Abstract
Free full text

KIS counteracts PTBP2 and regulates alternative exon usage in neurons
Abstract
Alternative RNA splicing is an essential and dynamic process in neuronal differentiation and synapse maturation, and dysregulation of this process has been associated with neurodegenerative diseases. Recent studies have revealed the importance of RNA-binding proteins in the regulation of neuronal splicing programs. However, the molecular mechanisms involved in the control of these splicing regulators are still unclear. Here, we show that KIS, a kinase upregulated in the developmental brain, imposes a genome-wide alteration in exon usage during neuronal differentiation in mice. KIS contains a protein-recognition domain common to spliceosomal components and phosphorylates PTBP2, counteracting the role of this splicing factor in exon exclusion. At the molecular level, phosphorylation of unstructured domains within PTBP2 causes its dissociation from two co-regulators, Matrin3 and hnRNPM, and hinders the RNA-binding capability of the complex. Furthermore, KIS and PTBP2 display strong and opposing functional interactions in synaptic spine emergence and maturation. Taken together, our data uncover a post-translational control of splicing regulators that link transcriptional and alternative exon usage programs in neuronal development.
Introduction
In the mammalian brain, alternative splicing (AS) is a key mechanism that contributes substantially to the enormous molecular diversity among neuronal cell types (Barbosa-Morais et al., 2012; Mazin et al., 2021; Merkin et al., 2012). Despite the similarity of neuronal gene expression programs, each neuronal subclass can be distinguished by unique alternative mRNA processing events. Moreover, AS is known to drive many aspects of neurogenesis, including neural progenitor cell proliferation and differentiation, axon guidance, synapse formation, and synaptic plasticity (Vuong et al., 2016b; Furlanis and Scheiffele, 2018). The relevance of AS as a regulatory mechanism is evidenced by numerous examples in which alterations in splicing are associated with pathological states (Gandal et al., 2018; LaForce et al., 2023; Schieweck et al., 2021). Although fine-tuning mRNA processing is essential for neuronal activity and maintenance, insights into its regulation are just starting to emerge. RNA-binding proteins (RBPs) play a critical role in regulating AS patterns. These proteins bind to pre-mRNAs and form ribonucleoprotein (RNP) complexes that can affect splicing by influencing the recognition of splicing signals or by blocking access of the splicing machinery to certain regions of the pre-mRNA. Some RBPs act as splicing enhancers, promoting the inclusion of specific exons, while others act as splicing repressors inhibiting the inclusion of exon inclusion (Fisher and Feng, 2022; Grosso et al., 2008; Traunmüller et al., 2023).
Among the most investigated RBPs are the polypyrimidine tract-binding proteins, PTBP1 and PTBP2, which are considered master regulators of neuronal fate (Keppetipola et al., 2012; Linares et al., 2015; Zhang et al., 2019). These two proteins exhibit tissue-specific expression patterns in which PTBP1 is expressed in most cell types and neuronal progenitor cells, whereas PTBP2 is expressed primarily in differentiating neurons and testis. During the differentiation of neuronal progenitor cells to postmitotic neurons a switch takes place from the predominant expression of PTBP1 to its neuronal paralog PTBP2, which is essential for the stem cell to neuron transition. PTBP2 null mice die shortly after birth and exhibit misregulation of AS in genes involved in cytoskeletal remodeling and cell proliferation (Boutz et al., 2007; Licatalosi et al., 2012) as well as in neurite growth and synaptic transmission (Li et al., 2014). Furthermore, PTBP2 knockout brains display reduced neural progenitor pools and premature neurogenesis (Licatalosi et al., 2012). PTBP2 also plays an important role in the embryonic-specific repression of alternative exons. As PTBP2 expression decreases from its highest levels in the embryonic brain to moderate levels during postnatal development, a cohort of exons switches from being skipped in embryos to displaying enhanced inclusion in adults (Licatalosi et al., 2012; Li et al., 2014; Zheng et al., 2012). Thus, sequential downregulation of PTBP1 followed by PTBP2 contributes to the activation of neural exon networks at the appropriate stages of development (Raj and Blencowe, 2015). AS of the DLG4 transcript, which encodes the excitatory postsynaptic protein PSD95, is an example of the relevance of the PTBP1/2 switch in neuronal differentiation (Zheng, 2016). Skipping of exon 18 in DLG4 transcripts causes premature translation termination and nonsense-mediated mRNA decay (NMD). The controlled induction of this exon during neuronal development is essential to ensure the timing of synapse formation (Zheng et al., 2012). This exon may also be dynamically regulated in mature neuronal circuits to adapt PSD95 expression to the requirements of synaptic remodeling (Lopez Soto et al., 2019).
PTBP1 and PTBP2 are 74% identical in amino acid sequence and have a similar domain organization of four RNA recognition motifs (RRMs) connected by three linker regions. Although primarily characterized as repressive splicing regulators, they can also activate some splice sites as a function of binding position relative to regulated exons (Corrionero and Valcárcel, 2009; Llorian et al., 2010; Xue et al., 2009). It has been shown that PTBP1/2 RRM2, along with the following linker sequence, is sufficient for splicing repressor activity (Robinson and Smith, 2006). RRM2 can interact with both RNA via its canonical β-sheet surface, and with other co-regulators such as Matrin3, Raver1, and hnRNPM (Coelho et al., 2015). Furthermore, Tyr247 of PTBP1 (PTBP2 Tyr244) within RRM2 is particularly critical for co-regulatory protein interactions (Joshi et al., 2011; Rideau et al., 2006). Regarding the regulation of PTBP proteins, different phosphate modifications are located in the unstructured regions, including the N-terminal, Linker 1, and Linker 2 regions (Pina et al., 2018). To date, the kinases involved are still under investigation as well as the role of these post-translational modifications in controlling PTBPs splicing activity.
KIS (encoded by the gene UHMK1) was first identified as a Kinase that Interacts with Stathmin (Maucuer et al., 1997), a phosphoprotein that controls microtubule dynamics (Watabe-Uchida et al., 2006). Curiously, KIS is the only known protein kinase that possesses a U2AF homology motif (UHM), an atypical RRM that has lost its RNA-binding ability, but mediates protein–protein interactions at the C-terminal region of KIS (Corsini et al., 2007; Kielkopf et al., 2004; Manceau et al., 2008). This motif shares 42% sequence similarity with U2AF65, a 65-kDa subunit of the splicing factor U2AF, suggesting that KIS could be involved in splicing-related processes. Various lines of evidence support the function of KIS in the modulation of gene expression at different levels such as transcription, splicing, mRNA stability, and protein translation (Arfelli et al., 2023; Manceau et al., 2008; Pedraza et al., 2014). Although KIS is ubiquitously expressed, higher levels are detected in the nervous system, and KIS protein is gradually upregulated during postnatal development, reaching its highest level in the mature brain (Bièche et al., 2003). In mature neurons, KIS has been implicated in dendritic spine morphogenesis and synaptic activity (Pedraza et al., 2014).
Here, we find that KIS kinase is involved in the control of alternative exon usage, a key mechanism for promoting functional gene expression patterns during neuronal differentiation. We show that AS regulators are the main phosphorylation targets of this UHM-containing kinase. Notably, we find that PTBP2, an essential AS factor for neuronal maturation, is the major phosphotarget of KIS. We demonstrate that phosphorylation by KIS dissociates PTBP2 protein complexes affecting their ability to bind RNA and therefore inhibiting PTBP2 activity. Furthermore, KIS counteracts PTBP2-mediated inhibition of dendritic spine maturation in a phosphosite-dependent manner, and downregulation of KIS partly rescues the negative effects caused by decreasing PTBP2 levels on spine maturation. Taken together, our data point to the notion that KIS phosphorylation counteracts PTBP2 activity and, in this way, alters the expression patterns of gene isoforms involved in neuronal differentiation and synaptic activity.
Results
KIS kinase regulates exon usage during neuronal differentiation
KIS contains a UHM domain frequently found in splicing regulatory proteins, and a GFP–KIS fusion accumulates in nuclear substructures adjacent to those formed by splicing factors (Figure 1A). Prompted by its role in proper expression of key postsynaptic proteins (Pedraza et al., 2014), we decided to analyze genome-wide exon usage during neuronal differentiation in vitro. First, immature neurons from mouse primary cortical neurons of 7 days in vitro (DIV) were compared with mature neurons from 18 DIV cultures when expression of UHMK1, the gene coding for KIS, reached maximal levels in cortical cultures (Figure 1B). Second, cortical neurons were infected at 7 DIV with a lentiviral construct co-expressing green fluorescent protein (GFP) and an shRNA targeted against UHMK1 (shKIS) and collected at 18 DIV (Figure 1C). Total RNA was purified from triplicate samples for deep sequencing analysis. When compared to control (shCtrl), shKIS-infected cells displayed a fivefold reduction in UHMK1 mRNA levels, similar to that at onset of neuronal differentiation (Figure 1D). Our analysis revealed that the impact of KIS on exon usage affected both exon inclusion and skipping, although with a clear bias toward exon inclusion 6.3% vs 2% skipping (Figure 1E). Notably, genes containing more than one exon with at least a twofold change coded for proteins significantly enriched in the synapse (Figure 1—figure supplement 1A), including two important activators of neurite outgrowth, CAMSAP1, and NCAM1 (Figure 1—figure supplement 1B). These data suggest that KIS promotes exon inclusion in genes related to neuronal differentiation, especially genes encoding synaptic proteins. Supporting this notion, the top 100 exons upregulated by KIS, that is downregulated by KIS knockdown, were strongly enriched among those increasingly used by cortical neurons from 7 to 18 DIV (Figure 1F). Among a total of 8457 exons that displayed low variability (Fano factor <5 × 10−4) within the KIS knockdown triplicate samples, 1141 and 1106 exons were found significantly upregulated and downregulated, respectively, by KIS (Figure 1G; Supplementary file 1). In addition, 28.0% of exons upregulated by KIS were also upregulated during differentiation, whereas only 18.8% decreased their usage from 7 to 18 DIV (Figure 1—figure supplement 1C). The opposite behavior was observed for exons downregulated by KIS in which 14.3% and 24.8% increased or decreased their usage during differentiation, respectively (Figure 1—figure supplement 1C). Finally, whereas genes containing KIS-downregulated exons did not retrieve specific gene ontology (GO) terms, exons upregulated by KIS encoded a higher percentage of protein disorder (Figure 1—figure supplement 1D) and were found in genes preferentially coding for RBPs (Figure 1H), key factors of RNA granule formation and transport along neurites. Overall, our data link KIS to the control of exon usage in cortical neurons and suggest a pivotal role of this UHM-containing kinase as a regulator of AS during neuronal differentiation.
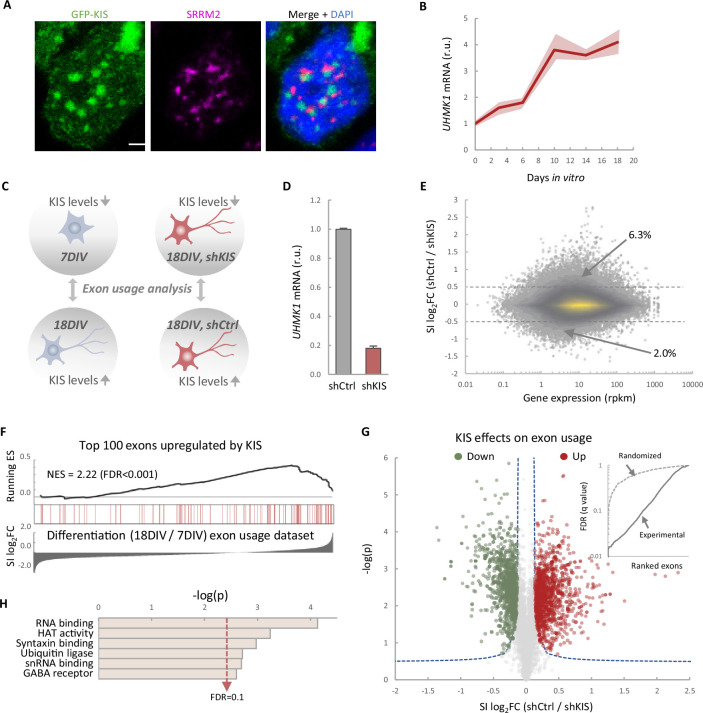
(A) Immunofluorescence staining of endogenous speckle marker SRRM2 (magenta) and 4',6-diamidino-2-phenylindole (DAPI) (bue) in HEK293T cells transfected with GFP-KIS (green). Scale bar, 2 μm. (B) Differential expression of UHMK1, the gene encoding KIS, in cultured cortical neurons from E17.5 mouse embryos. Samples were collected at different days in vitro (DIV), analyzed by qRT-PCR and made relative to day 0. Mean ± standard error of the mean (SEM) (n = 3) are plotted. (C) Experimental design of exon usage analysis. Cultured cortical neurons from E17.5 mice were collected at 7 or 18 DIV (left experimental set). Cortical neurons were transduced at 7 DIV with lentiviral vectors expressing shCtrl or shKIS and collected at 18 DIV (right experimental set). (D) UHMK1 mRNA levels in primary cortical neurons transduced at 7 DIV with lentiviral vectors expressing shCtrl or shKIS and collected at 18 DIV. Bars represent mean ± SEM (n = 3) (E) MA-plot shows splicing index (SI) fold change (log2FC) as a function of gene expression in rpkm (reads per kilobase million) in control neurons (shCtrl) compared to KIS knockdown neurons (shKIS) as in panel D. The percentage of exons displaying a log2FC above 0.5 or below −0.5 is indicated. (F) RNA-seq analysis from cortical neurons expressing shCtrl or shKIS as in panel D and from cortical neurons collected at 7 and 18 DIV for neuronal differentiation (n = 3). Barcode plot shows the position of exons upregulated by KIS, that is downregulated by KIS knockdown, within the transcriptomic dataset of exons upregulated during neuronal differentiation. Normalized enrichment scores (NES) and the corresponding false-discovery rate (FDR) values are shown. (G) Volcano plot shows t-test significance (−log2p) as a function of SI fold change (log2FC) in control neurons (shCtrl) compared to KIS knockdown neurons (shKIS). Significantly (FDR <0.05) downregulated (n = 1106, green) or upregulated (n = 1141, red) exons are highlighted. Inset shows the dependence of FDR on exon rank from experimental and randomized datasets. (H) Gene ontology (GO) term enrichment analysis of genes with exons upregulated by KIS. FDR value is shown.
Figure 1—figure supplement 1.
