Abstract
Free full text
Soybean steroids improve crop abiotic stress tolerance and increase yield
Summary
Sterols have long been associated with diverse fields, such as cancer treatment, drug development, and plant growth; however, their underlying mechanisms and functions remain enigmatic. Here, we unveil a critical role played by a GmNF‐YC9‐mediated CCAAT‐box transcription complex in modulating the steroid metabolism pathway within soybeans. Specifically, this complex directly activates squalene monooxygenase (GmSQE1), which is a rate‐limiting enzyme in steroid synthesis. Our findings demonstrate that overexpression of either GmNF‐YC9 or GmSQE1 significantly enhances soybean stress tolerance, while the inhibition of SQE weakens this tolerance. Field experiments conducted over two seasons further reveal increased yields per plant in both GmNF‐YC9 and GmSQE1 overexpressing plants under drought stress conditions. This enhanced stress tolerance is attributed to the reduction of abiotic stress‐induced cell oxidative damage. Transcriptome and metabolome analyses shed light on the upregulation of multiple sterol compounds, including fucosterol and soyasaponin II, in GmNF‐YC9 and GmSQE1 overexpressing soybean plants under stress conditions. Intriguingly, the application of soybean steroids, including fucosterol and soyasaponin II, significantly improves drought tolerance in soybean, wheat, foxtail millet, and maize. These findings underscore the pivotal role of soybean steroids in countering oxidative stress in plants and offer a new research strategy for enhancing crop stress tolerance and quality from gene regulation to chemical intervention.
Introduction
With the advent of climate change, abiotic stresses such as drought, salt, heat and cold are exerting increasingly severe influences on plant growth, leading to a reduction in grain production. However, being sessile organisms, plants have evolved a series of defence mechanisms to adapt to environmental stressors. For instance, extensive research has been conducted on the role of abscisic acid (ABA) in enhancing tolerance to abiotic stress conditions, such as drought (Zhang et al., 2022; Zhu, 2016). Cold stress triggers a Ca2+‐signal‐activated MAP‐kinase cascade that activates the expression of cold‐responsive genes in plants and can also modulate HSP gene expressions to enhance plant tolerance towards heat stress (Sangwan et al., 2002; Zhang et al., 2022; Zhu, 2016). Nevertheless, with advancements in metabolomics technology, several small molecular compounds have been discovered to play crucial roles in promoting plant stress tolerance. For example, recent studies have revealed that high levels of putrescine are induced during cold stress, and its exogenous application improves cold‐acclimated freezing tolerance in potato after acclimation to low temperatures (Kou et al., 2018). Additionally, flavonoid accumulation has been found beneficial for enhancing UV tolerance in rice (Liu et al., 2020). However, due to the inherent challenges associated with small metabolite techniques used for their characterisation and quantification purposes, our understanding of their precise roles under abiotic stress conditions remains limited.
One notable family of small metabolites is sterols. While extensive research has been conducted on these compounds in animal systems, progress regarding plant sterols has remained limited. Nonetheless, sterols have been implicated in numerous vital plant processes, encompassing cell division, elongation and polarity (He et al., 2018; Men et al., 2008; Schrick et al., 2000; Willemsen et al., 2003). They also play a crucial role in regulating auxin and ethylene signalling (Souter et al., 2002), modulating the activity of microRNAs (Brodersen et al., 2012), and enhancing tolerance to low‐temperature stress (Senthil‐Kumar et al., 2013). The biosynthesis of sterol requires the initial synthesis of squalene (SQ), a 30‐carbon intermediate, through the isopentenyl diphosphate (IPP) pathway (Phillips et al., 2006; Rasbery et al., 2007). Squalene is subsequently oxidised by squalene epoxidase (SQE, squalene monooxygenase), leading to the formation of 2,3‐oxidosqualene (2,3‐OSQ) (Phillips et al., 2006; Rasbery et al., 2007), which then undergoes cyclisation to generate sterols (Benveniste, 2004; Schaller, 2003, 2004; Spanova and Daum, 2011). SQE serves as a crucial rate‐limiting enzyme in the sterol biosynthesis pathway. In the model plant Arabidopsis thaliana, seven SQE proteins are identified; however, only SQE1, SQE2, and SQE3 exhibit catalytic activity in converting squalene to squalene‐2, 3‐epoxide (2,3‐OSQ) (Rasbery et al., 2007). The mutation of SQE1 results in plants exhibiting impaired root development, hypocotyl and stem elongation defects, as well as inviable seeds (Posé et al., 2009; Rasbery et al., 2007). The sqe1‐5 mutant exhibits altered sterol composition in roots and impaired stomatal responses (Posé et al., 2009). Loss of SQE2, which exhibits tissue‐specific tissue expression patterns, does not exert any discernible influence on the sterol composition at the whole plant level. However, depletion of SQE3 results in the squalene accumulation and consequential phenotypic defects (Laranjeira et al., 2015). These studies unveil the significance of phytosterols in plant growth and development, however, there remains a dearth of research on the molecular mechanisms governing plant sterol synthesis and their roles in abiotic stress.
