes is lacking, thus making it difficult to draw conclusions in this respect.
Despite the limited number of most likely candidate genes identified, our results appear globally in agreement with those obtained from similar metabolic GWAS analyses performed under abiotic stress in Arabidopsis thaliana [71] and maize [72].
It has also to be considered that our capability to identify the most likely candidate genes was reduced by some technical limitations and specific features of our study. On the one hand, gene functions are largely unknown in plants, particularly in a species such as sunflower. Indeed, 12 genes out of the 80 that were initially found associated with co-inherited SNPs sets, i.e. previous to the process of functional characterization, were annotated as ‘hypothetical’ or ‘putative’ proteins. On the other hand, our approach to link SNPs to genes was rather stringent, because it required the SNPs to directly land on exons in order to identify a potential candidate.
Conclusions
Our work represents the first characterization of the genetic control of abiotic stress-related specialized metabolites in sunflower. It provides hints concerning the importance of antioxidant compounds in this biological context, and it highlights some of the potential molecular mechanisms underlying their biosynthesis, thus paving the way for novel applications in sunflower breeding. Even if our capability to identify candidate genes was diminished by some technical limitations, our results were consistent with those obtained from similar metabolic GWAS performed under abiotic stress in plants such as Arabidopsis thaliana and maize. Although further analyses will be needed to obtain a deeper understanding of the topic, studying how antioxidants contribute to the tolerance to abiotic stresses in sunflower appears as a promising area of research.
Methods
Plant material and sampling
A panel of 475 sunflower hybrids, corresponding to an incomplete factorial design, was obtained by crossing 36 male and 36 female inbred lines as previously described [73, 74]. Each hybrid was named using its respective female and male parental lines and adding an underscore to separate them.
As already described [75], each hybrid was grown in a single 13 m2 plot, and all the plots were cultivated on the same field trial in Anais (Charente-Maritime, France) from 2 May 2015 to 29 September 2015. Four control hybrids, corresponding to 65 plots, were included in the field trial to allow for the subsequent adjustment of spatial biases. Therefore, the trial included a total number of 540 plots.
For each single plot, n-4 topmost leaves without petioles were sampled from four different plants on July 22 2015, i.e. 7 days (± 3 days according to genotypes) after blooming, between 11:00 to 12:30 (CET time), and then pooled. Each pool was immediately frozen in dry ice and stored at − 80
80 °C until grinding.
°C until grinding.
Metabolome profiling
Leaf samples were cryoground using a Retsch Mill MM 400 ball mixer (Thermo Scientific, Waltham, MA, USA) and lyophilized. Aliquots of 10 ±
± 1.0
1.0 mg of dry leaf powders were weighed in 1.1
mg of dry leaf powders were weighed in 1.1 mL Micronic tubes (Micronic, Lelystad, The Netherlands) and extracted at room temperature with a robotized Star/Starlet platform (Hamilton, Reno, NV, US) using ethanol/water (80:20, v/v) added with 0.1% formic acid and 1.37
mL Micronic tubes (Micronic, Lelystad, The Netherlands) and extracted at room temperature with a robotized Star/Starlet platform (Hamilton, Reno, NV, US) using ethanol/water (80:20, v/v) added with 0.1% formic acid and 1.37 mM methyl vanillate as solvent. Methyl vanillate was used as internal standard to verify the quality of injection for LC-MS.
mM methyl vanillate as solvent. Methyl vanillate was used as internal standard to verify the quality of injection for LC-MS.
Two successive extractions (1 min shaking followed by 15
min shaking followed by 15 min ultra-sonication) were performed with 300
min ultra-sonication) were performed with 300 μL of extraction solvent. The two supernatants were combined and filtered using 0.22
μL of extraction solvent. The two supernatants were combined and filtered using 0.22 μm hydrophilic Durapore filtering microplates (Merck Millipore, Carrigtwohill, Ireland). Several blank extracts were prepared using the same procedure and without sample powder. A quality control (QC) sample was prepared by pooling 10
μm hydrophilic Durapore filtering microplates (Merck Millipore, Carrigtwohill, Ireland). Several blank extracts were prepared using the same procedure and without sample powder. A quality control (QC) sample was prepared by pooling 10 μL of each sample extract.
μL of each sample extract.