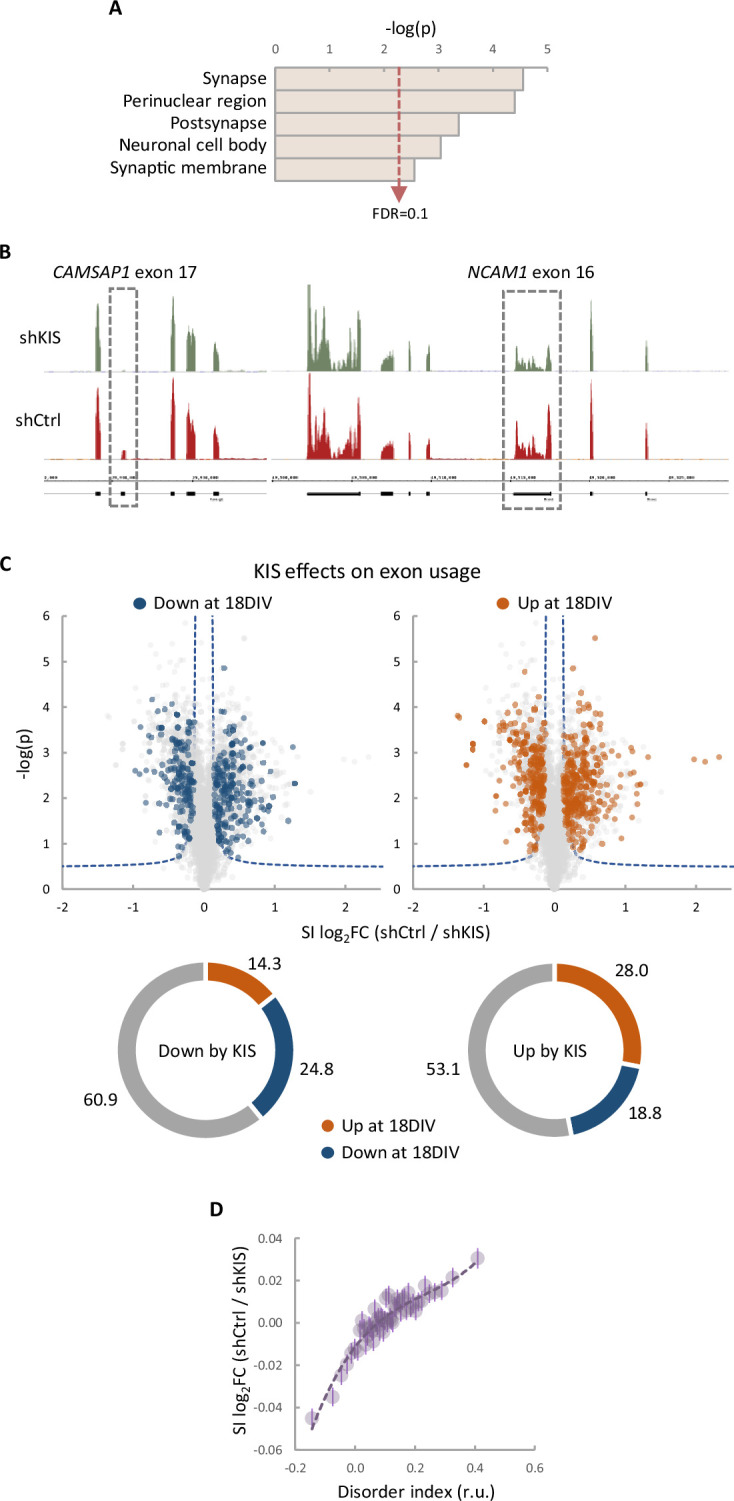
(A) Gene ontology (GO) term enrichment analysis of genes with more than one exon upregulated by KIS. False-discovery rate (FDR) value is shown. (B) Representative read profiles showing KIS-dependent exons in CAMSAP1 and NCAM1 genes (dashed-line rectangles). (C) Volcano plots show t-test significance (−log2p) as a function of splicing index (SI) fold change (log2FC) in control neurons (shCtrl) compared to KIS knockdown neurons (shKIS). Significantly (FDR <0.05) downregulated (blue) and upregulated (orange) exons during neuronal differentiation are highlighted. Sunburst charts show the percentage of exons differentially modulated during differentiation in sets downregulated (left) and upregulated (right) by KIS. (D) Exon usage fold change (log2FC) in control neurons (shCtrl) compared to KIS knockdown neurons (shKIS) as a function of peptide disorder index (see Methods). Mean values of binned data (bin size = 2500 exons, purple circles) with confidence limits for the mean (α = 0.05) and a regression line are shown.
Phosphoproteomics links KIS to RNA splicing and identifies PTBP2 as a novel phosphotarget
One possible way to control the use of alternative exons during neuronal differentiation is by changing the expression of splicing regulators. Our RNA-seq analysis showed that KIS knockdown did not affect transcript levels of splicing regulators whose expression underwent changes during cortical neurons differentiation (Figure 2—figure supplement 1A). Therefore, we tested whether the KIS phosphoproteome could reveal the mechanism by which KIS regulates exon usage. For this, we analyzed phosphorylation changes in the proteome of HEK 293T cells expressing KIS and KISK54A, a point mutant without kinase activity (Maucuer et al., 1997). We chose this cell line because of its high transfection efficiency and, although it is an embryonic kidney-derived line, there is evidence to suggest a neuronal lineage, which could explain the expression of neuron-specific genes (Lin et al., 2014). Quantitative phosphoproteomic analysis resulted in the identification of 9994 phospho-sites in a total of 2743 identified proteins (Supplementary file 2). As shown in the MA-plot (Figure 2A), proteins displaying the strongest differential phosphorylation by KIS are highly related to AS, that is PTBP2, RAVER1, hnRNPM, and MATR3. PTBP2 is considered one of the pivotal splicing regulators in neuronal development (Keppetipola et al., 2012) and, strikingly, phosphorylation at serine 308 of PTBP2 is among the top phosphosites in our KIS phosphoproteome (log2FC = 2.36). Furthermore, the kinase assay confirmed that among the three KIS serine targets on PTBP2 (Ser178, Ser308, and Ser434), Ser308 is key for PTBP2 phosphorylation by KIS (Figure 2—figure supplement 1B–D). The different levels of reduction in 32P incorporation displayed by the single phosphonull mutants (Figure 2—figure supplement 1C) suggests that phosphorylation follows a hierarchical pattern, P-Ser308 facilitating phosphorylation of the other two phosphosites. Ser178 is present in a very large tryptic peptide (37 aa) that was not detected in the KIS phosphoproteome but, as it fulfills the consensus site phosphorylated by KIS (Figure 2—figure supplement 1E), we decided to include this residue in our analysis. Note that the interactions between splicing factors that are KIS targets have been already shown except for SUGP1 (Figure 2B and see also Discussion).
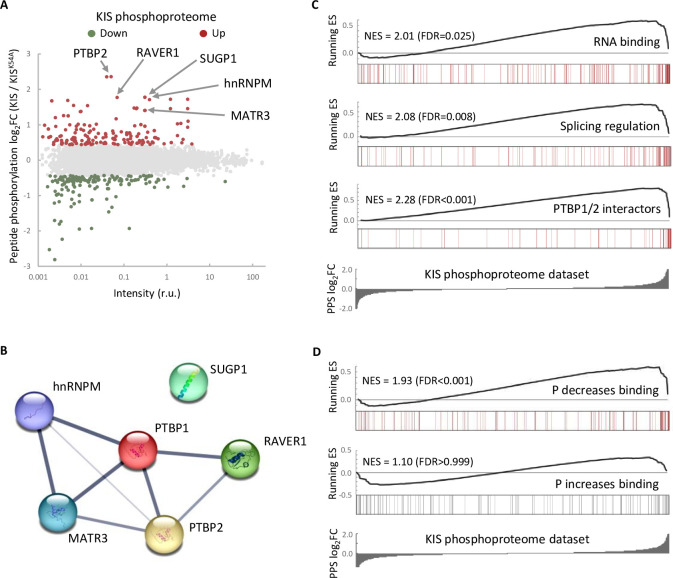
(A) MA-plot of phospho-sites found in the proteome of HEK293T cells transfected with KIS and KISK54A mutant (see also Supplementary file 3 for details). (B) STRING interaction map of top KIS-phosphorylated proteins. (C, D) Gene enrichment analysis of the KIS phosphoproteomic dataset. Barcode plots indicate the rank position of genes from (C) Gene ontology (GO) terms and PTBP1/2 interactors and (D) phosphorylated RNA-binding proteins (Vieira-Vieira et al., 2022) that were found significantly enriched within the KIS phosphoproteomic dataset. Normalized enrichment scores (NES) and the corresponding false-discovery rate (FDR) values are shown.
Figure 2—figure supplement 1.
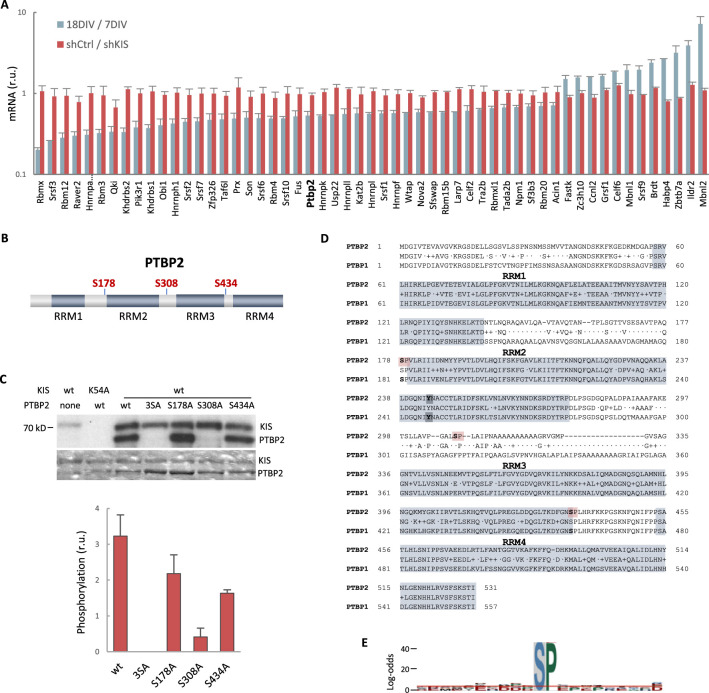
(A) mRNA levels of splicing regulators from gene ontology (GO) biological process in cortical neurons at 18 DIV relative to 7 DIV (blue) or expressing shKIS relative to shCtrl (red). Mean ± standard error of the mean (SEM) (n = 3) values from genes with log2FC >0.5 and false-discovery rate (FDR) <0.1 are shown. (B) PTBP2 protein scheme showing RNA recognition motifs (RRMs) and localization of KIS-phosphorylated serines within linker regions. (C) KIS in vitro kinase assay of indicated GST recombinant proteins purified from E. coli. In the phospho-null triple mutant (3SA), Ser178, Ser308, and Ser434 have been mutated to alanine. Representative autoradiography (top) and Coomassie staining (bottom) images are shown. The plot shows the quantification of phosphorylation signals as mean ± SEM (n = 3) values. (D) Aligned amino acid sequences of mouse PTBP2 and PTBP1. RRMs are highlighted in gray and KIS-phosphorylated serines in red. Tyr244 in RRM2 is highlighted in bold type. (E) pLogo analysis (O’Shea et al., 2013). KIS preferentially phosphorylates serines (S) with high frequency (89.38%) followed by proline (P) at position +1 with a frequency of 41.67% and statistical significance (p < 0.05). Input sequences = 143.
Figure 2—figure supplement 1—source data 1.
KIS in vitro kinase assays.
Next, to perform gene set enrichment analysis (GSEA), we ranked KIS targets using a Phospho Protein Score (PPS) that incorporates the fold change of all identified phosphopeptides per protein (see Methods). Proteins that are more phosphorylated in the presence of KIS have higher PPS, while those that are more phosphorylated in the presence of KISK54A have lower PPS. GSEA analysis confirmed that KIS-phosphorylated proteins are enriched in regulation of splicing, RNA binding, and PTBP1/2 interactors (Figure 2C). Interestingly, when we compared our dataset to a recent RNA-interactomic study, in which the pull-down efficiencies of phosphorylated and non-phosphorylated forms of mRNA-binding proteins were analyzed (Vieira-Vieira et al., 2022) (see also Methods), we found that among KIS-phosphorylated proteins there is a significant enrichment of RBPs that decrease their binding to mRNA upon phosphorylation (Figure 2D), but not the contrary, suggesting that KIS phosphorylation would likely act by promoting dissociation of protein complexes from mRNA. In all, KIS phosphoproteomics reveals a strong association of this kinase with AS regulators, PTBP2 being a relevant phosphotarget in neuronal differentiation.
KIS kinase activity counteracts PTBP2 function on exon usage
To define the role of KIS in AS in neurons, we decided to focus our study on the link between PTBP2 and KIS. It is known that loss of PTBP2 causes extensive changes in AS in brain (Vuong et al., 2016b). To assess whether these changes may be modified by KIS expression, we performed an enrichment analysis of PTBP2-regulated exons in our cortical samples to obtain population-wide data. We found that PTBP2-inhibited exons are significantly enriched (false-discovery rate [FDR] = 0.001) among KIS-activated exons, supporting the notion that KIS acts on AS, at least in part, by inhibiting PTBP2 activity. This analysis was also carried out with PTBP2-activated exons, but no significant enrichment was observed in this case (Figure 3A). As expected, randomization of our original RNA-seq dataset resulted in a total loss of enrichment (Figure 3—figure supplement 1A). In addition, applying our exon usage analysis to RNA-seq data from PTBP2 KO cells (Vuong et al., 2016b) produced high enrichment scores for both PTBP2-inhibited and activated exons as expected (Figure 3—figure supplement 1B), which supports our approach to analyzing exon inclusion from RNA-seq data.
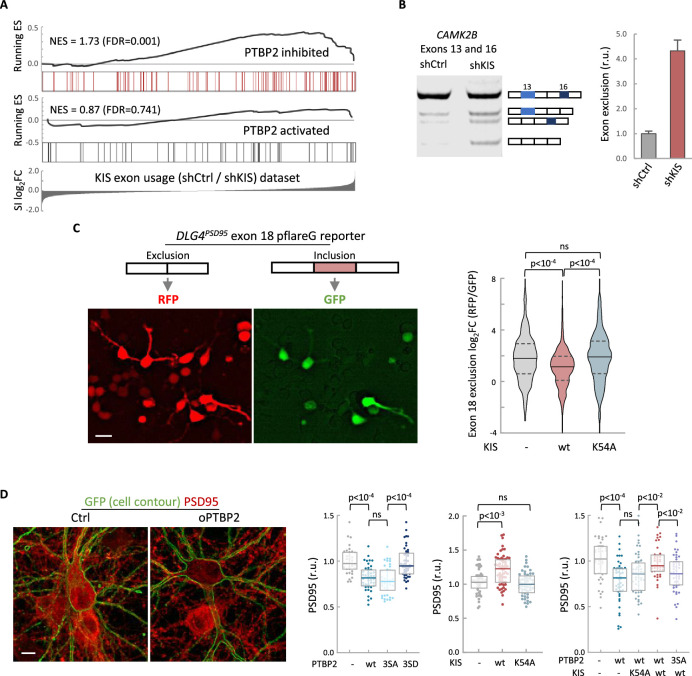
(A) Gene enrichment analysis of PTBP2-regulated exons. Barcode plots show the position of exons excluded (red) or included (gray) by PTBP2 within the transcriptomic dataset of exons upregulated by KIS. Normalized enrichment scores (NES) and the corresponding false-discovery rate (FDR) values are shown. (B) Semiquantitative RT-PCR of CAMK2B from control (shCtrl) and KIS knockdown neurons (shKIS). The level of isoforms displaying exon exclusion relative to the full-length isoform is plotted as mean ± standard error of the mean (SEM) (n = 3). (C) Neuroblastoma N2A cells co-transfected with dual-fluorescent reporter (Zheng et al., 2013) and KIS or KISK54A. DLG4 exon 18 exclusion is plotted as a function of the RFP to GFP fold change (log2FC) from ctrl (n = 672), KIS (n = 1054), or KISK54A (n = 970) expressing cells. Median ± Q values and the results of Mann–Whitney test are also shown. Scale bar, 20 µm. (D) PSD95 immunofluorescence of hippocampal neurons transfected at 5 DIV with vectors expressing PTBP2 and GFP as reporter, and fixed at 12 DIV. Plots show the quantification of PSD95 endogenous levels in single cells transfected with the indicated wild type and mutant proteins in three sets of experiments. Median ± Q values (n > 40) and the results of Mann–Whitney tests are also shown. Scale bar, 10 µm.
Figure 3—source data 1.
CAMK2B exon exclusion analysis.
Figure 3—figure supplement 1.