In human, SP1 and SREBF2 have been identified as two crucial transcription factors that regulate sterol synthesis by controlling the expression level of SQE (Zhang et al., 2019). In addition, research has demonstrated that WsWRKY1, a WRKY transcription factor in Withania somnifera, modulates biotic stress tolerance through the regulation of the SQE‐mediated triterpenoid system pathway (Singh et al., 2017). In Panax notoginseng, PnMYB4 exerts a repressive effect on saponin biosynthesis by modulating the transcript level of SQE (Man et al., 2023). These findings sequentially unveil the regulatory role of a transcription factor in sterol synthesis. However, among these transcription factors, the nuclear factor Y (NF‐Y) garners significant attention due to its distinctive structure and functionality. The NF‐Y transcription factor, composed of NF‐YA, NF‐YB, and NF‐YC subunits, exerts crucial regulatory functions in diverse plant developmental processes by binding to the CCAAT cis‐element in target gene promoters (Mantovani, 1999; Shen et al., 2020; Zanetti et al., 2010). Moreover, NF‐Y transcription factors also actively participate in enhancing plant stress tolerance. For instance, a recent study demonstrates that NF‐YB21 plays a crucial role in enhancing drought tolerance and modulating ABA‐mediated IAA transport to positively regulate root growth in Populus trichocarpa (Zhou et al., 2020). In our previous research, we discovered that GmNF‐YC14 conferred enhanced tolerance to drought and salt stresses by regulating the PYL‐mediated ABA signalling pathway in soybean (Yu et al., 2021). Recent research has unveiled the interaction between NF‐YCs and BIN2, which indirectly regulates the biosynthesis of brassinosteroids, crucial components of phytosterols. This interaction consequently influences light‐regulated hypocotyl elongation in Arabidopsis (Zhang et al., 2021). This highlights the significance of NF‐Y transcription factors in finely modulating phytosterol composition and plant development. However, the intricate interplay between the NF‐Y transcription factor, phytosterol, and abiotic stress remains shrouded in ambiguity.
In our previous study, we identified two NF‐YC family genes, GmNF‐YC9 and GmNF‐YC14, through a comprehensive analysis of the molecular characteristics of this gene family in soybean (Yu et al., 2021). These two genes exhibit significant induction under drought and salt stresses in soybean. Our current findings reveal that, unlike GmNF‐YC14, the GmNF‐YC9‐mediated NF‐Y transcription complex directly regulates the expression of GmSQE1 under stress conditions. This regulation leads to an enhancement in the squalene metabolic pathway and ultimately enhances plant abiotic stress responses. Our findings have revealed a previously unidentified regulatory module (NF‐Y‐SQE) that governs the biosynthesis pathway of plant sterols and confers stress tolerance, thereby establishing a theoretical basis for enhancing soybean variety improvement. Furthermore, our comprehensive analysis of transcriptome and metabolome association has demonstrated a significant upregulation of multiple soybean sterols under stressful conditions. Notably, exogenous application of soybean sterols has been shown to significantly enhance crop stress tolerance, highlighting the potential utility of plant steroids in improving crop resilience to environmental challenges. These results provide insights into a non‐GMO strategy for modulating crop stress tolerance through chemical intervention.
Results
GmNF‐YC9‐mediated CCAAT‐box transcriptional complex can improve stress tolerance in soybean
In our previous investigation, we observed a significant induction of GmNF‐YC9 expression in response to drought and salt treatments (Yu et al., 2021). To further elucidate the potential role of GmNF‐YC9 in drought tolerance, we generated transgenic soybean plants overexpressing GmNF‐YC9 (GmNF‐YC9‐OE1, GmNF‐YC9‐OE5, and GmNF‐YC9‐OE8) (Figure S1). Our findings revealed that the overexpression of GmNF‐YC9 in soybean resulted in increased biomass, longer root and hypocotyl lengths compared to wild‐type (WT) plants under stress conditions (Figure 1a–d). Microscopic analysis of root tip tissue sections further demonstrated an enhanced root tip differentiation rate in GmNF‐YC9‐OE plants subjected to 200 mM mannitol treatment (Figure 1e,f). These results highlighted the superior growth performance of GmNF‐YC9‐OE plants under stress conditions, which had been consistently observed in pot experiments (Figure 1g). Furthermore, GmNF‐YC9‐OE plants exhibited elevated proline levels, reduced malondialdehyde (MDA) content, and higher biomass compared to WT plants under drought and salt stresses (Figure 1h–j). Notably, these effects were more pronounced under stressful conditions, firmly establishing that the overexpression of GmNF‐YC9 enhances soybean's tolerance to osmotic and salt stresses.
mM mannitol treatment (Figure 1e,f). These results highlighted the superior growth performance of GmNF‐YC9‐OE plants under stress conditions, which had been consistently observed in pot experiments (Figure 1g). Furthermore, GmNF‐YC9‐OE plants exhibited elevated proline levels, reduced malondialdehyde (MDA) content, and higher biomass compared to WT plants under drought and salt stresses (Figure 1h–j). Notably, these effects were more pronounced under stressful conditions, firmly establishing that the overexpression of GmNF‐YC9 enhances soybean's tolerance to osmotic and salt stresses.