LC-MS profiling was performed using the ethanol supernatant extracts. The sample injection order was randomized, and QC samples were injected every 10 samples to correct for the signal intensity drift. The extracts were analyzed using an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific) interfaced to an UltiMate 3000 L UHPLC system (Thermo Scientific) using a C18 chromatographic column (C18-Gemini 2.0
L UHPLC system (Thermo Scientific) using a C18 chromatographic column (C18-Gemini 2.0 ×
× 150
150 mm, 3
mm, 3 μm, 110
μm, 110 Å, Phenomenex, Torrance, CA, USA). An 18-min acetonitrile gradient in acidified water (solvent A: ultrapure water +
Å, Phenomenex, Torrance, CA, USA). An 18-min acetonitrile gradient in acidified water (solvent A: ultrapure water + 0.1% formic acid, solvent B: LC-MS grade acetonitrile) was used with a 300
0.1% formic acid, solvent B: LC-MS grade acetonitrile) was used with a 300 μL/min flow rate and the following elution gradient: 0–0.5
μL/min flow rate and the following elution gradient: 0–0.5 min, 3% B; 0.5–1
min, 3% B; 0.5–1 min, 3–10% B; 1–9
min, 3–10% B; 1–9 min, 10–50% B; 9–13
min, 10–50% B; 9–13 min, 50–100% B; 13–14
min, 50–100% B; 13–14 min, 100% B; 14–14.5
min, 100% B; 14–14.5 min 100–3% B; 14.5–18
min 100–3% B; 14.5–18 min, 3% B. The column temperature was set at 30
min, 3% B. The column temperature was set at 30 °C and the injection volume was 5
°C and the injection volume was 5 μL. The LC-MS instrument was equipped with an electrospray ionization (ESI) source operated in the positive ion mode. Source parameters were set as follows: source voltage, 3.2
μL. The LC-MS instrument was equipped with an electrospray ionization (ESI) source operated in the positive ion mode. Source parameters were set as follows: source voltage, 3.2 kV; sheath gas, 45 arbitrary units (a.u.); auxiliary gas, 15
kV; sheath gas, 45 arbitrary units (a.u.); auxiliary gas, 15 a.u.; sweep gas, 0
a.u.; sweep gas, 0 a.u.; capillary temperature, 350
a.u.; capillary temperature, 350 °C; heater temperature, 350
°C; heater temperature, 350 °C. Full scan MS spectra were acquired at 240
°C. Full scan MS spectra were acquired at 240 k resolution power at 200
k resolution power at 200 m/z with a 50–1000
m/z with a 50–1000 m/z range. All the chemicals used for LC-MS were purchased from Sigma Aldrich (Saint Louis, MO, USA) and Extrasynthese (Genay, France).
m/z range. All the chemicals used for LC-MS were purchased from Sigma Aldrich (Saint Louis, MO, USA) and Extrasynthese (Genay, France).
LC-MS data were processed using the ‘XCMS’ R package [76]. Variables detected in blank extracts, with m/z values varying by more than 0.005 Da or with RT varying by more than 40
Da or with RT varying by more than 40 s between different samples were filtered out. Variables with intensity coefficients of variation in QCs greater than 20% were also removed. This resulted in a matrix of 3507 metabolite features. Intensity drift was corrected using support vector regression [77], and intensities were normalized according to the sample powder mass used for extraction. After a final step of quality assessment, the LC-MS data corresponding to 450 hybrids and 64 control hybrids were retained.
s between different samples were filtered out. Variables with intensity coefficients of variation in QCs greater than 20% were also removed. This resulted in a matrix of 3507 metabolite features. Intensity drift was corrected using support vector regression [77], and intensities were normalized according to the sample powder mass used for extraction. After a final step of quality assessment, the LC-MS data corresponding to 450 hybrids and 64 control hybrids were retained.
Processing, annotation and exploratory analysis of metabolome data
Biases occurring because of the spatial variation in the field trial were adjusted based on the information obtained from randomly replicated control hybrids using a script based on the ‘ASReml-R’ R package v 3.0 [78]. Data from control hybrids were discarded and the spatially corrected matrix was used for the subsequent steps of analysis.