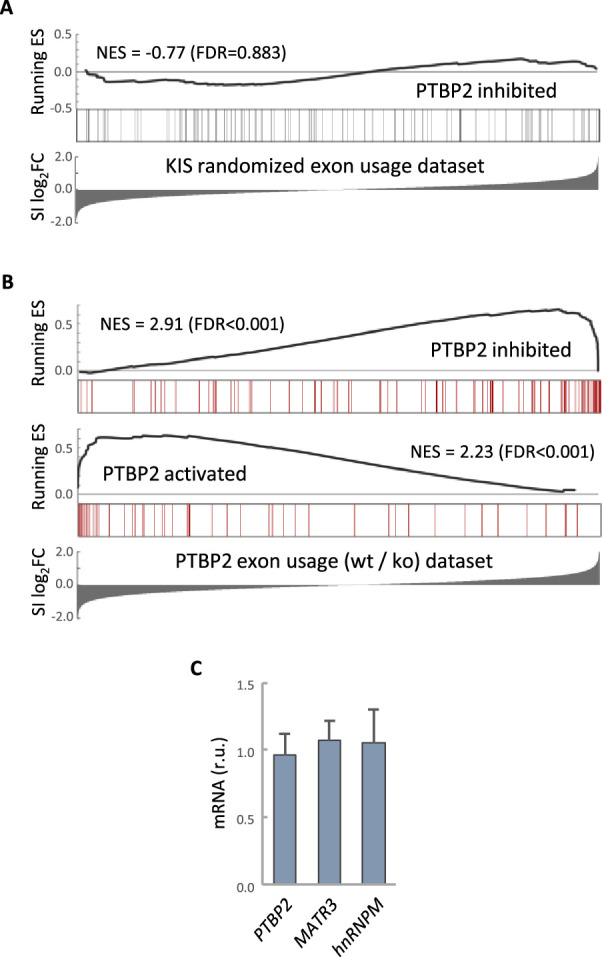
(A) Gene enrichment analysis of PTBP2-regulated exons. Barcode plots show the position of exons excluded by PTBP2 within a KIS randomized exon usage dataset. Normalized enrichment scores (NES) and the corresponding false-discovery rate (FDR) values are shown. (B) Gene enrichment analysis of PTBP2-regulated exons applying our exon usage analysis (see Methods). Barcode plots show the position of exons excluded and included by PTBP2 within the transcriptomic dataset from PTBP2 KO cells (Vuong et al., 2016a). NES and corresponding FDR values are indicated. (C) Quantification of mRNA levels from KIS knockdown cortical neurons relative to control neurons. Bars represent mean ± standard error of the mean (SEM) (n = 3).
To validate the effect of KIS on PTBP2 activity, we analyzed the AS of CAMK2B transcript in KIS knockdown samples (shCtrl vs shKIS). This transcript, which has important neuronal functions, undergoes exclusion of exons 13 and 16 in a PTBP2-dependent manner (Licatalosi et al., 2012; Li et al., 2014). Notably, KIS knockdown (shKIS) increased the exclusion of these exons about fourfold (Figure 3B). Since the observed changes could be due to changes in the expression of PTBP2 or in one of its interactors, we analyzed the expression levels of PTBP2, MATR3, and hnRNPM. No significant differences were observed between cortical neurons infected with shCtrl vs shKIS (Figure 3—figure supplement 1C).
To further confirm the effects of KIS on PTBP2 activity, we used a fluorescent reporter that monitors in vivo AS in cultured neuroblastoma N2A cells. The minigene reporter pFlareG-exon 18 of the DLG4 gene produces RFP and GFP from the exon-skipping and exon-including isoforms, respectively (Zheng et al., 2013). PTBP2 induces the exclusion of exon 18, which control DLG4 expression during neuronal development (Zheng et al., 2012). We found that KIS expression significantly decreased exon 18 exclusion levels compared to non-transfected cells or cells transfected with the K54A kinase dead mutant (Figure 3C). These results suggest that KIS inhibits PTBP2 activity on exon 18.
The DLG4 gene encodes PSD95, a key regulator of synaptic assembly, and the exclusion of exon 18 produces a change in the isoform expression pattern and induces mRNA degradation by NMD (Zheng et al., 2012). Thus, we decided to analyze the effects of PTBP2 and KIS on PSD95 endogenous levels in hippocampal neurons. Whereas transfection of wt or a phospho-null triple mutant (3SA) of PTBP2 produced a reduction in PSD95 levels when compared to control neurons, expression of a phosphomimetic mutant (3SD) had no significant effect (Figure 3D). In agreement with the above experiments using the splicing fluorescent reporter, KIS overexpression produced an increase in endogenous PSD95 levels compared to control neurons or neurons transfected with the K54A mutant (Figure 3D). Together, these results suggest that the phosphorylation state of PTBP2 strongly modulates its role on PSD95 expression. To test the effects of KIS on PTBP2 in this scenario, we co-transfected phosphorylation variants of KIS and PTBP2. First, KIS rescued the negative effects of PTBP2 on PSD95 expression levels. Second, this rescue was dependent on the kinase activity of KIS since co-transfection of PTBP2 with the inactive KISK54A mutant did not increase PSD95 expression. Finally, KIS did not rescue PSD95 levels when co-transfected with the phospho-null 3SA mutant of PTBP2 (Figure 3D). Taken together, our findings provide substantial evidence that KIS inhibits PTBP2 activity by phosphorylation.
KIS and PTBP2 display opposing functional effects on dendritic spine maturation
To study the functional relevance of KIS phosphorylation, we examined the physiological consequences of PTBP2 phosphomutants in differentiated neurons. We transfected dissociated hippocampal cultures with vectors expressing GFP and PTBP2 or PTBP2 phosphomutants and then examined the spine density of transfected neurons via GFP immunofluorescence. Consistent with data reported by other groups (Zheng et al., 2012), ectopic expression of PTBP2 caused a marked reduction in spine density compared with control neurons. Overexpression of phospho-null 3SA mutant had a similar effect to wild-type PTBP2, whereas transfection of the phospho-mimetic mutant 3SD resembled control neurons (Figure 4A; Figure 1—figure supplement 1A). These data support the notion that phosphorylation by KIS counteracts the negative effects of PTBP2 on synaptic spine maturation.
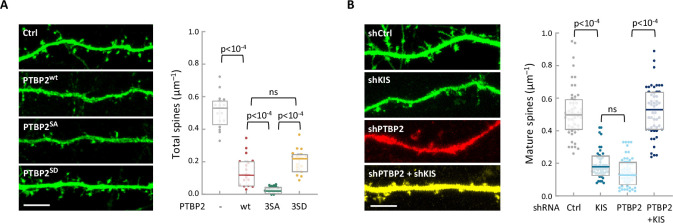
(A) Hippocampal neurons were co-transfected at 5 DIV with a GFP expression plasmid and either a control or PTBP2 expression plasmid. Spine morphology of transfected neurons at 14 DIV was visualized by GFP fluorescence (scale bar, 5 µm). Plot shows the number of spines/µm of 50 neurons per condition from three independent experiments. Median ± Q values and the results of Mann–Whitney tests are also shown. (B) Hippocampal neurons were transfected at 5 DIV with lentiviral vectors expressing GFP or RFP and control, KIS, or PTBP2 shRNAs. Spine morphology of transfected neurons at 14 DIV was visualized by GFP fluorescence (scale bar, 5 µm). Plot shows the number of mature spines/µm of 20 neurons/condition from three independent experiments. Median ± Q values and the results of Mann–Whitney tests are also shown.
Figure 4—figure supplement 1.
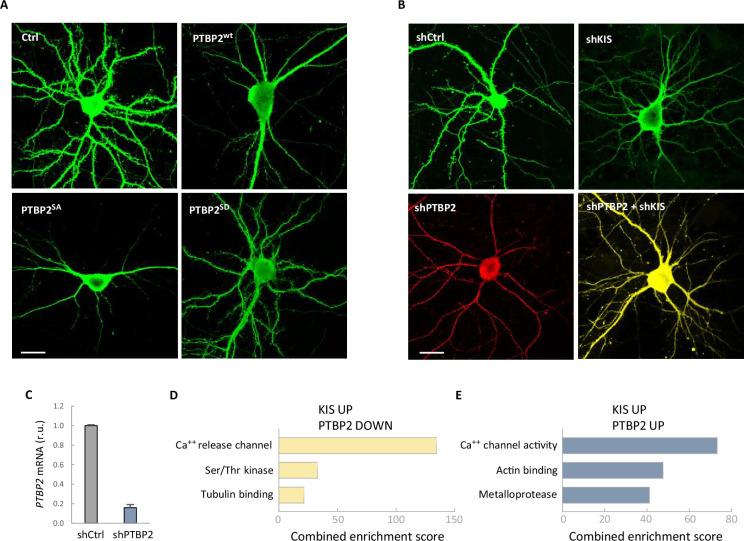
(A, B) Representative confocal images of neurons from the experiment shown in Figure 4. Hippocampal neurons were transfected at 5 DIV and analyzed at 14 DIV (scale bar, 20 µm). (C) PTBP2 mRNA levels in neuroblastoma N2A cells transfected with shCtrl or shPTBP2 and collected 4 days later. Bars represent mean ± standard error of the mean (SEM) (n = 3). Gene ontology (GO) terms significantly enriched (false-discovery rate [FDR] <0.05) of genes with exons upregulated by KIS and downregulated (D) or upregulated (E) by PTBP2.
To further confirm the functional relevance of PTBP2 inhibition by KIS we performed knockdown of the two proteins in hippocampal neurons. As seen previously (Pedraza et al., 2014), KIS depletion significantly hampered spine maturation. On the other hand, PTBP2 knockdown produced a drastic reduction on the number of mature spines, which was accompanied by a clear deterioration of the neurons transfected with the vector expressing shPTBP2 and DsRed. However, double-knockdown of KIS and PTBP2 attenuated the effects produced by loss of either PTBP2 or KIS (Figure 4B; Figure 4—figure supplement 1B, C), supporting the mutually counteracting nature of these two proteins in synaptic maturation.
As shown in Figure 3A, exons upregulated by KIS were clearly enriched in those inhibited by PTBP2. However, although not significantly enriched, we detected a different group of exons positively regulated by both PTBP2 and KIS. These two groups of exons upregulated by KIS belong to genes associated with calcium ion activity but with different specific functions. Genes with exons downregulated by PTBP2 are more enriched in transmembrane transfer of a calcium ion from intracellular stores whereas genes with exons upregulated by PTBP2 facilitate the diffusion of calcium ions through the postsynaptic membrane. Interestingly, with respect to cytoskeletal terms, the two groups showed clearly different term enrichments. Genes with exons downregulated by PTBP2 were significantly associated with the tubulin cytoskeleton, whereas genes with exons upregulated by PTBP2 were associated with the actin cytoskeleton (Figure 4—figure supplement 1C).
Phosphorylation by KIS dissociates PTBP2 protein complexes and impairs RNA binding
To further examine how phosphorylation by KIS modifies PTBP2 splicing activity, we decided to investigate whether the inhibition of PTBP2 produced by KIS could be due to alterations of protein–protein interactions within PTBP2 complexes. It is well known that PTBPs proteins are able to bind to different proteins acting synergistically (Figure 2B) and, in some cases, modify its specificity for some mRNAs (Attig et al., 2018; Coelho et al., 2016). On the other hand, in the present study we have shown that key PTBP2 interactors are also KIS phosphotargets. Therefore, it is reasonable to speculate that the association between PTBP2 and its co-regulators could be modulated as a result of phosphorylation by KIS. To assess interactions in vivo, HEK293T cells were co-transfected with FLAG-PTBP2 and HA-KIS or KISK54A, and endogenous levels of MATR3 and hnRNPM proteins were analyzed in FLAG immunoprecipitates. Co-transfection with KIS resulted in a significant decrease in PTBP2 binding to MATR3 and hnRNPM. In contrast, this decrease did not occur when the K54A mutant was co-transfected, indicating that the kinase activity of KIS is required to cause dissociation of protein complexes (Figure 5A). Similar results were obtained in binding assays in vitro, in which purified KIS forms were added to FLAG-PTBP2 immunoprecipitates, evidencing a direct participation of KIS in the dissociation of PTBP2–MATR3–hnRNPM complexes (Figure 5—figure supplement 1A). in our experiments co-expressing KIS and PTBP2 in HEK293 cells we did not observe any reduction in splicing factor levels. Next, we wanted to confirm these results using an orthogonal approach, by measuring Förster resonance energy transfer (FRET) between mGFP-PTBP2 and mScarlet-hnRNPM in the nucleus of hippocampal neurons transfected at 6 DIV, when KIS expression has not yet reached maximal levels (Figure 1B). In agreement with the data from immunoprecipitation experiments, KIS transfected neurons showed a significant reduction in FRET levels compared to the KISK54A mutant (Figure 5B). Similar results were obtained for both hnRNPM and MATR3 when FRET was analyzed in the nuclei of HEK293T transfected cells (Figure 5—figure supplement 1B). Thus, the two orthogonal approaches show that phosphorylation by KIS disturbs protein–protein interactions within splicing regulatory complexes.
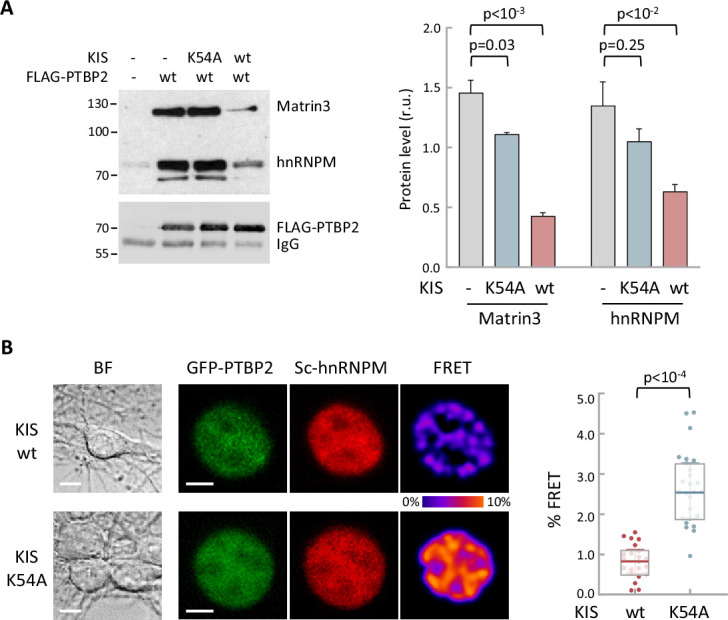
(A) Constructs expressing FLAG-PTBP2 and KIS, KISK54A were transiently transfected into HEK293T cells, and after 24 hr, cell lysates were subject to FLAG immunoprecipitation. Immunoprecipitates were analyzed by immunoblotting to detect endogenous Matrin3 and hnRNPM proteins. A representative blot is shown with empty vector as control (−). Plots show the quantification of endogenous protein levels relative to FLAG-PTBP2 in immunoprecipitates as mean ± standard error of the mean (SEM) (n = 3) values. (B) Bright-field (BF), fluorescence and Förster resonance energy transfer (FRET) images of representative nuclei from hippocampal neurons co-expressing GFP-PTBP2 and mScarlet-hnNRPM in combination with KIS or KISK54A proteins (scale bar, 5 µm). BF images are also shown (scale bar, 10 µm). Plot shows FRET levels from single neuron nuclei expressing KIS (n = 22) or KISK54A (n = 26) as median ± Q values. The results of Mann–Whitney test are also shown.
Figure 5—source data 1.
hnRNPM and Matrin3 in FLAG-PTBP2 immunoprecipitates in vivo.
Figure 5—figure supplement 1.
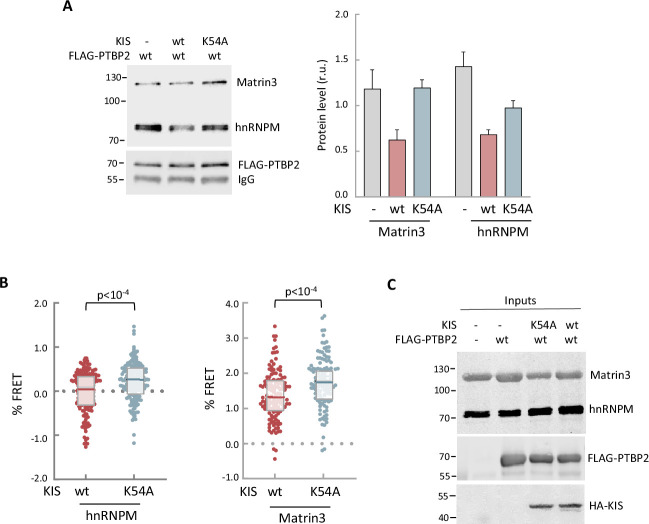
(A) Cell lysates from HEK293T cells expressing FLAG-PTBP2 protein were incubated with KIS recombinant proteins purified from bacteria as His fusions. FLAG immunoprecipitates were analyzed by immunoblotting to detect endogenous Matrin3 and hnRNPM proteins. A representative blot is shown with empty vector as control (−). Plots show the quantification of endogenous protein levels relative to FLAG-PTBP2 in immunoprecipitates as mean ± standard error of the mean (SEM) (n = 3). (B) HEK293T cells co-expressing GFP-PTBP2 and mScarlet-hnNRPM or mScarlet-Matrin3 in combination with HA-KIS or KISK54A proteins. Plots show the Förster resonance energy transfer (FRET) levels from single neuron nuclei as median ± Q values (n >100). The results of Mann–Whitney test are also shown. (C) A representative blot of input samples from the experiment shown in Figure 5A. Constructs expressing FLAG-PTBP2, HA-KIS, and HA-KISK54A were transiently co-transfected into HEK293T cells, and after 24 hr, cell lysates were subject to FLAG immunoprecipitation. Immunoprecipitates were analyzed by immunoblotting to detect endogenous Matrin3 and hnRNPM proteins.