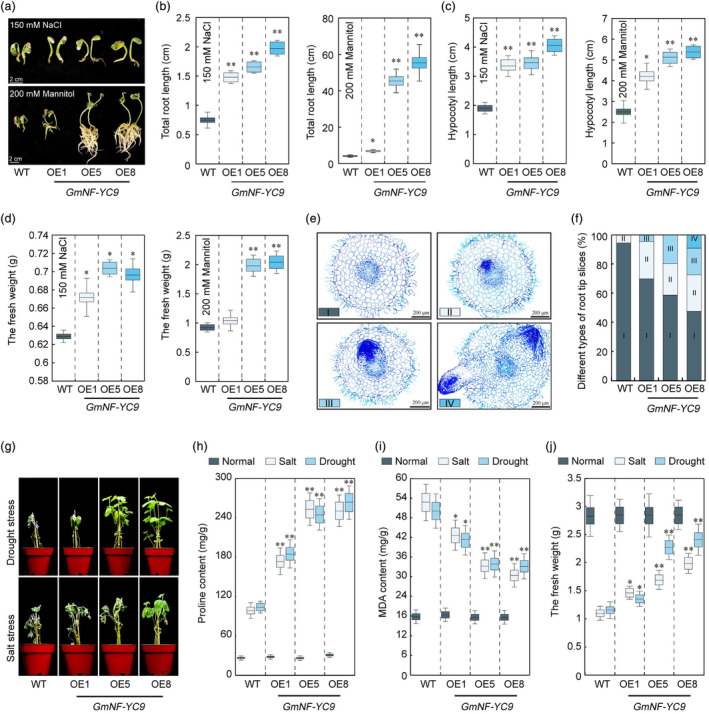
Stress tolerance analysis of GmNF‐YC9 overexpressing soybean plants. (a) Phenotype analysis of 7‐day‐old WT and GmNF‐YC9‐OE soybean seedling plants after 7 days of different stress treatments. (b–d) Total root length (b), hypocotyl (c), and fresh weight (d) analysis of 7‐day‐old WT and GmNF‐YC9‐OE soybean plants after 7
days of different stress treatments. (b–d) Total root length (b), hypocotyl (c), and fresh weight (d) analysis of 7‐day‐old WT and GmNF‐YC9‐OE soybean plants after 7 days of different stress treatments. (e) The different slice samples of the root tip elongation zone after 7
days of different stress treatments. (e) The different slice samples of the root tip elongation zone after 7 days of 200
days of 200 mM mannitol treatment. (f) The percentage of different types of root tip slice samples. (g) Phenotypic analysis of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions. (h, i) Proline (h) and MDA (i) content analysis of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions. (j) The fresh weight of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions.
mM mannitol treatment. (f) The percentage of different types of root tip slice samples. (g) Phenotypic analysis of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions. (h, i) Proline (h) and MDA (i) content analysis of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions. (j) The fresh weight of GmNF‐YC9‐OE and WT plants at seedling stage under drought and salt stress conditions.
To further elucidate the function of GmNF‐YC9, we conducted a screening of a soybean cDNA library to identify candidate proteins. This screening led to the identification of several positive clones, including two additional subunits of the CCAAT‐box complex, GmNF‐YA2 and GmNF‐YB24. Subsequent yeast two‐hybrid (Y2H) assays confirmed interactions among protein pairs GmNF‐YC9/GmNF‐YA2, GmNF‐YC9/GmNF‐YB24, and GmNF‐YA2/GmNF‐YB24 (Figure 2a). These interactions were further validated through bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplast cells (Figure 2b–d). Additionally, luciferase complementary imaging (LCI) assays detected LUC signal in tobacco leaf areas co‐expressing GmNF‐YA2‐cLUC/GmNF‐YC9‐nLUC, GmNF‐YB24‐cLUC/GmNF‐YC9‐nLUC, or GmNF‐YB24‐cLUC/GmNF‐YA2‐nLUC (Figure 2e–g). GST‐pulldown assays demonstrated physical interactions among GmNF‐YC9, GmNF‐YB24, and GmNF‐YA2 proteins (Figure 2h–j). These results unequivocally indicated that GmNF‐YC9, GmNF‐YB24, and GmNF‐YA2 proteins interacted with each other both in vivo and in vitro. Subsequent analysis of GmNF‐YA2 and GmNF‐YB24, through the generation of transgenic hairy‐root composite plants, confirmed their involvement in abiotic stress tolerance (Figure S2a–e). Under stress conditions, the transgenic hairy roots expressing either GmNF‐YA2 or GmNF‐YB24 exhibited improved growth characteristics, elevated proline content, reduced MDA content, and decreased relative electrical conductivity compared to those transformed with an empty vector control (Figure S2f–m). These findings collectively suggested that, similar to GmNF‐YA9, GmNF‐YA2 and GmNF‐YB24 actively participate in abiotic stress responses.
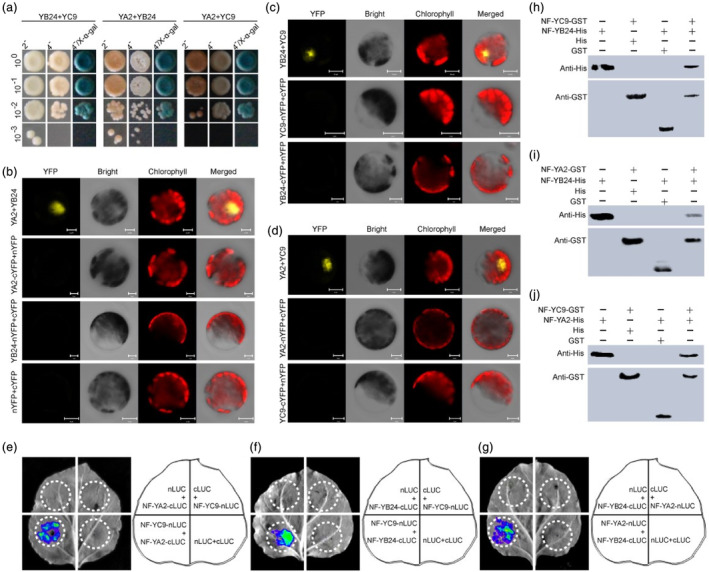
Interaction verification of GmNF‐YC9 and its candidate proteins. (a–j) Interaction verification between GmNF‐YC9 and its candidate proteins (GmNF‐YB24 and GmNF‐YA2) by yeast two‐hybrid (a), BiFC (b–d), LCI (e–g), and pull‐down (h–j) assays. YB24 and NF‐YB24, GmNF‐YB24; YA2 and NF‐YA2, GmNF‐YA2; YC9 and NF‐YC9, GmNF‐YC9.