Isotopes and adducts were searched for among the initial 3507 LC-MS features using the ‘Binner’ software [79]. A total of 950 redundant variables were removed, thus bringing the final LC-MS dataset to 2557 features. The most intense ions were then annotated using RT and accurate m/z values and the information available from previous studies [16, 18, 27]. This resulted in the putative annotation of 21 compounds (Table S02), whose MSI levels were attributed according to [80]. Eventually, to gather information about the structure of metabolome data, a PCA was carried out after scaling and mean centering using the ‘pca’ function of the ‘mixOmics’ R package v 6.16.3 [81].
Genotyping
The genotyping of the hybrids was carried out within the frame of the sunflower genome sequencing project [74]. Briefly, the parent lines were genotyped by whole genome resequencing, and the genotype of each hybrid was then obtained from those of its parents [74].
Initially, 14,127,553 SNPs were detected using the XRQ v1.0 assembly of the sunflower genome. Then, all the sets of SNPs in complete linkage disequilibrium among them, called ‘co-inherited SNPs sets’, were identified, and only one SNP was kept for each set. Subsequently, SNPs presenting a minor allelic frequency (MAF) <
< 0.1 or only detected in the male or female panel were filtered out. A final number of 350,052 SNPs, referred to as ‘reference SNPs’, were used for GWAS and are available through the Heliagene XRQ v1.0 genome portal (https://www.heliagene.org/HanXRQ-SUNRISE/). The genotypes of hybrids were coded as ‘0’, ‘1’, or ‘2’ for homozygous XRQ, heterozygous and variant homozygous, respectively. Both the additive (A) and the dominant (D) centered genotyping matrices were produced [73].
0.1 or only detected in the male or female panel were filtered out. A final number of 350,052 SNPs, referred to as ‘reference SNPs’, were used for GWAS and are available through the Heliagene XRQ v1.0 genome portal (https://www.heliagene.org/HanXRQ-SUNRISE/). The genotypes of hybrids were coded as ‘0’, ‘1’, or ‘2’ for homozygous XRQ, heterozygous and variant homozygous, respectively. Both the additive (A) and the dominant (D) centered genotyping matrices were produced [73].
Association analysis
Association analysis was performed in two steps (Fig. (Fig.2).2). First, a multi-locus with forward selection GWAS was carried out with the ‘mlmm.gwas’ R package v 1.0.6 [82] and using the 2557 filtered LC-MS features and the 350,052 reference SNPs. Both the additive (A) and the dominant (D) effects of SNP markers were considered and 20 maximum steps were imposed for the fitting of the linear mixed model, which had an equation of this form:
Where
A
i is the random additive effect of the ith hybrid with the vector A ~
The best GWAS model was chosen using the extended Bayesian information criterion (eBIC) as proposed by [84]. The value of genomic heritability (h2g) for each LC-MS feature was calculated using the general purpose solver function ‘mixed.solve’ of the ‘rrBLUP’ R package v 4.6.1 [85].
The second step of the analysis consisted in linking reference SNPs to their corresponding co-inherited SNPs sets, which included all the SNPs in complete linkage disequilibrium with them (Fig. (Fig.2).2). This step was similar to the ‘block analysis’ conducted by Temme and coworkers [25], although in our case blocks were defined by requiring complete LD among the different SNPs.
Unsupervised identification and enrichment analysis of putative candidate genes
To identify the candidate genes putatively involved in the biosynthesis of metabolites in an unsupervised way, i.e. without any further biological information, SNPs from co-inherited sets were mapped to exons, introns, and intergenic regions using a gft file corresponding to the annotation of the XRQ v1.0 sunflower genome (http://www.heliagene.org/HanXRQ-SUNRISE/). A gene was then considered associated to an LC-MS feature if at least one SNP from a co-inherited set mapped to one of its exons. The corresponding XRQ v2.1 genes were identified using the synonymy table available at the ‘Download’ section of the XRQ v2.1 portal at (https://www.heliagene.org/HanXRQr2.0-SUNRISE/).
The putative candidate XRQ v2.1 genes were used to perform enrichment analyses using the software ClueGO 2.5.8 [86]. A two-tailed hypergeometric test was performed to identify enriched ontology terms. Significance was set at a Benjamini-Hochberg-adjusted p-value of 0.05, the ‘GO fusion’ option was used and the k-score was fixed at 0.4. Three custom sunflower ontologies were used for the analysis, corresponding to the two Gene Ontology (GO) subsets ‘biological process’ and ‘molecular function’ and to the KEGG pathways.