Figure 5—figure supplement 1—source data 1.
hnRNPM and Matrin3 in FLAG-PTBP2 immunoprecipitates in vitro.
Figure 5—figure supplement 1—source data 2.
Representative blot of input samples from Figure 5A.
Finally, we wanted to test whether phosphorylation by KIS could also affect the RNA-binding capacity of PTBP2. For this purpose, we applied the crosslinking and immunoprecipitation cross-linking and immunoprecipitation (CLIP) method to HEK293 cells transfected with FLAG-PTBP2 wt or phosphorylation mutants. Notably, we immunoprecipitated a much lower amount of endogenous RNA crosslinked to the phospho-mimetic mutant 3SD compared to FLAG-PTBP2 wt or 3SA mutant proteins (Figure 6A). Next, we used the same CLIP approach with HEK293 cells transfected with the pFlareG-exon 18 reporter (as in Figure 3C), and analyzed levels of the reporter mRNA in PTBP2 immunoprecipitates. In agreement with the previous CLIP experiment, the reporter mRNA levels were strongly reduced in phospho-mimetic 3SD pulldown samples. Moreover, we observed that the reporter mRNA levels in FLAG-PTBP2 immunoprecipitates were reduced when KIS was co-expressed, whereas the KISK54A mutant had no effect (Figure 6B). These results reinforce the notion that phosphorylation of linker regions within PTBP2 may affect the interactions between RRM domains and RNA. Interestingly, AlphaFold2 predicts important structural alterations in the phosphomimetic S178D and triple 3SD mutants particularly affecting RRM2, which has been shown to function as the minimal splicing repressor domain of PTBP2 (Robinson and Smith, 2006; Figure 6C; Figure 6—figure supplement 1A).
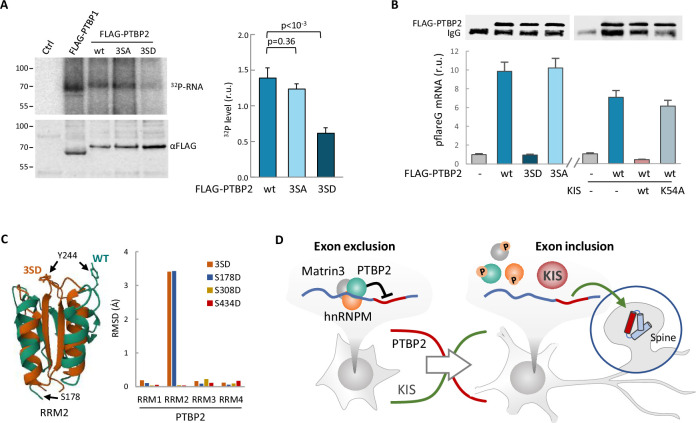
(A) PTBP1 and PTBP2 CLIPs were performed in HEK293T cells expressing FLAG-PTBP1 and FLAG-PTBPs proteins. UV RNA crosslinked and co-precipitated with FLAG-PTBP1 and FLAG-PTBP2s was labeled with 32P-ATP. A representative radiogram gel is shown in the upper image, while the FLAG immunoprecipitates blot is shown in the bottom image. Plot shows the quantification of 32P signal relative to FLAG-PTBP2 immunoprecipitates. Bars represent mean ± standard error of the mean (SEM) (n = 3). (B) PTBP2 CLIP was carried out in HEK293T cells transfected with the pFlareG-PSD95 reporter in combination with plasmid vectors expressing the indicated PTBP2 and KIS proteins. UV-crosslinked pFlareG mRNA co-precipitated with FLAG-PTBP2 was analyzed by RT-qPCR and normalized relative to FLAG-PTBP2 in immunoprecipitates (upper blots). Bars represent mean ± SEM (n = 3). (C) AlphaFold2 prediction of the RRM2 domain from wild-type (green) and 3SD (orange) PTBP2. The different orientation of Tyr244 is highlighted. Plot shows the root-mean-squared deviation (RMSD) values of the four RRM domains in the different phosphomimetic PTBP2 mutants. (D) PTBP2 protein complexes inhibit gene expression through exon exclusion in neuronal precursors whereas, in differentiated neurons, phosphorylation by KIS dissociates PTBP2 protein complexes, which, by being released from the pre-mRNA, allow inclusion of the exon. Since KIS and PTBP2 expression levels are relatively inverted during brain development, phosphorylation-dependent inhibition of PTPB2 by KIS would generate a molecular switch linking transcriptional and alternative exon usage programs during neuronal differentiation.
Figure 6—source data 1.
PTBP2 RNA binding by CLIP.
Figure 6—source data 2.
FLAG-PTBP2 immunoprecipitated levels for CLIP analysis in Figure 6B.
Figure 6—figure supplement 1.
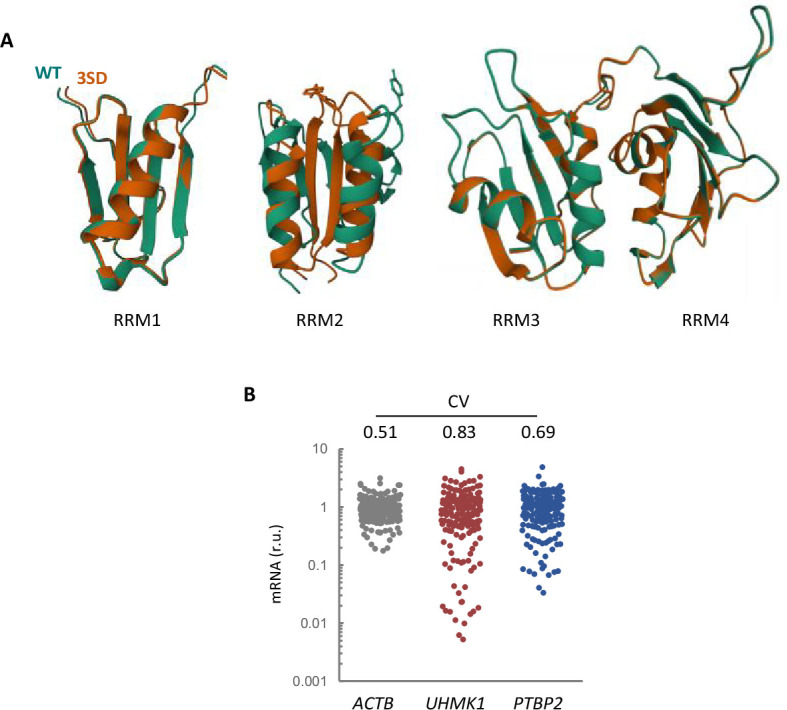
(A) AlphaFold2 prediction of the four RRM domains from wild-type (green) and 3SD (orange) PTBP2. (B) Expression variability of KIS and PTBP2 in single-cell transcriptomic data from hippocampal neurons (Perez et al., 2021). The plot shows relative levels of ACTB, UHMK1, and PTBP2 mRNAs from single-cell somas. The coefficient of variation (CV) is indicated at the top.
Discussion
Alternative splicing has emerged as a critical and widespread gene regulatory mechanism across organisms and tissues. Among the broad tissue types present in metazoans, the nervous system contains some of the highest levels of AS. AS controls multiple steps of neuronal development and plasticity and is a causal agent of pathology (Furlanis and Scheiffele, 2018). We had previously found that KIS is required for the proper expression of postsynaptic proteins at the level of translation (Cambray et al., 2009; Pedraza et al., 2014) and here we show that KIS plays a key additional role in the regulation of AS, pointing to the existence of concerted mechanisms to fine tune postsynaptic protein expression during neuronal differentiation and synaptogenesis.
Although many documented examples of AS regulation exist, much remains to be understood about the control of specific splicing regulators originating tissue and neuron subtype-specific splicing patterns (Raj and Blencowe, 2015). In the present study, we show that phosphorylation by KIS counteracts the activity of the splicing regulator PTBP2 and, as a consequence, controls gene isoform expression during neuronal differentiation and synaptogenesis (Figure 6D). Splicing site recognition and pre-mRNA splicing are dynamic processes involving constant rearrangement of ribonuclear proteins on the pre-mRNA being processed. Splicing regulatory proteins contain RNA-binding and protein–protein interaction domains. They bind with low specificity to mostly single-stranded parts of the pre-mRNA. To overcome the low RNA-binding specificity, splicing regulatory proteins use their protein interaction domains to bind to each other. Most splicing factors are post-translationally modified, phosphorylation being an important contributor to the splicing code that governs the fate of a pre-mRNA. Among the best studied classes of splicing proteins regulated by phosphorylation are the SR proteins (serine-/arginine-rich splicing factors) (Stamm, 2008). For example, phosphorylation of the SR protein SF2/ASF increases its binding to U1 snRNP and decreases its binding to the RNA export factor TAP/NXF1. Moreover, the ability of its arginine/serine domain (RS) to bind to RNA also depends on its phosphorylation state (Huang et al., 2004). The RS domain is highly disordered and switches to a compact, partly ordered arched shape upon phosphorylation (Thapar, 2015). Bioinformatics studies have shown that phosphorylation of intrinsically disordered regions occurs more frequently than in folded domains. This could be due to their increased flexibility and therefore improved kinase accessibility. In accordance with this idea, recombinant PTBP1 and PTBP2 under in vitro splicing conditions present different phosphate modifications located in the unstructured N-terminal and linker regions (Pina et al., 2018). Although the underlying structural mechanism is not clear, the presence of a phosphoryl group introduces electrostatic charges that can affect the structural stability of binding regions or their ability to interact with RNA or other proteins (Newcombe et al., 2022). Supporting this idea, we show that the phosphorylation of PTBP2 linker regions by KIS dissociates both protein–protein and protein–RNA interactions in the PTBP2 complex. Interestingly, AlphaFold2 predicts that a mutation mimicking Ser178 phosphorylation, located at the end of linker 1, would produce major structural changes in the RRM2 domain (Figure 6C; Figure 6—figure supplement 1A). This domain is of particular relevance for the interaction with RNA and splicing co-regulators, such as Matrin3, Raver1, or hnRNPM (Coelho et al., 2015). Within the RRM2 domain, Tyr244 is particularly critical for these interactions (Joshi et al., 2011). The AlphaFold2 prediction shows that a phosphomimetic mutation in Ser178 completely changes the spatial orientation of this Tyr244. AlphaFold2 is not yet reliable enough to predict structures with posttranslational modifications, such as glycosylation, methylation, and phosphorylation (Bertoline et al., 2023). Thus, we cannot rule out that phosphorylation of other residues (Ser308 and Ser434) causes important structural alterations in PTBP2. In particular, AlphaFold2 fails when the modifications lie within unstructured regions, as is the case of Ser308 (Chakravarty and Porter, 2022). The UHM acts as a protein–protein interaction domain (Kielkopf et al., 2004) recognizing short peptides called UHM-ligand motifs (ULM). However, among the KIS phosphotargets identified by us, only SUGP1 contained clear ULM candidates (KRKRKSRW385 and KMGW573K). The absence of ULM in PTBP2, Matrin3, and hnRNPM would indicate that phosphorylation is not mediated by stable protein–protein interactions.
It is worth pointing out that Ser178 and Ser434 are also present in PTPB1, suggesting that phosphorylation of PTBP1 by KIS may have a role in tumorigenesis. In gastric cancer, KIS upregulation promotes growth and migration, indicating that KIS functions as an oncoprotein (Feng et al., 2020). In hepatocellular cancer, KIS stimulates the expression of specific cell cycle regulation genes. Furthermore, the KIS interactome in liver cancer cells revealed a significant enrichment of potential KIS interactors in mRNA splicing, including hnRNPM and PTBP1 (Wei et al., 2019). Finally, KIS has been associated with the epithelial–mesenchyme transition by phosphoproteomic and RNA-seq analyses (Arfelli et al., 2023). Thus, in a non-neuronal context, it is plausible that phosphorylation by KIS modulates the capacity of PTBP1 to interact with RNA and co-regulatory proteins to control cell proliferation. One of the new PTBP1 interactors is SUGP2 (SURP and G-patch domain containing 2), which shares high homology with SUGP1. SUGP2 has been shown to interact with the RRM2 region of PTBP1 in HeLa nuclear extracts (Coelho et al., 2016; Coelho et al., 2015), and SUGP1 interacts and is phosphorylated by KIS in NIH3T3 cells (Arfelli et al., 2023). It is worth mentioning that SUGP1 is one of the most enriched proteins in our KIS phosphoproteome (see Figure 2A). It forms part of the spliceosome complex, interacts with the general splicing factor U2AF2 and has been reported to play an important role in branch-site recognition by its association with SF3B1. Altered levels of SUGP1 have been found in a large number of uveal melanoma and breast cancer cells (Zhang et al., 2019). In the future, it would be interesting to explore whether phosphorylation by KIS interferes with SUGP–PTBP interactions and the functional relevance of this mechanism in cancer and neurogenesis.
The PTBP family represents one of the most salient regulators of splicing in neurogenesis. Sequential changes in the expression of PTBP1 and PTBP2 allow for the establishment of distinct splicing regulatory patterns during neuronal maturation (Fisher and Feng, 2022; Zheng, 2016). PTBP2 expression decreases during the first postnatal week of brain development, maintaining a moderate level in adulthood (Fisher and Feng, 2022; Li et al., 2007). Accordingly, our RNA-seq analysis showed significant PTBP2 expression in cortical neurons at 18 DIV (Figure 2—figure supplement 1A). Strikingly, PTBP1 knockdown stimulates PTBP2 expression and is sufficient to induce neuronal differentiation of fibroblasts (Xue et al., 2013). On the other hand, PTBP2 depletion in developing mouse cortex causes degeneration of this tissue over the first three postnatal weeks, a time when the normal cortex expands and develops mature circuits. Cultured PTBP2−/− neurons fail to mature and start to die after 2 weeks in culture (Li et al., 2014). However, despite all these data on the physiological relevance of PTBP2 in neurogenesis, the role of this splicing regulator in differentiated neurons and how its activity is regulated were not understood. Regarding neuronal differentiation, it is worth noting that KIS expression increases during brain development (Bièche et al., 2003) and in vitro differentiation of hippocampal cultures (Figure 1B), coinciding with postnatal decrease of PTBP2 levels (Zheng et al., 2012). Therefore, the phosphorylation-dependent inhibition of PTPB2 by KIS and the concerted relative inversion in their levels would generate a molecular switch linking transcriptional and alternative exon usage programs in neuronal development.
In mature neurons, AS has a well-established role in expanding proteome diversity (Mauger and Scheiffele, 2017). Although the connection between synaptic activity and the control of KIS expression and/or kinase activity is not yet established, the contraposition of PTBP2 and KIS in splicing may constitute a fine-tuning mechanism to modulate proteome diversity as a function of plasticity-inducing signals. In this regard, single-cell transcriptomic data from hippocampal neurons show that expression variability of KIS and PTBP2 is much higher compared to actin (Perez et al., 2021; Figure 6—figure supplement 1B). Thus, differences in the expression of these two splicing regulators at a single neuron level would increase protein isoform variability and expand diversity in neural circuits, a crucial property in information processing (Miller et al., 2019).