GmNF‐YC9‐mediated CCAAT‐box transcriptional complex can activate the expression of GmSQE1
To elucidate the molecular mechanism underlying GmNF‐YC9‐mediated stress tolerance in soybean, we conducted RNA‐seq assays on GmNF‐YC9‐OE and WT plants. The analysis revealed upregulation of numerous stress‐related genes in GmNF‐YC9‐OE plants. Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis identified significant enrichment in genes associated with sterol metabolism, including biosynthetic processes, transporters, and responses to steroid hormones (Figure 3a,b, Figure S3a,b). Among these genes, squalene monooxygenase (GmSQE1), which encodes the enzyme responsible for the rate‐limiting step in sterol synthesis, displayed significant induction in GmNF‐YC9‐OE plants under stress conditions (Figure 3b). This suggested that the GmNF‐YC9 transcription factor regulated the sterol synthesis pathway by modulating GmSQE1 activity. Our qRT‐PCR results further confirmed elevated transcript levels of GmSQE1 in GmNF‐YC9‐OE plants (Figure S3c), which were significantly induced by drought and salt stress treatments (Figure S3d). Notably, GmSQE1 exhibited higher expression in GmNF‐YA2 and GmNF‐YB24 transgenic hairy roots (Figure S4a,b), suggesting that it could be a potential target gene of GmNF‐YC9‐mediated CCAAT‐box transcriptional complex.
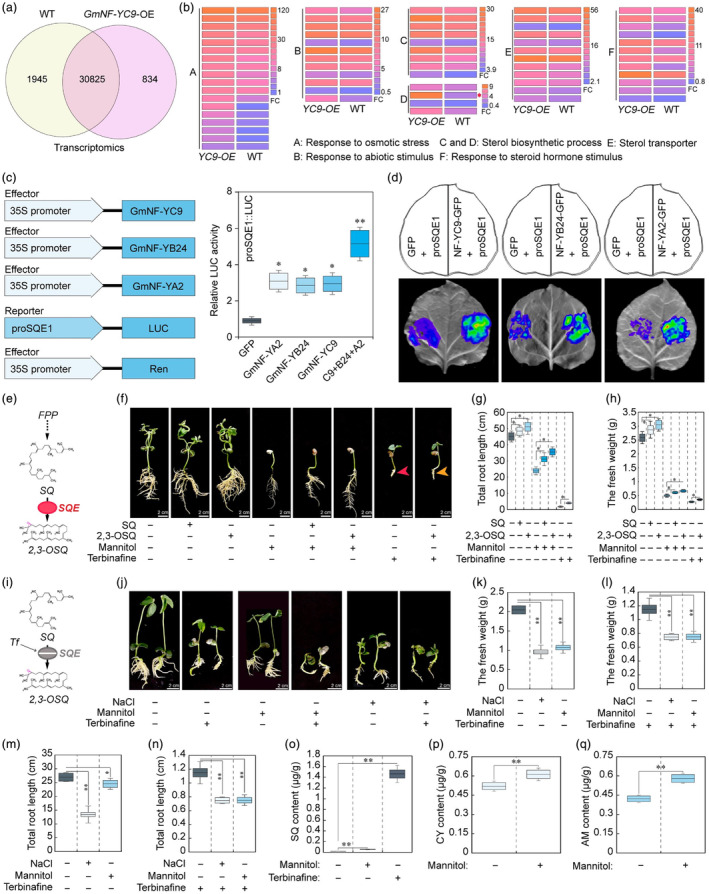
Downstream target gene and its regulation pathway analysis of GmNF‐YC9 transcription factor. (a) The Venn diagram shows gene co‐expression analysis of WT and GmNF‐YC9‐OE soybean plants under drought conditions. (b) GO enrichment analysis of differentially expressed genes (DEGs) in GmNF‐YC9‐OE soybean plants. Red asterisk indicates squalene monooxygenase (GmSQE1). (c, d) Interaction analysis between the NF‐Y transcription factor and GmSQE1 promoter by LUC activity assay. (e) The module of SQE‐mediated catalysation to squalene. (f) Phenotype analysis of 7‐day‐old soybean seedling plants after 10 days of SQ (600
days of SQ (600 μg/L), 2,3‐OSQ (300
μg/L), 2,3‐OSQ (300 μg/L), mannitol (200
μg/L), mannitol (200 mM), and terbinafine (100
mM), and terbinafine (100 μM) treatments, respectively. (g, h) Total root length (g) and fresh weight (h) of soybean seedlings under different treatments. (i) The inhibition of terbinafine on SQE‐mediated catalysation to SQ. (j) Phenotype of 7‐day‐old soybean seedlings after 10
μM) treatments, respectively. (g, h) Total root length (g) and fresh weight (h) of soybean seedlings under different treatments. (i) The inhibition of terbinafine on SQE‐mediated catalysation to SQ. (j) Phenotype of 7‐day‐old soybean seedlings after 10 days of 100
days of 100 mM NaCl, 100
mM NaCl, 100 mM mannitol, and 25
mM mannitol, and 25 μM terbinafine treatments. (k–n) Fresh weight (k, l) and total root length (m, n) of 7‐day‐old soybean seedlings after 10
μM terbinafine treatments. (k–n) Fresh weight (k, l) and total root length (m, n) of 7‐day‐old soybean seedlings after 10 days of 100
days of 100 mM NaCl, 100
mM NaCl, 100 mM mannitol, and 25
mM mannitol, and 25 μM terbinafine treatment conditions. (o) Squalene content analysis of soybean seedling roots under different treatment conditions (25
μM terbinafine treatment conditions. (o) Squalene content analysis of soybean seedling roots under different treatment conditions (25 μM terbinafine and 100
μM terbinafine and 100 mM mannitol). (p, q) Cycloartenol (p) and beta‐amyrin (q) content analysis of soybean seedling roots under 100
mM mannitol). (p, q) Cycloartenol (p) and beta‐amyrin (q) content analysis of soybean seedling roots under 100 mM mannitol treatment conditions. AM, beta‐amyrin; CY, cycloartenol.