The GO sub-ontology files were built using the Blast2GO output files available on the Heliagene website (https://www.heliagene.org/HanXRQr2.0-SUNRISE/), while the KEGG ontology file was created by performing a double best hit search using the XRQ v2.1 sunflower protein sequences on the KAAS automatic annotation server (https://www.genome.jp/kegg/kaas/). The KO codes thus obtained were then manually inspected in order to remove ontologies spuriously related to bacteria, fungi and animals.
Hot spots of metabolite-associated SNPs and genes
To describe the patterns of localization of metabolite-associated SNPs and genes along the sunflower genome and detect the potential presence of hot spots, a sliding windows approach was chosen. The window length was set at 5 Mb, and the window was incrementally advanced along the chromosomes using a pass of 1
Mb, and the window was incrementally advanced along the chromosomes using a pass of 1 Mb. For each window, the following measures were calculated: (i) the absolute number of metabolite-associated SNPs and their frequency respect to the total number of SNPs, and (ii) the absolute number of metabolite-associated genes and their frequency respect to the total number of genes. The values of the frequencies thus obtained were then plotted against the sunflower chromosomes and visualized with the ‘Circlize’ R package v 0.4.14 [87].
Mb. For each window, the following measures were calculated: (i) the absolute number of metabolite-associated SNPs and their frequency respect to the total number of SNPs, and (ii) the absolute number of metabolite-associated genes and their frequency respect to the total number of genes. The values of the frequencies thus obtained were then plotted against the sunflower chromosomes and visualized with the ‘Circlize’ R package v 0.4.14 [87].
A sliding window was considered as being part of a hot spot if it presented a frequency of metabolite-associated genes higher than 0.075 and an absolute number of genes higher than 10, and adjacent windows were merged in order to obtain the final hot spots. Eventually, the identification of potential metabolic clusters as defined by [33] was performed by visually inspecting the identified regions.
Identification of the SNPs co-localizing with known abiotic stress-related QTLs and of the associated LC-MS features
To study the genetic control of specialized metabolites linked to abiotic stresses, we followed a strategy based on co-localization with QTLs of interest that had been discovered in prior works. First, SNPs from co-inherited sets obtained from our GWAS analysis were tested for co-localization with two groups of QTLs, i.e.: (i) QTLs related to drought, cold, and nutrient stress tolerance which had been detected on other field trials; (ii) QTLs related to productivity and development traits which had been detected either on the same field trial or on other trials [15, 34]. An SNP was considered as co-localizing with a QTL if it fell in an interval of 50 kb downstream or upstream respect to an SNP belonging to the QTL itself. Second, all the SNPs in complete LD with co-localizing SNPs were identified and subsequently used to identify the putative candidate genes for metabolite biosynthesis. Eventually, using the previously defined associations among co-inherited SNPs and reference SNPs, reference SNPs co-localizing with QTLs were found. These reference SNPs were then used to determine which LC-MS features could be considered as co-localizing with QTLs, and which therefore had to be annotated.
kb downstream or upstream respect to an SNP belonging to the QTL itself. Second, all the SNPs in complete LD with co-localizing SNPs were identified and subsequently used to identify the putative candidate genes for metabolite biosynthesis. Eventually, using the previously defined associations among co-inherited SNPs and reference SNPs, reference SNPs co-localizing with QTLs were found. These reference SNPs were then used to determine which LC-MS features could be considered as co-localizing with QTLs, and which therefore had to be annotated.
Annotation of LC-MS features co-localizing with known QTLs
None of the LC-MS features co-localizing with known QTLs possessed an annotation based on previous works (see previous paragraph). Therefore, the tentative annotation of these features was based on their raw chemical formulas and on the comparison with MS-related information available from KNApSAcK (http://www.cheminfo.org/Chemistry/Database/Knapsack/index.html) and Dictionary of natural products (https://dnp.chemnetbase.com). This resulted in the putative annotation of 30 other compounds belonging to 11 compound families (Table (Table11).
Identification of the most likely candidate genes
The putative candidate genes previously identified through GWAS and co-localization analysis were further characterized in order to identify the most likely candidates for the associations with putative metabolites. This was carried out in a functional perspective and following a two-step procedure. The first step focused on the annotated metabolites and consisted in a systematic bibliographic search through the NCBI PubMed database (https://pubmed.ncbi.nlm.nih.gov). The names of the metabolites and of the corresponding biochemical families were used as inputs.