Materials and methods
Key resources table
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus) | RjOrl:SWISS | Janvier Labs | Pregnant female | |
| Cell line (H. sapiens) | HEK 293T | ATCC | CRL-11268 | From human embryo kidney tissue |
| Cell line (M. musculus) | Neuro-2a | ATCC | CCL-131 | Mouse neuroblasts isolated from brain tissue |
| Recombinant DNA reagent | pcDNA3-FLAG-PTBP1 | This work | FLAG tag fused to PTBP1 in pcDNA3 | |
| Recombinant DNA reagent | pcDNA3-FLAG-PTBP2 | This work | FLAG tag fused to PTBP2 in pcDNA3 | |
| Recombinant DNA reagent | pEGFP-C3 | Addgene | 6082-1 | N-terminal tagging vector with GFP |
| Recombinant DNA reagent | pEGFP-PTBP2 | This work | PTBP2 fused to EGFP | |
| Recombinant DNA reagent | mScarlet-C1 | Addgene | 85044 | N-terminal tagging vector with Scarlet |
| Recombinant DNA reagent | mScarlet-Matr3 | This work | Matr3 fused to Scarlet | |
| Recombinant DNA reagent | mScarlet-hnRNPM | This work | hnRNPM fused to Scarlet | |
| Recombinant DNA reagent | pGEX-KG | ATCC | 77103 | Bacterial expression vector expressing GST tag |
| Recombinant DNA reagent | pET28a | Novagen | 69864-3 | Bacterial expression vector expressing His tag |
| Recombinant DNA reagent | pcDNA3-FLAG-PTBP1 | This work | PTBP1 fused to FLAG tag | |
| Recombinant DNA reagent | pcDNA3-FLAG-PTBP2 | This work | PTBP2 fused to FLAG tag | |
| Recombinant DNA reagent | pFlareG-PSD95 exon 18 | Addgene | 90266 | Minigene reporter for alternative splicing analysis |
| Recombinant DNA reagent | pCMV-dR8.2 dvpr | Addgene | 8455 | 2nd generation lentiviral packaging plasmid |
| Recombinant DNA reagent | pCMV-VSV-G | Addgene | 8454 | Lentiviral packaging plasmid expressing envelope protein |
| Recombinant DNA reagent | pLKO.3G | Addgene | 14748 | Lentiviral plasmid expressing GFP |
| Recombinant DNA reagent | pLKO.3-DsRed shPTBP2 | This work | Lentiviral vector expressing DsRed and shRNA sequence against PTBP2 | |
| Chemical compound, drug | α-FLAG M2 (monoclonal mouse) affinity gel beads | Merck-Sigma | A2220 | |
| Antibody | α-FLAG M2 (monoclonal mouse) | Merck-Sigma | F3165 | 1:500 (WB) |
| Antibody | α-Matr3 (rabbit polyclonal) | Bionova | A300-591A-T | 1:500 (WB) |
| Antibody | α-hnRNP M3/4 (rabbit polyclonal) | Bionova | A303-910A-T | 1:500 (WB) |
| Antibody | α-mouse IRDye 680RD (goat polyclonal) | LI-COR Biosciences | 926-68070 | 1:1000 (WB) |
| Antibody | α-rabbit HRP (goat polyclonal) | Thermo Fisher | 31460 | 1:1000 (WB) |
| Antibody | α-PSD95 (mouse monoclonal) | Millipore | MABN68 | 1:400 (IF) |
| Antibody | α-SRRM2 (rabbit polyclonal) | Merck-Sigma | HPA066181 | 1:250 (IF) |
| Antibody | α-GFP Alexa 488 (rabbit polyclonal) | Thermo Fisher | A21311 | 1:1000 (IF) |
| Antibody | α-mouse Alexa 568 (donkey polyclonal) | Life Technologies | A10037 | 1:1000 (IF) |
| Antibody | α-rabbit Alexa 568 (rabbit polyclonal) | Life Technologies | A10042 | 1:1000 (IF) |
| Antibody | α-rat Alexa 568 (goat polyclonal) | Invitrogen | A11077 | 1:1000 (IF) |
| Commercial assay or kit | CalPhos mammalian transfection kit | Clontech | 631312 | |
| Commercial assay or kit | Lipofectamine 2000 | Invitrogen | 11668030 | |
| Commercial assay or kit | FuGene HD | Promega | E2311 | |
| Commercial assay or kit | NucleoSpin Plasmid Kit | Macherey-Nagel | 740588 | |
| Commercial assay or kit | NucleoBond Xtra Midi Plus EF Kit | Macherey-Nagel | 740422 | |
| Commercial assay or kit | EZNA total RNA purification Kit | Omega | R6834-01 | |
| Software, algorithm | FRETmapJ | This work | https://www.ibmb.csic.es/en/department-of-cells-and-tissues/control-of-local-mrna-expression/ | |
| Software, algorithm | SpineJ | Pedraza et al., 2014 | https://www.ibmb.csic.es/en/department-of-cells-and-tissues/control-of-local-mrna-expression/ | |
| Software, algorithm | Prism (version 9.5.1) | GraphPad | RRID: SCR_002798 | https://graphpad.com/features |
Primary cultures
Animal experimental procedures were approved by the ethics committee of the National Research Council of Spain (approval number: B9900083). We have used hippocampal neurons for imaging experiments in which it is important to have isolated mature neurons with well-defined dendritic spines, whereas cortical neuron cultures have been used for biochemical experiments where a larger amount of material is required. Hippocampi and cortex were dissected from E17.5 embryos of Swiss mice (undetermined sex) in Hank's Balanced Salt Solution (HBSS) containing 0.6% glucose and 10 mM 4-(2-hydroxyethyl)-1-piperazineethane sulfonic acid (HEPES). After dissection, tissues were trypsinized (Fisher, 15090046) in HBSS at 37°C for 15 min. Enzymatic digestion was stopped by washing the tissue three times with Minimum Essential Medium Eagle (MEM; Fisher, 31095029) supplemented with 10% fetal bovine serum (FBS) and 0.6% glucose. Hippocampi were left to sediment between washes, and centrifugation was avoided to keep cell viability. Trypsin-treated tissue was then mechanically disaggregated by passing through a flame-polished Pasteur pipette (~10 times). Cells were plated at desired density on poly-D-lysine (Merck-Sigma, P7886)-coated plates (0.5 mg/ml poly-L-lysine in borate buffer, pH 8.5) and maintained in MEM, 10% FBS and 0.6% glucose for 2–4 hr. Culture medium was then substituted for Neurobasal (Fisher, 11570556) supplemented with 2% B27 (Fisher, 11530536) and 1% GlutaMAX (Fisher, 35050-038). Primary hippocampal cultures were transfected using CalPhos mammalian transfection kit (Clontech, 631312) as previously (Jiang and Chen, 2006).
Cell line culture and transfection
HEK293T cells and neuroblastoma N2A cells were cultured in Dulbecco’s modified Eagle medium (DMEM) medium with 10% FBS. HEK293T transfection was carried out with Lipofectamine 2000 (Invitrogen, 11668030) and N2A transfection was performed with FuGene HD (Promega, E2311), according to the manufacturer’s instructions. Cells were routinely tested for mycoplasma contamination.
DNA constructs
Site-directed mutagenesis in PTBP2 and UHMK1 cDNAs was performed by In-Fusion HD (Takara, 638909). pcDNA3-FLAG, pEGFP-C3, mScarlet-C1, pGEX-KG, pET28a, and pcDNA3-HA were used as host vectors. Plasmids were prepared using NucleoSpin Plasmid Kit (Macherey-Nagel, 740588) for cell line transfections and NucleoBond Xtra Midi Plus EF Kit (Macherey-Nagel, 740422) for neuron transfections.
Lentivirus production and infection
shRNA sequences in pLKO.3 lentiviral vectors were as follows: GAGTGCGGAGAATGAGTGTTT (TRCN0000027622) against Mm UHMK1, and GCTGTACCCTAAGGATTGATT (TRCN0000109231) against Mm PTBP2. We used MISSION pLKO.1-puro non-mammalian shRNA control (Merck-Sigma, SHC002). HEK293FT cells were transfected with Lipofectamine 2000 with lentiviral vector, envelope plasmid pVSV-G, and packaging plasmid pCMV-dR8.2 dvpr, and cultured in DMEM 10% FBS. Lentiviruses were harvested 1, 2, and 3 days after transfection, filtered through 0.45 μm cellulose-acetate syringe filters, and concentrated by centrifugation at 26,000 rpm for 2 hr at 4°C. Titration was performed by serial dilution infection in HEK293T and checking the transduction efficiency by fluorescence as described (Ritter et al., 2017). Cortical neurons cultured for 7 DIV were infected with 2 Multiplicity of Infection (MOI) lentiviruses for 3 hr, and harvested at 18 DIV.
RT-PCR and semiquantitative PCR and qPCR
RNAs were extracted from indicated samples using EZNA total RNA purification KIT (Omega, R6834-01), digested with RNase-free DNase (Roche, 11119915001), and 1 µg of RNA was reverse-transcribed into cDNAs using the Prime Script RT Reagent Kit (Takara, RR037B) according to the manufacturer’s protocol. RT-PCR primers designed to amplify two spliced isoforms with different sizes are shown in Supplementary file 3 was performed with Taqman probes (6xFAM-BQ1) on LightCycler@96 Real-Time PCR system (Roche) according to manufacturer’s instructions. qPCR primers are shown in Supplementary file 3.
Co-immunoprecipitation, in vitro phosphorylation, and western blot
HEK293T cells were transfected using Lipofectamine 2000 with a proportion 1:3:9 of pEGFP, pcDNA-FLAG/FLAG-PTBP2, and pcNBM470-HA-KIS/HA-KISK54A. After 48 hr post-transfection, cells were washed with cold phosphate-buffered saline (PBS) and homogenized with lysis buffer (50 mM Tris–HCl pH 8, 150 mM KCl, 5 mM MgCl2, 0.25% NP-40, 0.05% sodium deoxycholate, 5% glycerol, 1 mM Ethylene Glycol-bis-(beta-aminoethylether)-N,N,N',N'-tetraacetic Acid [EGTA], 1 mM Ethylenediaminetetraacetic Acid [EDTA], and protease and phosphatase inhibitor). To preserve only direct interactions, RNA was digested using RNase I (Fisher, 10330065). After sonication, lysates were clarified by centrifugation at 12,000 × g and incubated 2 hr at 4°C with pre-equilibrated α-FLAG M2 affinity gel beads (Merck-Sigma, A2220). Beads were washed three times with lysis buffer. After washes, beads were eluted for western blot analysis or equilibrated to perform in vitro phosphorylation assay. For in vitro assay, FLAG immunoprecipitates were incubated in BK buffer (50 mM HEPES pH 7.6, 10 mM MgCl2, 2 mM dithiothreitol [DTT], 10 µM adenosine triphosphate [ATP]) with 200 ng of His-KIS or His-KISK54A purified from E. coli for 30 min at 30°C. After three washes with lysis buffer, beads were eluded with sodium dodecyl sulfate (SDS) sample buffer. Antibodies used for immunoblotting were: α-FLAG M2 (1:500; Merck-Sigma, F3165), α-Matr3 (1:500; BioNova, A300-591A-T), α-hnRNP M3/4 (1:500; BioNova, A303-910A-T), IRDye 680RD (1:10,000; LI-COR Biosciences, 926-68070), and α-rabbit HRP (1:10,000; Fisher, 31460, RRID: AB_228341).
In vitro kinase assay
Kinase reactions were carried out in 20 μl of BK buffer (50 mM HEPES pH 7.6, 10 mM MgCl2, 2 mM DTT, 10 µM ATP), 10 μCi of [γ-32P]-ATP, 0.2 μg of GST-KIS (or GST-KISK54A) (Cambray et al., 2009), and 2 μg of the corresponding GST-PTBP2 proteins. Kinase reactions were incubated for 20 min at 30°C. Phosphorylated products were separated by SDS–polyacrylamide gel electrophoresis, stained with Coomassie Brilliant Blue, dried, and analyzed by autoradiography.
PSD95 and SSRM2 immunofluorescence
Hippocampal neurons were fixed at different DIVs using 4% paraformaldehyde and 4% sucrose in PBS for 30 min at 4°C and then washed three times with PBS. Neurons were permeabilized for 5 min with 0.1% Triton X-100 and blocked with 5% Normal Goat Serum (NGS) in PBS (blocking solution). Primary antibody α-PSD95 (EMD Millipore, MABN68, RRID: AB_10807979) were diluted 1:400 in blocking solution. Proteins were detected by incubation with secondary antibodies Alexa 568 donkey anti-mouse (Life Technologies, A10037, RRID: AB_11180865) diluted 1:1000 in blocking solution. Images were acquired with a Zeiss LSM780 confocal microscope using the following parameters: ~15-μm-thick stacks were imaged every 0.5 μm (pinhole set at 1 Airy unit), under ×40 objective (0.13 μm/pixel). Laser power and PMT values were kept constant throughout images and conditions. PSD95 somatic quantification was performed using ImageJ by measuring fluorescent intensity on transfected neurons and relativizing it to the somatic expression of the non-transfected neurons in the same field (Zheng et al., 2012). For nuclear SRRM2 immunofluorescence, HEK293T were transfected with GFP or GFP-KIS plasmids. 24 hr post-transfection, cells were fixed with 4% paraformaldehyde for 20 min. Plates were washed twice with PBS and permeabilized with cold 100% methanol for 2 min, before sequential staining with primary and secondary antibodies. Cells were washed three times with PBS and imaged within 24 hr. Primary immunofluorescence was staining with α-SRRM2 (1:250; Merck-Sigma, HPA066181) and secondary with Alexa 568 α-rabbit (1:1000; Life Technologies, A10042; RRID: AB_2534017).
Spine analysis
Hippocampal neurons were seeded in 4-well plates with 40,000 cells per well, transfected at 4 DIV and fixed at 14 DIV. Primary antibodies α-GFP-Alexa Fluor 488 (Fisher, A21311) and α-Red Rat (Gift from Dr Pons) were diluted 1:1000 in blocking solution. α-Rat Goat Polyclonal Antibody, Alexa Fluor 568 (Invitrogen, A11077) diluted 1:1000 in blocking was used as secondary antibody. Number of fluorescent spines were determined as previously described (Mendoza et al., 2021). Images were acquired using a Zeiss LSM780 confocal microscope. Stacks of 10 slices were imaged every 0.37 µm, with a pinhole value of 1 airy unit under a ×63 objective at 0.11 µm per pixel.
Dual fluorescence labeling
N2A cells were reverse transfected with a proportion 3:9 of pFlareG-PSD95-e18 (Zheng et al., 2013) and HA-KIS or HA-KISK54A. After 72 hr fluorescent levels were recorded with a Leica Thunder Imager. Led power and exposition time were kept constant for all images and conditions. After image-deconvolution process, single-cell RFP and GFP were measured with ImageJ. The ratio of exclusion/inclusion of exon 18 in pFlareG was measured as the log2(RFP/GFP).
PTBP1 and PTBP2 CLIP assay
HEK293T cells were transfected with FLAG-PTBPs proteins. 48 hr post-transfection, cells were washed once with PBS and crosslinked with 150 mJ/cm2 UV light at 254 nm in Stratalinker 2400. Cells were collected by centrifugation and resuspended on lysis buffer (50 mM Tris–HCl pH 7.4, 100 mM NaCl, 1% IGEPAL CA-630, 0.1% SDS, 0.5% sodium deoxycholate, protease and phosphatase inhibitors). After sonication, lysates where digested with 4 U/ml Turbo DNase (Fisher, AM2238) and 1.5 U/μl RNase I (Fisher, 10330065). Lysates were centrifugated and supernatants incubated with pre-equilibrated α-FLAG M2 affinity gel beads for 2 hr at 4°C. Beads were washed twice with high-salt wash buffer (50 mM Tris–HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 0.1% SDS, 0.5% sodium deoxycholate, protease and phosphatase inhibitors). At that point, beads were equilibrated twice with PNK buffer (20 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 0.2% Tween-20) and RNA 5′ ends where labeled with [γ-32P]-ATP and T4 PNK (NEB, M0201L) at 37°C for 5 min. Beads were eluded at 70°C for 5 min with NuPAGE loading buffer (Fisher, 11549166) and loaded in 4–12% NuPAGE Bis-Tris gel (Fisher, 10247002) for subsequent electrophoresis and transference to nitrocellulose membrane (Merck-Sigma, GE10600003). Precipitated RNA was analyzed by autoradiography and FLAG-PTBP protein levels were measured by western blot.
pFlareG CLIP assay
Protocol used for CLIP assay from pFlareG plasmid was essentially as in the previous section with some modifications. Briefly, HEK293T cells were transfected with pFlareG reporter in combination with plasmid vectors expressing FLAG-PTBP2 proteins and HA-KIS proteins. After crosslinking, cells were resuspended in lysis buffer plus 40 U/ml RNase inhibitor (Attendbio, RNI-G), sonicated and incubated with α-FLAG M2 affinity gel beads overnight at 4°C. Beads were washed twice with lysis buffer, twice with lysis buffer without detergents and once with DNase buffer (10 mM Tris–HCl, pH 7.4, 2.5 mM MgCl2, 0.5 mM CaCl2). Beads were digested with DNase I (Merck-Sigma, DN25) for 15 min and inactivated. RNAs bound to the beads were used as a template for cDNA synthesis using Maxima H Minus cDNA first strand synthesis Kit (Thermo Fisher, 10338179) and random hexamers as primers. 2 µl of cDNA was used as a template in a real-time qPCR assay. pFlareG mRNA was amplified with GFP annealing primers (see Supplementary file 3). Relative levels of pFlareG were calculated for each condition using GAPDH as a housekeeping gene and normalized relative to FLAG-PTBP2 protein levels immunoprecipitated checked by western blot using α-FLAG M2 antibody (1:500; Merck-Sigma, F3165) and IRDye 800RD (1:10,000; LI-COR Biosciences, 926-32210).