mM mannitol treatment conditions. AM, beta‐amyrin; CY, cycloartenol.
To validate the regulatory role of GmNF‐YC9 on GmSQE1 expression, luciferase (LUC) activity analysis assays were conducted. These assays revealed a substantial increase in GmSQE1 promoter‐induced LUC activity in the presence of GmNF‐YA2, GmNF‐YB24, or GmNF‐YC9 proteins (Figure 3c,d). Electrophoretic mobility shift assays (EMSA) further demonstrated the binding of the GmNF‐YA2/GmNF‐YB24/GmNF‐YC9 transcription complex to the CCAAT‐box of the GmSQE1 gene promoter (Figure S4c). These findings conclusively established that GmSQE1 was a downstream target gene of a trimeric nuclear transcriptional activation complex. Moreover, enzyme‐linked immunosorbent assay (ELISA) was employed to analyse SQE enzyme activity and its catalytic product, 2,3‐oxidosqualene (2,3‐OSQ), in both GmNF‐YC9‐OE and WT plants. The results demonstrated a significant increase in SQE enzyme and 2,3‐OSQ content in salt and drought‐treated GmNF‐YC9‐OE plants compared to WT plants (Figure S4d–g), indicating that the regulation of GmSQE1 activity by GmNF‐YC9 can affect the sterol synthesis pathway.
Squalene and squalene‐2, 3‐epoxide, two key components in SQE‐mediated catalytic pathway, can affect plant stress tolerance
To elucidate the contribution of enhanced GmSQE1 expression to plant stress tolerance, we investigated the correlation between stress tolerance and two key components of the SQE‐mediated catalytic pathway: squalene (SQ) and 2,3‐oxidosqualene (2,3‐OSQ) (Figure 3e). 7‐day‐old soybean seedlings were subjected to different treatments. After a treatment period of 10 days, we observed the phenotypes of the soybean seedlings under different treatment conditions. The results revealed that exogenous application of SQ (600
days, we observed the phenotypes of the soybean seedlings under different treatment conditions. The results revealed that exogenous application of SQ (600 μg/L) and 2,3‐OSQ (300
μg/L) and 2,3‐OSQ (300 μg/L) promoted soybean growth under normal conditions (Figure 3f–h). Moreover, these compounds significantly enhanced soybean stress tolerance under 200
μg/L) promoted soybean growth under normal conditions (Figure 3f–h). Moreover, these compounds significantly enhanced soybean stress tolerance under 200 mM mannitol conditions (Figure 3f–h). In contrast, terbinafine, a specific inhibitor of SQE widely used to inhibit sterol biosynthesis pathways (Figure 3i), conditionally impeded the development of normal root structures in soybean seedlings exposed to mannitol‐induced stress (Figure 3f). Notably, this inhibitory effect was alleviated by providing exogenous concentrations of 2,3‐OSQ at various levels, which partially prevented the occurrence of short‐thick root phenotype (Figure 3f–h, Figure S5a–d). These results underscored the crucial role played by SQE‐mediated plant sterol synthesis pathways in both growth and stress tolerance.
mM mannitol conditions (Figure 3f–h). In contrast, terbinafine, a specific inhibitor of SQE widely used to inhibit sterol biosynthesis pathways (Figure 3i), conditionally impeded the development of normal root structures in soybean seedlings exposed to mannitol‐induced stress (Figure 3f). Notably, this inhibitory effect was alleviated by providing exogenous concentrations of 2,3‐OSQ at various levels, which partially prevented the occurrence of short‐thick root phenotype (Figure 3f–h, Figure S5a–d). These results underscored the crucial role played by SQE‐mediated plant sterol synthesis pathways in both growth and stress tolerance.