The second step focused on the genes, and consisted in (i) retrieving the gene descriptions available for each gene using a gft file corresponding to the XRQ v2.1 of the sunflower genome; (ii) carrying out tBLASTn searches through the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi); (iii) performing a systematic bibliographic search through the NCBI PubMed database, using both the gene descriptions and the gene names obtained with the tBLASTn search as inputs.
Genotypic boxplots were produced for all the reference SNPs associated to the tentatively annotated LC-MS features using the ‘genotypes.boxplot’ function of the ‘mlmm.gwas’ R package v 1.0.6. Eventually, we characterized all the co-inherited SNPs found in exons by determining if they represented synonymous or missense mutations using the information available for the XRQ v1.0 sunflower genome (http://www.heliagene.org/HanXRQ-SUNRISE/).
Supplementary Information
Acknowledgments
We would like to thank the RAGT group and Innolea for the help provided in setting up the experimental field and in collecting the plant material used for our study.
Abbreviations
| ROS | Reactive oxygen species |
| LC-MS | Liquid chromatography coupled to mass spectrometry |
| RT | Retention time |
| m/z | Mass over charge ratio |
| SNP | Single nucleotide polymorphism |
| GWAS | Genome-wide association study |
| PCA | Principal component analysis |
| QTL | Quantitative trait locus |
| LD | Linkage disequilibrium |
Authors’ contributions
NBL, AM and YG conceived and designed the experiment. NB and NBL participated to the sampling. HD supervised and coordinated the production of data. SB, TB and OF performed the LC-MS analyses. MM performed the GWAS and all of the other statistical analyses. MM and NBL interpreted the results and wrote the original draft. AM and SB critically read and edited the manuscript. All the authors reviewed and accepted the manuscript.
Funding
This work was supported by the French National Research Agency (SUNRISE ANR-11-BTBR-0005, PHENOME ANR-11-INBS-0012 and MetaboHUB ANR-11-INBS-0010 projects). This research was part of the French Laboratory of Excellence project “TULIP” (ANR-10-LABX-41; ANR-11-IDEX-0002-02).
Availability of data and materials
The genotype and the kinship matrices needed to perform GWAS and the information concerning previously known QTLs are found in the corresponding cited references. All the genomic information concerning candidate genes are available on the Heliagene portal (http://www.heliagene.org). LC-MS data are reported in Tables S01-S02. The results of all the other analyses are reported in Tables S03-S10 and in Figs. S01-S02.
Declarations
All the methods used in this study on field-grown sunflowers complied with national regulations.
Not applicable.
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Articles from BMC Genomics are provided here courtesy of BMC
Full text links
Read article at publisher's site: https://doi.org/10.1186/s12864-024-10104-9
Read article for free, from open access legal sources, via Unpaywall:
https://bmcgenomics.biomedcentral.com/counter/pdf/10.1186/s12864-024-10104-9
Citations & impact
Impact metrics
Article citations
Genome-Wide Identification and Expression Analysis of TGA Family Genes Associated with Abiotic Stress in Sunflowers (Helianthus annuus L.).
Int J Mol Sci, 25(7):4097, 07 Apr 2024
Cited by: 0 articles | PMID: 38612905 | PMCID: PMC11012525
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome-wide investigation and expression analysis of membrane-bound fatty acid desaturase genes under different biotic and abiotic stresses in sunflower (Helianthus annuus L.).
Int J Biol Macromol, 175:188-198, 04 Feb 2021
Cited by: 11 articles | PMID: 33549671
Identification of differentially expressed genes in salt-tolerant oilseed sunflower (Helianthus annuus L.) genotype by RNA sequencing.
Mol Biol Rep, 49(5):3583-3596, 04 Feb 2022
Cited by: 2 articles | PMID: 35119610
Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.).
Gene, 910:148336, 04 Mar 2024
Cited by: 10 articles | PMID: 38447680
Review
Genome-wide identification and in-silico expression analysis of CCO gene family in sunflower (Helianthus annnus) against abiotic stress.
Plant Mol Biol, 114(2):34, 03 Apr 2024
Cited by: 10 articles | PMID: 38568355 | PMCID: PMC10991017

 1
1 