FRET imaging
Hippocampal cultures were transfected at 6 DIV with FRET biosensor plasmids expressing mGFP-PTBP2 proteins and pmScarlet-hnRNPM, together with HA-KIS or HA-KISK54A, as indicated. Live imaging was conducted 16 hr after transfection. Neurons were live-imaged using a Zeiss LSM780 confocal microscope with a ×40 1.2 NA water-immersion objective. Images were 1024 × 1024 pixels, with a pixel width of 65 nm. Briefly, donor (mGFP-PTBP2) protein was excited at 488 nm, and its emission was measured at 490–532 nm (ID). Excitation of the acceptor (pmScarlet-hnRNPM) was at 561 nm, and emission was measured at 563 to 695 nm (intensity of acceptor [IA]) . We also measured the total signal emitted at 563–695 nm when excited at 488 nm (intensity of FRET [IF]) to obtain the FRET efficiency as F% = 100 * (IF − kD*ID − kA*IA)/IA, kD, and kA, correcting acceptor cross-excitation and donor bleed-through, respectively, with the aid of FRETmapJ, a plugin that also provides maps with the FRET signal as pixel value for local quantification. For HEK293T experiments, cells were transfected with plasmids expressing mGFP-PTBP2 and pmScarlet-hnRNPM or pmScarlet-Matr3, together with HA-KIS or HA-KISK54A, as indicated.
Mass spectrometry-based phosphoproteomics analysis
HEK293T cells were transfected with plasmids expressing KIS and KISK54A. Methods used for sample preparation and data acquisition were as previously described (Azman et al., 2023) with some modifications. Briefly, 100 µg of total lysate were used for filter aided sample preparation (iFASP). Digestion with trypsin and chymotrypsin was performed in two different experiments before TMT isobaric labeling (TMT-126TM and TMT-131TM Thermo Fisher). Trypsin (Promega, V5280) and chymotrypsin (Promega, V106A) digested eluates were then pooled independently. The two pooled mixtures were fractionated into four fractions using PierceTM High pH reverse-phase fractionation kit (Life Technologies, 84868), according to the manufacturer’s instructions. Fractions samples were subjected to TiO phosphopeptide enrichment using TiO enrichment kit (GL Sciences, 5010-21308), according to the manufacturer’s instructions. Phosphopeptides were analyzed using a Q Exactive plus Orbitrap mass spectrometer (Barts Cancer Institute, London). MaxQuant (version 1.6.3.3) software was used for database search and label-free quantification of mass spectrometry raw files. The search was performed against a FASTA file of Homo sapiens proteome, extracted from Uniprot (2016). All downstream data analysis was performed using Perseus (version 1.6.2.3). To perform enrichment analysis, using GSEA software (Mootha et al., 2003), phospho-site information was summarized at protein level by PPS. PPS was calculated as the sum of the log2FC value for each phospho-site on a given protein. For GO analysis, the c5.all.v2023.1.Hs.symbols.gmt list provided by GSEA was utilized. Top 50 PTBP2 interactors were obtained from the STRING database by full string network.
RNA extraction, sequencing, and data analysis
Total RNA was obtained from 18 DIV infected cortical neurons and 7 and 18 DIV of untreated neurons using EZNA total RNA purification kit (Omega, R6834-01), and used to prepare polyA+ enriched libraries for paired-end (2 × 100 bp) sequencing (BGI DNBSEQ platform). More than 90 million reads were obtained per sample, which were aligned to the Mm10 mouse genome using HISAT2 (Kim et al., 2015) in the Galaxy platform (Boekel et al., 2015). Assignment to transcriptional units, standard filtering steps and basic statistical analysis of triplicate samples was done using Rsubread (version 3.6) (Liao et al., 2019) in Bioconductor (Huber et al., 2015) and custom R scripts. Genes with a single exon were removed and only exons with more than 5 reads in all samples of one condition were considered. For every annotated exon a splicing index (SI) (Mauger et al., 2016) was calculated as the ratio of exon to gene rpkm. GO term and specific GSEA was performed using Enrichr (Chen et al., 2013) and GSEA (Mootha et al., 2003) tools, respectively. A subset of 8457 exons with low variability (Fano factor <0.0005) within the KIS knockdown triplicate samples was selected, and the significance of differences to control triplicate samples was assessed by calculating the FDR with a modified t-test statistic that incorporates a background variance parameter s0 = 0.1 (Tusher et al., 2001). In this manner, a total of 2247 exons offered q < 0.05. In contrast, when the same analysis was applied to the experimental dataset randomized by Gaussian attribution or permutation, less than 5 exons displayed q < 0.05.
Statistical analyses
Statistical analyses were performed using GraphPad PRISM (version 9). Pair comparisons were performed with the nonparametric Mann–Whitney test, and the resulting pairwise p values are shown in the corresponding figure panels. Data are displayed as median and quartile (Q) values, and the number of samples is described in the figure legends. Protein levels by immunoblotting and mRNA levels by RT-PCR were determined in triplicate samples, and mean ± standard error of the mean values are shown.
Software
FRETmapJ can be obtained as an ImageJ (Wayne Rasband, NIH) plugin from https://www.ibmb.csic.es/en/department-of-cells-and-tissues/control-of-local-mrna-expression/.
Acknowledgements
We thank E Rebollo for technical assistance, M Aldea for the helpful comments, and C Rose for editing the manuscript. We also thank members of M Aldea and C Gallego laboratories for the stimulating discussions and ideas. Funding This work was funded by a grant from the Ministry of Science and Innovation of Spain and the European Union (FEDER) (PID2020-113231GB-I00) to CG Medical Research Council UK (MR/P009417/1) and Barts Charity (MGU0346) grants to FKM.
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
Jens Lüders, Institute for Research in Biomedicine Spain,
Amy H Andreotti, Iowa State University United States,
Funding Information
This paper was supported by the following grants:
Ministerio de Ciencia e Innovación PRE2018-083268 to Mónica B Mendoza.
Ministerio de Ciencia e Innovación PID2020-113231GB-I00 to Carme Gallego.
Medical Research Council MR/P009417/1 to Faraz K Mardakheh.
Barts Charity MGU0346 to Faraz K Mardakheh.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Methodology.
Formal analysis, Methodology.
Formal analysis, Methodology.
Methodology.
Formal analysis, Methodology.
Conceptualization, Methodology.
Conceptualization, Formal analysis, Supervision, Funding acquisition, Investigation, Methodology, Writing - original draft, Writing - review and editing.
Ethics
Animal experimental procedures were approved by the ethics committee of the Research Council of Spain (CSIC).
Additional files
Supplementary file 1.
RNA-seq datasets from cortical neurons.Supplementary file 2.
Phosphoproteomic data from HEK293T expressing KIS and KISK54A.Supplementary file 3.
Primers and vectors used in this study.MDAR checklist
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository (PXD050320). RNA-seq fastq files and read summaries are available from GEO (GSE260790).
The following datasets were generated:
Moreno-Aguilera M, Neher AM, Mendoza MB, Dodel M, Ortiz R, Gallego C. 2024. KIS counteracts PTBP2 and regulates alternative exon usage in neurons. PRIDE. PXD050320
Moreno-Aguilera M, Neher AM, Mendoza MB, Dodel M, Ortiz R, Gallego C. 2024. KIS counteracts PTBP2 and regulates alternative exon usage in neurons. NCBI Gene Expression Omnibus. GSE260790
References
- Arfelli VC, Chang YC, Bagnoli JW, Kerbs P, Ciamponi FE, Paz L, Pankivskyi S, de Matha Salone J, Maucuer A, Massirer KB, Enard W, Kuster B, Greif PA, Archangelo LF. UHMK1 is a novel splicing regulatory kinase. The Journal of Biological Chemistry. 2023;299:103041. 10.1016/j.jbc.2023.103041. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Attig J, Agostini F, Gooding C, Chakrabarti AM, Singh A, Haberman N, Zagalak JA, Emmett W, Smith CWJ, Luscombe NM, Ule J. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell. 2018;174:1067–1081. 10.1016/j.cell.2018.07.001. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Azman MS, Alard EL, Dodel M, Capraro F, Faraway R, Dermit M, Fan W, Chakraborty A, Ule J, Mardakheh FK. An ERK1/2-driven RNA-binding switch in nucleolin drives ribosome biogenesis and pancreatic tumorigenesis downstream of RAS oncogene. The EMBO Journal. 2023;42:e110902. 10.15252/embj.2022110902. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. 10.1126/science.1230612. [Abstract] [CrossRef] [Google Scholar]
- Bertoline LMF, Lima AN, Krieger JE, Teixeira SK. Before and after AlphaFold2: An overview of protein structure prediction. Frontiers in Bioinformatics. 2023;3:1120370. 10.3389/fbinf.2023.1120370. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bièche I, Manceau V, Curmi PA, Laurendeau I, Lachkar S, Leroy K, Vidaud D, Sobel A, Maucuer A. Quantitative RT-PCR reveals a ubiquitous but preferentially neural expression of the KIS gene in rat and human. Brain Research. Molecular Brain Research. 2003;114:55–64. 10.1016/s0169-328x(03)00132-3. [Abstract] [CrossRef] [Google Scholar]
- Boekel J, Chilton JM, Cooke IR, Horvatovich PL, Jagtap PD, Käll L, Lehtiö J, Lukasse P, Moerland PD, Griffin TJ. Multi-omic data analysis using Galaxy. Nature Biotechnology. 2015;33:137–139. 10.1038/nbt.3134. [Abstract] [CrossRef] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Development. 2007;21:1636–1652. 10.1101/gad.1558107. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cambray S, Pedraza N, Rafel M, Garí E, Aldea M, Gallego C. Protein kinase KIS localizes to RNA granules and enhances local translation. Molecular and Cellular Biology. 2009;29:726–735. 10.1128/MCB.01180-08. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chakravarty D, Porter LL. AlphaFold2 fails to predict protein fold switching. Protein Science. 2022;31:e4353. 10.1002/pro.4353. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. 10.1186/1471-2105-14-128. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Coelho M, Attig J, Bellora N, König J, Hallegger M, Kayikci M, Eyras E, Ule J, Smith CWJ. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. The EMBO Journal. 2015;34:653–668. 10.15252/embj.201489852. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Coelho MB, Ascher DB, Gooding C, Lang E, Maude H, Turner D, Llorian M, Pires DEV, Attig J, Smith CWJ. Functional interactions between polypyrimidine tract binding protein and PRI peptide ligand containing proteins. Biochemical Society Transactions. 2016;44:1058–1065. 10.1042/BST20160080. [Abstract] [CrossRef] [Google Scholar]
- Corrionero A, Valcárcel J. RNA processing: Redrawing the map of charted territory. Molecular Cell. 2009;36:918–919. 10.1016/j.molcel.2009.12.004. [Abstract] [CrossRef] [Google Scholar]
- Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcárcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nature Structural & Molecular Biology. 2007;14:620–629. 10.1038/nsmb1260. [Abstract] [CrossRef] [Google Scholar]
- Feng X, Ma D, Zhao J, Song Y, Zhu Y, Zhou Q, Ma F, Liu X, Zhong M, Liu Y, Xiong Y, Qiu X, Zhang Z, Zhang H, Zhao Y, Zhang K, Hong X, Zhang Z. UHMK1 promotes gastric cancer progression through reprogramming nucleotide metabolism. The EMBO Journal. 2020;39:e102541. 10.15252/embj.2019102541. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fisher E, Feng J. RNA splicing regulators play critical roles in neurogenesis. Wiley Interdisciplinary Reviews. RNA. 2022;13:e1728. 10.1002/wrna.1728. [Abstract] [CrossRef] [Google Scholar]
- Furlanis E, Scheiffele P. Regulation of neuronal differentiation, function, and plasticity by alternative splicing. Annual Review of Cell and Developmental Biology. 2018;34:451–469. 10.1146/annurev-cellbio-100617-062826. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y, Shieh AW, Haney J, Parhami S, Belmont J, Kim M, Moran Losada P, Khan Z, Mleczko J, Xia Y, Dai R, Wang D, Yang YT, Xu M, Fish K, Hof PR, Warrell J, Fitzgerald D, White K, Jaffe AE, PsychENCODE Consortium. Peters MA, Gerstein M, Liu C, Iakoucheva LM, Pinto D, Geschwind DH. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. 10.1126/science.aat8127. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Research. 2008;36:4823–4832. 10.1093/nar/gkn463. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. PNAS. 2004;101:9666–9670. 10.1073/pnas.0403533101. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 2015;12:115–121. 10.1038/nmeth.3252. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature Protocols. 2006;1:695–700. 10.1038/nprot.2006.86. [Abstract] [CrossRef] [Google Scholar]
- Joshi A, Coelho MB, Kotik-Kogan O, Simpson PJ, Matthews SJ, Smith CWJ, Curry S. Crystallographic analysis of polypyrimidine tract-binding protein-Raver1 interactions involved in regulation of alternative splicing. Structure. 2011;19:1816–1825. 10.1016/j.str.2011.09.020. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical Reviews in Biochemistry and Molecular Biology. 2012;47:360–378. 10.3109/10409238.2012.691456. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kielkopf CL, Lücke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes & Development. 2004;18:1513–1526. 10.1101/gad.1206204. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. 10.1038/nmeth.3317. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- LaForce GR, Philippidou P, Schaffer AE. mRNA isoform balance in neuronal development and disease. Wiley Interdisciplinary Reviews. RNA. 2023;14:e1762. 10.1002/wrna.1762. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nature Reviews. Neuroscience. 2007;8:819–831. 10.1038/nrn2237. [Abstract] [CrossRef] [Google Scholar]
- Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. 10.7554/eLife.01201. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research. 2019;47:e47. 10.1093/nar/gkz114. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes & Development. 2012;26:1626–1642. 10.1101/gad.191338.112. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lin Y-C, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, Chen J, Speleman F, Lambrechts D, Van de Peer Y, Tavernier J, Callewaert N. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nature Communications. 2014;5:4767. 10.1038/ncomms5767. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Linares AJ, Lin CH, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:e09268. 10.7554/eLife.09268. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan L-Y, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, Smith CWJ. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nature Structural & Molecular Biology. 2010;17:1114–1123. 10.1038/nsmb.1881. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lopez Soto EJ, Gandal MJ, Gonatopoulos-Pournatzis T, Heller EA, Luo D, Zheng S. Mechanisms of neuronal alternative splicing and strategies for therapeutic interventions. The Journal of Neuroscience. 2019;39:8193–8199. 10.1523/JNEUROSCI.1149-19.2019. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Manceau V, Kielkopf CL, Sobel A, Maucuer A. Different requirements of the kinase and UHM domains of KIS for its nuclear localization and binding to splicing factors. Journal of Molecular Biology. 2008;381:748–762. 10.1016/j.jmb.2008.06.026. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Maucuer A, Ozon S, Manceau V, Gavet O, Lawler S, Curmi P, Sobel A. KIS is a protein kinase with an RNA recognition motif. The Journal of Biological Chemistry. 1997;272:23151–23156. 10.1074/jbc.272.37.23151. [Abstract] [CrossRef] [Google Scholar]
- Mauger O, Lemoine F, Scheiffele P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016;92:1266–1278. 10.1016/j.neuron.2016.11.032. [Abstract] [CrossRef] [Google Scholar]
- Mauger O, Scheiffele P. Beyond proteome diversity: alternative splicing as a regulator of neuronal transcript dynamics. Current Opinion in Neurobiology. 2017;45:162–168. 10.1016/j.conb.2017.05.012. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mazin PV, Khaitovich P, Cardoso-Moreira M, Kaessmann H. Alternative splicing during mammalian organ development. Nature Genetics. 2021;53:925–934. 10.1038/s41588-021-00851-w. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mendoza MB, Gutierrez S, Ortiz R, Moreno DF, Dermit M, Dodel M, Rebollo E, Bosch M, Mardakheh FK, Gallego C. The elongation factor eEF1A2 controls translation and actin dynamics in dendritic spines. Science Signaling. 2021;14:eabf5594. 10.1126/scisignal.abf5594. [Abstract] [CrossRef] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. 10.1126/science.1228186. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Miller DJ, Bhaduri A, Sestan N, Kriegstein A. Shared and derived features of cellular diversity in the human cerebral cortex. Current Opinion in Neurobiology. 2019;56:117–124. 10.1016/j.conb.2018.12.005. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. PNAS. 2003;100:605–610. 10.1073/pnas.242716699. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Newcombe EA, Delaforge E, Hartmann-Petersen R, Skriver K, Kragelund BB. How phosphorylation impacts intrinsically disordered proteins and their function. Essays in Biochemistry. 2022;66:901–913. 10.1042/EBC20220060. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. pLogo: A probabilistic approach to visualizing sequence motifs. Nature Methods. 2013;10:1211–1212. 10.1038/nmeth.2646. [Abstract] [CrossRef] [Google Scholar]