Furthermore, we employed the SQE inhibitor terbinafine to investigate the significance of squalene production in plant stress tolerance. Similarly, 7‐day‐old soybean seedlings were exposed to various stress treatments. After a duration of 10 days, the phenotypes of soybean seedlings were observed under different treatments. Our findings clearly demonstrated that the application of terbinafine compromised soybean's tolerance to stress (Figure 3j–n). Additionally, we performed liquid chromatography‐mass spectrometry (LC–MS) analysis to examine changes in key metabolites from the SQE‐mediated plant sterol synthesis pathway in soybean roots under 100
days, the phenotypes of soybean seedlings were observed under different treatments. Our findings clearly demonstrated that the application of terbinafine compromised soybean's tolerance to stress (Figure 3j–n). Additionally, we performed liquid chromatography‐mass spectrometry (LC–MS) analysis to examine changes in key metabolites from the SQE‐mediated plant sterol synthesis pathway in soybean roots under 100 mM mannitol‐induced stress. These analyses revealed a significant increase in squalene content in soybean roots upon exposure to 100
mM mannitol‐induced stress. These analyses revealed a significant increase in squalene content in soybean roots upon exposure to 100 mM mannitol (Figure 3o, Figure S5e). Interestingly, treatment with terbinafine led to a substantial accumulation of squalene in soybean roots (Figure 3o, Figure S5e). Moreover, there was a notable elevation in the levels of cycloartenol and β‐amyrin in soybean roots‐ two crucial products derived from 2,3‐OSQ – after treatment with 100
mM mannitol (Figure 3o, Figure S5e). Interestingly, treatment with terbinafine led to a substantial accumulation of squalene in soybean roots (Figure 3o, Figure S5e). Moreover, there was a notable elevation in the levels of cycloartenol and β‐amyrin in soybean roots‐ two crucial products derived from 2,3‐OSQ – after treatment with 100 mM mannitol (Figure 1p,q, Figure S5f), further highlighting their role in plant stress tolerance. Collectively, these results underscored the indispensable function of SQE in both plant growth and stress tolerance.
mM mannitol (Figure 1p,q, Figure S5f), further highlighting their role in plant stress tolerance. Collectively, these results underscored the indispensable function of SQE in both plant growth and stress tolerance.
Overexpression of the NF‐Y target gene GmSQE1 in soybean improves drought and salt stress tolerance
To assess the potential role of squalene‐derived compounds in enhancing plant stress tolerance, we generated three distinct GmSQE1 overexpression lines (GmSQE1‐OE1, GmSQE1‐OE3, and GmSQE1‐OE6) (Figure S6a–d). The impact of GmSQE1 overexpression on drought and salt stress tolerance paralleled the observations made for GmNF‐YC9‐OE plants during the seedling stage (Figure S6e–j). Under stress conditions, GmSQE1‐OE plants exhibited enhanced characteristics such as increased biomass and longer hypocotyl lengths compared to their WT counterparts (Figure S6e–j). These findings were further supported by pot experiments, highlighting the robust adaptation of GmSQE1‐OE plants to drought and salt stresses (Figure 4a). Moreover, elevated proline and chlorophyll contents along with reduced malondialdehyde (MDA) levels were observed in GmSQE1‐OE lines compared to WT plants (Figure 4b), providing evidence for the enhanced stress resilience conferred by overexpressing GmSQE1.
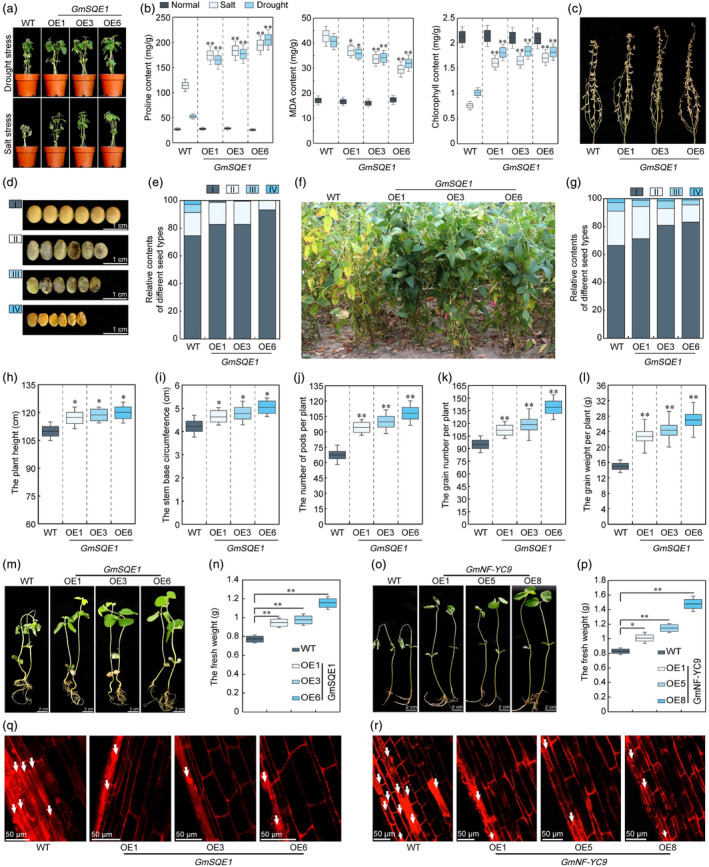