- Pedraza N, Ortiz R, Cornadó A, Llobet A, Aldea M, Gallego C. KIS, A kinase associated with microtubule regulators, enhances translation of AMPA receptors and stimulates dendritic spine remodeling. The Journal of Neuroscience. 2014;34:13988–13997. 10.1523/JNEUROSCI.1573-14.2014. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Perez JD, Dieck S, Alvarez-Castelao B, Tushev G, Chan IC, Schuman EM. Subcellular sequencing of single neurons reveals the dendritic transcriptome of GABAergic interneurons. eLife. 2021;10:e63092. 10.7554/eLife.63092. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pina JM, Reynaga JM, Truong AAM, Keppetipola NM. Post-translational modifications in polypyrimidine tract binding proteins PTBP1 and PTBP2. Biochemistry. 2018;57:3873–3882. 10.1021/acs.biochem.8b00256. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. 10.1016/j.neuron.2015.05.004. [Abstract] [CrossRef] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Hüttelmaier S, Singer RH, Matthews S, Curry S, Smith CWJ. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nature Structural & Molecular Biology. 2006;13:839–848. 10.1038/nsmb1137. [Abstract] [CrossRef] [Google Scholar]
- Ritter B, Ferguson SM, De Camilli P, McPherson PS. A lentiviral system for efficient knockdown of proteins in neuronal cultures [version 1; referees: 2 approved] MNI Open Research. 2017;1:2. 10.12688/mniopenres.12766.1. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Robinson F, Smith CWJ. A splicing repressor domain in polypyrimidine tract-binding protein. The Journal of Biological Chemistry. 2006;281:800–806. 10.1074/jbc.M510578200. [Abstract] [CrossRef] [Google Scholar]
- Schieweck R, Ninkovic J, Kiebler MA. Rna-binding proteins balance brain function in health and disease. Physiological Reviews. 2021;101:1309–1370. 10.1152/physrev.00047.2019. [Abstract] [CrossRef] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. The Journal of Biological Chemistry. 2008;283:1223–1227. 10.1074/jbc.R700034200. [Abstract] [CrossRef] [Google Scholar]
- Thapar R. Structural basis for regulation of RNA-binding proteins by phosphorylation. ACS Chemical Biology. 2015;10:652–666. 10.1021/cb500860x. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Traunmüller L, Schulz J, Ortiz R, Feng H, Furlanis E, Gomez AM, Schreiner D, Bischofberger J, Zhang C, Scheiffele P. A cell-type-specific alternative splicing regulator shapes synapse properties in A trans-synaptic manner. Cell Reports. 2023;42:112173. 10.1016/j.celrep.2023.112173. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. 10.1073/pnas.091062498. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vieira-Vieira CH, Dauksaite V, Sporbert A, Gotthardt M, Selbach M. Proteome-wide quantitative RNA-interactome capture identifies phosphorylation sites with regulatory potential in RBM20. Molecular Cell. 2022;82:2069–2083. 10.1016/j.molcel.2022.03.024. [Abstract] [CrossRef] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nature Reviews. Neuroscience. 2016a;17:265–281. 10.1038/nrn.2016.27. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vuong JK, Lin CH, Zhang M, Chen L, Black DL, Zheng S. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal Pre-mRNA splicing. Cell Reports. 2016b;17:2766–2775. 10.1016/j.celrep.2016.11.034. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. 10.1016/j.neuron.2006.07.020. [Abstract] [CrossRef] [Google Scholar]
- Wei T, Weiler SME, Tóth M, Sticht C, Lutz T, Thomann S, De La Torre C, Straub B, Merker S, Ruppert T, Marquardt J, Singer S, Gretz N, Schirmacher P, Breuhahn K. YAP-dependent induction of UHMK1 supports nuclear enrichment of the oncogene MYBL2 and proliferation in liver cancer cells. Oncogene. 2019;38:5541–5550. 10.1038/s41388-019-0801-y. [Abstract] [CrossRef] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, Fu XD, Zhang Y. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Molecular Cell. 2009;36:996–1006. 10.1016/j.molcel.2009.12.003. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. 10.1016/j.cell.2012.11.045. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, Raza A, Mukherjee S, Manley JL. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Molecular Cell. 2019;76:82–95. 10.1016/j.molcel.2019.07.017. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zheng Sika, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nature Neuroscience. 2012;15:381–388. 10.1038/nn.3026. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zheng S, Damoiseaux R, Chen L, Black DL. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Research. 2013;23:998–1007. 10.1101/gr.147546.112. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zheng S. Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. International Journal of Developmental Neuroscience. 2016;55:102–108. 10.1016/j.ijdevneu.2016.03.003. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
This well-presented and concise study provides important new insights into the regulation of alternative splicing in neurons by the kinase KIS. The provided data convincingly demonstrates how KIS-mediated phosphorylation of the splicing regulator PTB2 controls exon usage during neuronal differentiation. The findings will be of interest to investigators of splicing regulation, neuronal differentiation and maturation.
Decision letter
[Editors' note: this paper was reviewed by Review Commons.]
Author response
Reviewer #1 (Evidence, reproducibility and clarity (Required)):
Summary
In this manuscript, the authors characterized the molecular function of the brain-enriched kinase KIS by combining transcriptome-wide approaches with molecular and functional studies. They uncover that KIS regulates isoform selection of genes involved in neuronal differentiation and inhibits through phosphorylation the capacity of the splicing regulator PTB2 to interact with both target RNAs and protein partners.
Major comments
– This is a very clear and well-written manuscript presenting high-quality and carefully controlled experimental results. The authors used an impressive range of approaches (transcriptome-wide exon usage, phospho-proteomic, imaging, biochemical assays..) to profile exon usage alterations upon KIS knock down and provide a mechanistic understanding of how KIS regulate the splicing activity of PTBP2. Specifically, they convincingly demonstrate that the phosphorylation of PTBP2 by KIS leads to both dismantling of PTBP2 protein complexes and impaired RNA binding.
My only main concerns relate to the understanding of the biological context in which the mechanism studied may be at play. That KIS can counteract PTB2 activity through direct phosphorylation has been very clearly shown by the authors using overexpression of KIS and /or PTB constructs in different contexts (HEK293T cells, N2A cell line, hippocampal neurons). Whether this occurs endogenously in the context of neuronal differentiation, and how much this contributes to the overall phenotypes induced by KIS inactivation, is less clear. While fully investigating the interplay between KIS and PTB2 in the context of neuronal differentiation is beyond the scope of this study, the three following points could be addressed to provide some evidence in this direction.
1. Building on the experiments they perform in a KIS knock-down context (e.g. Figure 3B, or previously described spine phenotype), the authors should investigate whether inhibiting PTBP2 in this context (through shRNA or expression of a phospho-mimetic construct) might suppress the phenotypes observed when inactivating KIS.
As suggested by the reviewer, we have added a new Results section showing the effects on spine maturation in hippocampal neurons expressing PTBP2 phosphomutants and in a PTBP2-KIS double knockdown scenario (Figure 4; Figure 4—figure supplement 1; Results section: P6 L15-P7 L26). First, PTBP2-overexpression effects on post-synaptic protrusion density are exacerbated by the phosphoablated mutant. Intriguingly, the phosphomimetic mutant still has a negative impact in spine formation, suggesting either a residual ability of this protein to interact with its normal partners or the existence of additional roles of PTBP2 in spine development that are Matrin3 and hnRNPM independent. Second, KIS knockdown partially suppresses the defects in mature spine formation produced by the loss of PTBP2. In all, these data support the notion of KIS being a phosphorylation-mediated inhibitor of PTBP2 activity during neuronal differentiation.
2. Based on Figures 1E and 3A, it seems that KIS downregulation affects both exon inclusion and exon skipping, and that its function in exon usage is only partly explained by modulation of PTBP2-dependent exons. Have the authors analyzed the populations of PTBP2-dependent exons that are regulated by KIS in an opposite manner? This may point to specific classes of transcripts (in terms of expression pattern, function, molecular signature) important in the context of endogenous neuronal differentiation.
We have analyzed the GO terms of genes with KIS-upregulated exons by that are either downregulated or upregulated by PTBP2. In both groups we found an enrichment of genes in terms associated with calcium ion activity but with different specific functions. Genes with exons downregulated by PTBP2 are more involved in transmembrane transfer of calcium ions from intracellular stores whereas genes with exons upregulated by PTBP2 facilitate the diffusion of calcium ions through transmembrane postsynaptic. Interestingly, with respect to the cytoskeleton, the two groups show a clearly different term enrichment. Genes with exons downregulated by PTBP2 are significantly associated with the tubulin cytoskeleton, whereas genes with exons upregulated by PTBP2 are associated with the actin cytoskeleton. We have added a paragraph in the Results section (P8 L11-21) and a new panel in Figure 4—figure supplement 1C.
3. The authors should better discuss when and where they think PTBP2 phosphorylation by KIS might be relevant. Is there evidence that this process (or PTBP2 complex assembly) might be regulated upon differentiation or plasticity?
We have modified the Discussion section (P12 L5-L22) as follows:
“Regarding neuronal differentiation, it is worth noting that KIS expression increases during brain development (Bièche et al., 2003) and in vitro differentiation of hippocampal cultures (Figure 1B), coinciding with postnatal decrease of PTBP2 levels (Zheng et al., 2012). Therefore, the phosphorylation-dependent inhibition of PTPB2 by KIS and the concerted relative inversion in their levels would generate a molecular switch linking transcriptional and alternative exon usage programs in neuronal development (Figure 6D).
In mature neurons, alternative splicing has a well-established role in expanding proteome diversity (Mauger & Scheiffele, 2017). Although the connection between synaptic activity and the control of KIS expression and/or kinase activity is not yet established, the contraposition of PTBP2 and KIS in splicing may constitute a fine-tuning mechanism to modulate proteome diversity as a function of plasticity-inducing signals. In this regard, single-cell transcriptomic data from hippocampal neurons show that expression variability of KIS and PTBP2 is much higher compared to actin (Perez et al., 2021) (Figure 6—figure supplement 1B). Thus, differences in the expression of these two splicing regulators at a single neuron level would increase protein isoform variability and expand diversity in neural circuits, a crucial property in information processing (Miller et al., 2019).”
Minor comments
1. Figures and associated legends are overall very clear and well-organized. Addressing the following points would however help improving the clarity of some Figures:
– In Figure 2EV2C legend, the characteristics of the 3SA constructs are not described
We have modified the legend of Figure 2EV2C (Figure 2—figure supplement 1C Figure in revised version) to clarify this point.
– The difference between Figure EV1A and Figure 1H classifications is unclear, nor the interpretation regarding the different GO classes identified
The gene lists used for the two GO term analyses are different. In Figure EV1A (now Figure 1—figure supplement 1A) the gene list is more restrictive than in Figure 1H as we choose genes with more than one exon upregulated by KIS. In contrast, the analysis in Figure 1H includes all genes with one exon upregulated by KIS.
2. Whether PTBP2 is endogenously the major target of KIS explaining transcriptome-wide changes in exon selection is a possibility that remains to be demonstrated. Thus, the authors should correct and tune down the following sentences:
"KIS phosphorylation counteracts PTBP2 activity and thus alters isoform expression patterns …" (end of introduction)
"PTBP2 being one of the most relevant phosphotargets" (results, end of the second section)
We agree with the reviewer and we have modified the two sentences (P4 L6-8) and (P6 L13-14) in the revised version of the paper.
Reviewer #1 (Significance (Required)):
– The splicing regulator PTBP2 is a known master regulator of neuronal fate whose tightly controlled expression drives the progenitor-to-neuron transition as well as the establishment of neuronal differentiation programs. How this protein is regulated at the post-translational level has so far remains poorly investigated. In this manuscript, the authors provide a thorough mechanistic understanding of how KIS-mediated phosphorylation of PTB2 impacts on its regulation of exon usage. They also provide a transcriptome-wide view on the function of the brain-enriched KIS kinase in exon usage, uncovering its broad functions in alternative splicing.
If the physiological context in which KIS-mediated phosphorylation of PTB2 is induced remains to be precisely defined, this work opens interesting new perspectives on regulatory mechanisms at play during neuronal differentiation. Providing extra lines of evidence indicating that KIS acts on neuronal functions through PTBP2 phosphorylation will help further strengthen this aspect.
– This manuscript will be of interest to different large communities interested on one hand on the regulation of gene expression programs underlying neuronal differentiation and on the other hand on the molecular regulation of major complexes involved in alternative splicing and isoform selection. It opens new perspectives related to the spatiotemporal regulation of neuronal isoform selection.
Reviewer #2 (Evidence, reproducibility and clarity (Required)):
Summary
The manuscript by Moreno-Aguilera et al. shows that the brain enriched protein kinase KIS targets the well known neuronal splicing regulator PTBP2 and several of its interaction partners. As a consequence, PTBP2 activity is down-regulated. Using cultured primary immature neurons they show that KIS expression increases during differentiation and that shRNA knockdown of KIS alters the splicing of many alternative exons. Phosphoproteomic analysis of HEK293 cells transfected with KIS or a kinase dead mutant (K545A) show that it phosphorylates both PTBP2 as well as a cluster of proteins that are known to interact with PTBP2 or its paralog PTBP1. By comparing the new data on KIS-dependent splicing with previous data-sets on PTBP2-dependent splicing targets they show that KIS appears to act antagonistically with PTBP2 when it acts as a repressive regulator, but not when it is an activator. Using combinations of wt and kinase-dead KIS with PTBP2 mutants in the 3 main phosphorylation sites (3SA – non-phosphorylatable, S3D – phosphomimetic) to look at the effects on a known PTBP2 functional target, PSD95, they show that the likely effect of KIS is to antagonise PTBP2 function by phosphorylation at one or more of three residues (S178, S308, S434). Finally, they show that transfected KIS (but not K54A) reduces known protein-protein interactions of PTBP2 and that the triple phosphomimetic PTBP2 mutant shows reduced binding to RNA. Alphafold2 predictions show that the S178 phosphomimetic mutant might alter the conformation of the RRM2 domain, in particular altering the environment of Y244, which has been shown in PTBP1 to be critical for interaction with MATR3 and other coregulators.
Major points
In general, the conclusions drawn are consistent with the data. I have a few suggestions where the authors could either extend their findings with a few straightforward additional experiments, or clarify some of the existing data.