Stress tolerance identification analysis of GmSQE1 overexpressing soybean plants. (a) Phenotype of WT and GmSQE1‐OE seedling soybean plants under drought and 200 mM NaCl treatment conditions. (b) Proline, MDA, and chlorophyll content analysis of WT and GmSQE1‐OE soybean plants under different stress conditions. (c) Phenotype analysis of GmSQE1‐OE soybean plants at adult stage under drought conditions in the field in the year 2020. (d, e) Phenotype (d) and relative content (e) of different seed types in GmSQE1‐OE soybean plants under drought conditions in the field in the year 2020. (f) Phenotype analysis of GmSQE1‐OE soybean plants at adult stage under drought conditions in the field in the year 2021. (g) Relative contents of different seed types in GmSQE1‐OE soybean plants under drought conditions in the field in the year 2021. (h–l) Agronomic trait analysis of WT and GmSQE1‐OE soybean plants under drought conditions in the field in the year 2021. (m) Oxidative stress tolerance analysis of GmSQE1 transgenic soybean plants under 50
mM NaCl treatment conditions. (b) Proline, MDA, and chlorophyll content analysis of WT and GmSQE1‐OE soybean plants under different stress conditions. (c) Phenotype analysis of GmSQE1‐OE soybean plants at adult stage under drought conditions in the field in the year 2020. (d, e) Phenotype (d) and relative content (e) of different seed types in GmSQE1‐OE soybean plants under drought conditions in the field in the year 2020. (f) Phenotype analysis of GmSQE1‐OE soybean plants at adult stage under drought conditions in the field in the year 2021. (g) Relative contents of different seed types in GmSQE1‐OE soybean plants under drought conditions in the field in the year 2021. (h–l) Agronomic trait analysis of WT and GmSQE1‐OE soybean plants under drought conditions in the field in the year 2021. (m) Oxidative stress tolerance analysis of GmSQE1 transgenic soybean plants under 50 μM MV treatment. (n) Fresh weight analysis of GmSQE1 transgenic soybean plants under 50
μM MV treatment. (n) Fresh weight analysis of GmSQE1 transgenic soybean plants under 50 μM MV treatment. (o) Oxidative stress tolerance analysis of GmNF‐YC9 transgenic soybean plants under 50
μM MV treatment. (o) Oxidative stress tolerance analysis of GmNF‐YC9 transgenic soybean plants under 50 μM MV treatment. (p) Fresh weight analysis of GmNF‐YC9 transgenic soybean plants under 50
μM MV treatment. (p) Fresh weight analysis of GmNF‐YC9 transgenic soybean plants under 50 μM MV treatment. (q, r) 200
μM MV treatment. (q, r) 200 mM H2O2‐induced cell death analysis of GmSQE1 (q) and GmNF‐YC9 (r) transgenic soybean plant roots by Pi staining.
mM H2O2‐induced cell death analysis of GmSQE1 (q) and GmNF‐YC9 (r) transgenic soybean plant roots by Pi staining.
Furthermore, the stress tolerance effects of GmSQE1 overexpression were also observed during the flowering stage, as evidenced by increased biomass in GmSQE1‐OE and GmNF‐YC9‐OE plants compared to WT plants under drought and salt stress conditions (Figure S6k–p). These results confirmed that the high expression of GmSQE1 and GmNF‐YC9 in soybean provided benefits for enhancing plant stress tolerance. To validate the durability of these effects, we conducted drought tolerance tests in field conditions over two seasons. Under normal irrigation conditions, no discernible differences in agronomic performance were observed between GmSQE1‐OE, GmNF‐YC9‐OE, and WT plants (Figure S7). However, under drought stress, both GmSQE1‐OE and GmNF‐YC9‐OE plants exhibited significantly improved agronomic performance compared to WT plants. This was evident through the increases in stem base circumference, grain count per plant, and grain weight per plant (Figure 4c–l, Figure S8a–t). Furthermore, an analysis of seed traits indicated that under drought conditions, GmSQE1‐OE and GmNF‐YC9‐OE plants produced more high‐quality seeds and fewer low‐quality seeds than WT plants (Figure 4d–g, Figure S8l–t, Table S1). Importantly, GmSQE1‐OE and GmNF‐YC9‐OE plants maintained these drought tolerance characteristics and improved agronomic traits at the adult stage under drought stress conditions (Figure 4c–l, Figure S8).