FigEV4 (also introductory text on p3): RRMs 3 and 4 of PTBP1/2/3 fold as a single back to back packed didomain – with the so-called linker contributing to the didomain fold (e.g. PMID: 24688880, PMID: 16179478) and also extending the RNA binding surface by creating a positive patch (e.g. PMID:20160105 PMID: 24957602). AlphaFold successfully predicts the didomain in full length PTBP2 (https://alphafold.ebi.ac.uk/entry/A0A7I2RVZ4). The authors should therefore use AlphaFold2 to predict the RRM3-4 di-domain structure of wt and phosphomimetic mutant PTBP2s. Phosphorylation of S434 or S434D, which is on the C-terminal end of RRM3 may have no predicted effect on RRM3 alone (FigEV4), but it could conceivably disrupt didomain packing, which could itself have important knock-on consequences for RNA binding. In addition, the inrtoduction of negative charges at S434 might affect the ability of R438, K440 & K441 to interact with RNA. An image of the didomain charge density of WT and mutant PTBP2 would be useful to address this.
As suggested by the reviewer, we have considered the di-domain structure of RRM3 and RRM4, and AlphaFold2 predicted no effects by the phosphomimetic residues. We have added the di-domain predictions to Figure 6—figure supplement 1B.
S434 lies at the very end of RRM3 and limits with a basic region that would not bind RNA in a canonical RRM-dependent manner. In addition, as predicted by AlphaFold2, this basic region is not structured and the effects of a nearby negative charge may be difficult to predict.
Figure 4 could also easily go further in experimentally testing the effects of individual phosphomimetic mutations upon protein-protein interactions (Alphafold predicts that S178D, but not S308 or S434D, should affect Y244 mediated interactions, such as MATR3). The co-IP approach in Figure 4A could readily be used with FLAG-PTBP2 mutants. Likewise, consequences of individual mutations upon RNA binding (Figure 4D) could be tested. The use of a Y244N mutant here would test whether the loss of RNA binding is a consequence of the loss of protein-protein interactions. Such experiments are not essential, but they are readily carried out and have the potential to unravel the consequences of the individual phosphorylation events (more correctly of phosphomimetic mutants).
After building the Y244N mutant we tested PTBP2 interactions with protein partners and observed no significant changes in the levels of Matrin3 and hnRNPM proteins in FLAG-PTBP2 immunoprecipitates nor in the RNA binding ability of PTBP2. These data suggest that, although Y244 is involved in the interaction between PTBP1 and PRI-containing proteins such as Raver1, the interaction between Matrin3 and PTBP2 would involve structural determinants other than the Matrin3 PRI and the PTPB2 Y244 residue. Compared to PTBP1, the nearby flexible loop between RRM2 and RRM3 in PTBP2 is very different and could accommodate specific interaction determinants with Matrin3.
Minor points
Do KIS regulated exons show enrichment of motifs associated with PTBP2, consistent with the proposed model – particularly CU-rich motifs upstream of exons that are more repressed upon KIS shRNA treatment.
We have not observed a significant enrichment of CU-rich sequences upstream of the top 100 exons upregulated by KIS. Indeed, our data suggest that only a fraction of exons upregulated by KIS are inhibited by PTBP2.
For the splicing analysis pipeline, how were exon-exon junction reads treated? If "only exons with more than 5 reads in all samples" were considered, will this not exclude highly regulated exons that are completely skipped under one condition?
This sentence has been corrected as "only exons with more than 5 reads in all samples of one condition…" (P17 L15)
The Introduction mentions U2AF homology (UHM) domains, but neglects to discuss their known binding partners – ULMs (UHM ligand motifs), which contain an essential tryptophan. It would be useful for the discussion to highlight whether any direct KIS interactors possess ULMs and how this relates to the phospho-targets identified here. The authors may wish to draw the parallel with the structurally analogous way that PTBP1 (and presumably PTBP2) interact with their short peptide ligand motifs.
As suggested by the reviewer, we have searched for ULM sequences in the identified KIS phosphotargets, but we only found clear ULMs in SUGP1, which contains KRKRKSRW385 and KMGW573K. The absence of ULM motifs in most of the proteins identified in the KIS phosphoproteome would indicate that phosphorylation does not require stable protein-protein interactions. We have added these lines to the Discussion section (P11 L5-L9). We completely agree with the reviewer that, in future work, it would be very interesting to test the possibility that KIS binding modulates the composition and functional properties of splicing complexes through ULM domains.
Figure EV2C. The S3A and S308A mutations clearly reduce phosphorylation. However, the effects of S178A and S434A are far less clear. Presumably the quantitation shown in the lower panel of EV2C relies on normalization to PTBP2 protein input, which appears quite variable in the Coomassie gel. It might be better to repeat the experiment with uniform protein inputs. Minimally, details of the quantitation approach should be added to Materials and methods.
The different levels of reduction in 32P incorporation displayed by the single phosphonull mutants suggests that phosphorylation follows a hierarchical pattern, S308-P facilitating phosphorylation of the other two phosphosites. We have added this comment to the revised version of the paper (P5 L27-31). As mentioned by the reviewer, 32P incorporation was made relative to the total amount of PTBP2 present in the assay, which was deduced from cold Coomassie-stained gels run in parallel to the radioactive gels with same amount of proteins. We have added details of the quantification in the Methods section from 3 independent experiments
Figure 3D shows PTBP2 overexpression, but the main text (p7) states KIS overexpression.
The text and panel order in Figure 3D were misleading and have been corrected (P7 L14-19).
Figure 4B should have a scale bar for the FRET signal
Done (now Figure 5B)
Figure 4E should indicate the location of S178
Done (now Figure 6C)
Reviewer #2 (Significance (Required)):
Significance
This interesting, clear and concise manuscript provides important new insights into the way that a neuron specific kinase can regulate neuronal splicing networks by phosphorylating and thereby downregulating the known neuronal splicing regulator PTBP2. Alternative splicing is known to play a particularly important role in neurons, so this demonstration of an additional layer of regulation by post-translational modification should make the manuscript of wide interest to investigators of splicing regulation, neuronal differentiation and maturation.
Issues that are not addressed in the manuscript include; (i) how does KIS specifically target PTBP2 and related proteins? The UHM domain can mediate interaction with ULM containing splicing factors (such as U2AF2, SF3B1), but none of the identified targets have known ULMs. (ii) the consequences of individual phoshomimetic mutants upon protein-protein interactions and RNA binding could readily be explored further using computational and experimental methods already used in the manuscript.
For context, this reviewer has a direct interest in the mechanisms of regulation of alternative splicing, but not in the context of neurons (though I am familiar with a lot of the relevant literature), and I do not have expertise in neuronal cell biology.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
In this manuscript, the authors explored the function of the protein kinase KIS in splicing regulation associated with neuronal differentiation in vitro. KIS is a serine threonine kinase known to phosphorylate splicing factors such as SF1 and SUGP1, and to be preferentially expressed in adult brain in mammals. Using an shRNA based approach, the authors characterize cassette exon usage upon partial KIS depletion in cultured mouse cortical neurons.
In parallel using mass spectrometry of proteins in KIS overexpressing HEK293 cells, they identify potential KIS substrates including the splicing regulator PTBP2. They confirm that recombinant KIS can phosphorylates PTBP2 in vitro. They show a correlation between KIS-activated and PTBP2-inhibited exons using published data for this factor. They report opposite effects of KIS and PTBP2 on CamKIIB splicing and Finally, coimmunoprecipitation and FRET experiments suggest that KIS inhibits the interactions of PTBP2 with known protein binders, hnRNPM and Matrin3 as well as with RNA. Altogether these data suggest that KIS downregulates PTBP2 during neuronal differentiation.
Major comments:
Overall the manuscript is well written and the data are interesting.
However several points could have been more extensively studied or discussed to achieve a stronger demonstration of the role of KIS in PTBP2 phosphorylation and neuronal differentiation.
1) To minimize possible off target problems, the RNAseq analysis would be more convincing if replicated with a second shRNA to knockdown KIS.
The efficiency of the selected shRNA had been validated both by the supplier (Merck-Σ) and in our previous work, which also included a complementation assay (see Figure 4A-C in Pedraza et al. 2014).
2) Part of the reported splicing changes might reflect an indirect consequence of an altered differentiation contributing to the correlation observed in figure 1F. It would be interesting to confirm splicing changes using shorter incubation times with the shRNA compared to the 11 days used in this study.
The levels of splicing regulators such as PTBP1 and PTBP2 change quite markedly during the initial phases of neuronal differentiation (Zheng et al. 2012). However, we observed no change in their levels when comparing KIS knockdown to control conditions, suggesting no major upstream effects on the differentiation program per se. But, more important, whereas the splicing pattern of CamKIIβ transcript was clearly affected by KIS knockdown at 18 DIV (Figure 3B), we observed no changes at 14 DIV.
3) Standard deviation is more relevant to describe data dispersion in all figures.
For non-parametric statistics we prefer the interquartile range as a measure of dispersion. For the parametric statistics of 3 independent experiments we show the standard error of the mean as a measure of dispersion.
4) Previous papers of the group described a function of KIS in translation (Cambray et al. 2009, Pedraza et al. 2014). This is not discussed here. For example, the possibility that RBPs are regulated by KIS at the translation level is not excluded by the analysis in Figure EV2a.
It is an interesting comment by the reviewer that we have considered during the course of this work. Nevertheless, in our experiments coexpressing KIS and PTBP2 in HEK293 cells we did not observe any reduction in splicing factor levels. We have included a representative immunoblot (Figure 5—figure supplement 1C) of input samples from the experiments shown in the corresponding main figure (Figure 5A in the revised version).
Minor comments:
Figure 1: The authors state that "KIS…accumulates in nuclear sub-structures adjacent to those formed by splicing factors". As the figure presents in fact GFP-KIS, it should be mentioned, and how this localisation is relevant for endogenous KIS should be addressed.
We have corrected the text to mention that GFP-KIS was used to analyse its nuclear localization pattern as shown in Figure 1A (P4 L12-13). We had previously validated the nuclear localization (Boehm et al., 2002) of an N-terminal fusion of KIS to a fluorescent protein (Cambray et al., 2009).
Figure EV1: SI range in panel D is very different from that in panel C and Fig1E.
In this figure we plot the average SI obtained from bins with 2500 exons, which would necessarily narrow the SI range obtained from individual exons. Our data indicate that protein disorder would only constitute a minor, but significant, factor in exon usage.
On page 4 "KIS expression reached maximal levels in hippocampal cultures (Figure 1B)." However the figure legend indicates that this analysis was performed with cortical neurons. The use of cortical or hippocampal neurons along the manuscript should be clarified.
We apologize for the typing mistake, and it has been corrected in the revised version (P4 L17-18)
page 4 " KISK54A, a point mutant without kinase activity" The authors should indicate the reference.
The reference to Maucuer et al. (1997) has been added (P5 L17)
Figure EV2C: It is not clear whether the Coomassie staining and autoradiography do correspond to the same gel.
32P incorporation was made relative to the total amount of PTBP2 present in the assay, which was deduced from cold Coomassie-stained gels run in parallel to the radioactive gels with same amount of proteins.
Figure 3C. The authors use a dual fluorescence reporter to analyse PSD95 exon 18 splicing. However the well to well variability in such experiments might be elevated. Not only the cells number in a single well but also the number of replicates should be indicated and well to well variability reported.
As stated in the figure legend, the dual fluorescence reporter experiment has been analyzed at a single-cell level. Using ImageJ software, we analyzed the fluorescence of 1054, 970 and 672 cells expressing KIS, KISK54A or none, respectively, from 3 independent experiments.
Figure 3D. The precise timing for the transfection and culture of cells before staining is unclear.
Hippocampal neurons were transfected at 5DIV and fixed at 12 DIV. This description has been added to the legend in Figure 3D.
Figure 4A. The input should be loaded to evaluate the coIP efficiencies and ascertain that KIS does not downregulate Matrin3 and hnRNPM levels.
We agree with the reviewer. We have included a representative immunoblot (Figure 5—figure supplement 1C) of input samples from the experiments shown in the corresponding main figure (Figure 5A in the revised version).
Figure EV4A. No difference of Matrin3 binding is to be seen on the gel. In addition, the authors should confirm that PTBP2 or binders are phosphorylated by recombinant KIS. The preparation of GST-KIS is not described.
We agree with the reviewer that in the in vitro assays the differences in phosphorylation are not as clear as in the in vivo experiments. Figure S2C shows an in vitro kinase assay of PTBP2 by recombinant KIS. Finally, we include a reference (Pedraza et al., 2009) for the preparation of recombinant GST-KIS (P14 L14)
Page 6: "We found that PTBP2-inhibited exons are significantly (FDR=0.001) enriched in KIS knockdown neurons, supporting the notion that KIS acts on AS, at least in part, by inhibiting PTBP2 activity." This should be rephrased as in fact PTBP2-inhibited exons are enriched among KIS activated exons.
The sentence has been rephrased as “We found that PTBP2-inhibited exons are significantly (FDR=0.001) among KIS activated exons…” (P6 L20-21)
Page 10: "SUGP1 is one of the most enriched proteins in our KIS phosphoproteome (see Figure 2A)". Phosphorylation and interaction with KIS was already reported by Arfelli and coll. 2023 supplementary figure 2.
We have modified the Discussion section to add this information (P11 L21-22)
Page 10: " It forms part of the spliceosome complex, interacts with the general splicing factor U2AF2 and has been reported to play an important role in branch recognition by its association with SF3B1." A reference is needed there.
A reference to Zhang et al. (2019) has been added.
Page 10: The authors previously reported a differentiation defect in cultured neurons 'Cambray et al., 2008' that was not observed by another group (Manceau et al., PLOS One 2012). This should be discussed in view of these more recent results. Is there any differentiation defect in the experiments reported there?
We have added a new Results section showing the effects on spine maturation in hippocampal neurons expressing PTBP2 phosphomutants and in a PTBP2-KIS double knockdown scenario (Figure 4 and Figure 4—figure supplement 1; Results section: P6 L15-P7 L26). First, PTBP2-overexpression effects on post-synaptic protrusion density are exacerbated by the phosphoablated mutant. Related to the point raised by the reviewer, KIS knockdown also decreased spine emergence and maturation, but partially suppressed the defects produced by the loss of PTBP2. In all, these data support the notion of KIS being a phosphorylation-mediated inhibitor of PTBP2 activity during neuronal differentiation.
Statistical values are difficult to read in the figures. Please use larger fonts.
Done
Reviewer #3 (Significance (Required)):
This manuscript brings new elements supporting the function of the protein kinase KIS in splicing regulation in neurons. In particular it identifies for the first time the splicing regulator PTBP2 as a substrate for KIS.
It will be of interest to a specialized audience of researchers interested in splicing regulators in neuronal differentiation.
Articles from eLife are provided here courtesy of eLife Sciences Publications, Ltd
Citations & impact
Impact metrics
Article citations
KIS counteracts PTBP2 and regulates alternative exon usage in neurons.
Elife, 13:e96048, 10 Apr 2024
Cited by: 1 article | PMID: 38597390 | PMCID: PMC11045219
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Polypyrimidine tract binding proteins PTBP1 and PTBP2 interact with distinct proteins under splicing conditions.
PLoS One, 17(2):e0263287, 03 Feb 2022
Cited by: 7 articles | PMID: 35113929 | PMCID: PMC8812845
PTBP1 and PTBP2 Repress Nonconserved Cryptic Exons.
Cell Rep, 17(1):104-113, 01 Sep 2016
Cited by: 42 articles | PMID: 27681424 | PMCID: PMC5082185
Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain.
Genes Dev, 26(14):1626-1642, 01 Jul 2012
Cited by: 138 articles | PMID: 22802532 | PMCID: PMC3404389
The latest role of nerve-specific splicing factor PTBP1 in the transdifferentiation of glial cells into neurons.
Wiley Interdiscip Rev RNA, 14(2):e1740, 16 May 2022
Cited by: 3 articles | PMID: 35574699
Review
Funding
Funders who supported this work.
Barts Charity (1)
Grant ID: MGU0346
Medical Research Council (1)
The role of RNA Binding Proteins (RBPs) in breast cancer progression and metastasis
Dr Faraz Mardakheh, Queen Mary University of London
Grant ID: MR/P009417/1
Ministerio de Ciencia e Innovación (2)
Grant ID: PID2020-113231GB-I00
Grant ID: PRE2018-083268