GmSQE1 and GmNF‐YC9 can enhance tolerance to abiotic stress‐induced oxidation
A recent study has demonstrated that the depletion of SQE in cholesterol auxotrophic lymphomas leads to an accumulation of squalene, thereby preventing oxidative cell death (Garcia‐Bermudez et al., 2019). This finding suggests that the involvement of SQE in mediating cellular antioxidant process. However, our results and previous studies indicate that abiotic stress induces increased levels of reactive oxygen species (ROS) in plants (Figure S9a,b). Consequently, we hypothesised that GmSQE1 may enhance plant stress tolerance by improving soybean's resilience to oxidative stress. To test this hypothesis, GmSQE1‐OE, GmNF‐YC9‐OE, and WT plants were subjected to methyl viologen (MV) and hydrogen peroxide (H2O2). After 7 days of treatment, MV induced wilting in WT plants, whereas GmSQE1‐OE and GmNF‐YC9‐OE plants displayed varying degrees of resistance to this effect (Figure 4m–p). These transgenic plants exhibited a higher biomass compared to WT plants under MV treatment conditions (Figure 4n,p). Additionally, after a 1.5‐h exposure to 200
days of treatment, MV induced wilting in WT plants, whereas GmSQE1‐OE and GmNF‐YC9‐OE plants displayed varying degrees of resistance to this effect (Figure 4m–p). These transgenic plants exhibited a higher biomass compared to WT plants under MV treatment conditions (Figure 4n,p). Additionally, after a 1.5‐h exposure to 200 mM H2O2, cell damage was evaluated using propidium iodide (PI) staining. While WT plant root cells exhibited PI red fluorescence in their cytoplasm, only a few GmSQE1‐OE and GmNF‐YC9‐OE plant root cells displayed such fluorescence (Figure 4q,r). This result indicated that GmSQE1‐OE and GmNF‐YC9‐OE plants experienced less cell damage compared to WT plants under 200
mM H2O2, cell damage was evaluated using propidium iodide (PI) staining. While WT plant root cells exhibited PI red fluorescence in their cytoplasm, only a few GmSQE1‐OE and GmNF‐YC9‐OE plant root cells displayed such fluorescence (Figure 4q,r). This result indicated that GmSQE1‐OE and GmNF‐YC9‐OE plants experienced less cell damage compared to WT plants under 200 mM H2O2 treatment conditions (Figure 4q,r). Moreover, high‐temperature (43
mM H2O2 treatment conditions (Figure 4q,r). Moreover, high‐temperature (43 °C) stress, known to induce ROS production (Pospíšil, 2016), was applied to four‐week‐old seedlings. Following a recovery period after 12
°C) stress, known to induce ROS production (Pospíšil, 2016), was applied to four‐week‐old seedlings. Following a recovery period after 12 h of exposure, both GmSQE1‐OE and GmNF‐YC9‐OE plants survived at higher rates and accumulated more shoot biomass than the control group, which further highlighted their enhanced tolerance towards oxidative stress (Figure S9c–h). Interestingly, the application of terbinafine compromised soybean's oxidative stress tolerance (Figure S10a–c). Collectively, these results suggested that overexpressing both GmSQE1 and GmNF‐YC9 significantly improved plant responses to oxidative stress across various contexts.
h of exposure, both GmSQE1‐OE and GmNF‐YC9‐OE plants survived at higher rates and accumulated more shoot biomass than the control group, which further highlighted their enhanced tolerance towards oxidative stress (Figure S9c–h). Interestingly, the application of terbinafine compromised soybean's oxidative stress tolerance (Figure S10a–c). Collectively, these results suggested that overexpressing both GmSQE1 and GmNF‐YC9 significantly improved plant responses to oxidative stress across various contexts.
Exogenous application of soybean steroids can enhance crop stress tolerance
To further investigate the underlying mechanisms of GmSQE1 and GmNF‐YC9‐mediated plant stress tolerance, we performed metabolomic analyses on GmSQE1‐OE, GmNF‐YC9‐OE, and WT plants subjected to 100 mM mannitol treatment. The correlation analysis between transcriptome and metabolome data unveiled 134 KEGG pathways enriched in GmNF‐YC9‐OE compared to WT plants, with 132 pathways enriched in the transcriptomics data and 47 pathways in the metabolomics data (Figure 5a–c). Among these, 45 KEGG pathways overlapped (Figure 5c), primarily focus on metabolism and secondary metabolite biosynthesis (Figure 5d). Notably, genes and metabolites related to steroid biosynthesis, including brassinosteroid, were identified. In total, there was a total of 493 upregulated metabolites in GmNF‐YC9‐OE plants and 435 in GmSQE1‐OE plants, with an overlap of 267 metabolites (Figure 5e). Since SQE catalysis is a rate‐limiting step in the synthesis of steroids, their derivatives, and triterpenoids (Figure 5f), we specifically examined these compound classes. Our analysis revealed that a total of 20 upregulated steroids and steroid derivatives along with 22 triterpenoids under stress conditions in either GmNF‐YC9‐OE or GmSQE1‐OE plants (Figure 5g,h). Notably, fucosterol and soyasaponin II displayed significant upregulation under stress conditions in both GmNF‐YC9‐OE and GmSQE1‐OE plants (Figure 5g,h), highlighting their significance for plant stress tolerance.
mM mannitol treatment. The correlation analysis between transcriptome and metabolome data unveiled 134 KEGG pathways enriched in GmNF‐YC9‐OE compared to WT plants, with 132 pathways enriched in the transcriptomics data and 47 pathways in the metabolomics data (Figure 5a–c). Among these, 45 KEGG pathways overlapped (Figure 5c), primarily focus on metabolism and secondary metabolite biosynthesis (Figure 5d). Notably, genes and metabolites related to steroid biosynthesis, including brassinosteroid, were identified. In total, there was a total of 493 upregulated metabolites in GmNF‐YC9‐OE plants and 435 in GmSQE1‐OE plants, with an overlap of 267 metabolites (Figure 5e). Since SQE catalysis is a rate‐limiting step in the synthesis of steroids, their derivatives, and triterpenoids (Figure 5f), we specifically examined these compound classes. Our analysis revealed that a total of 20 upregulated steroids and steroid derivatives along with 22 triterpenoids under stress conditions in either GmNF‐YC9‐OE or GmSQE1‐OE plants (Figure 5g,h). Notably, fucosterol and soyasaponin II displayed significant upregulation under stress conditions in both GmNF‐YC9‐OE and GmSQE1‐OE plants (Figure 5g,h), highlighting their significance for plant stress tolerance.
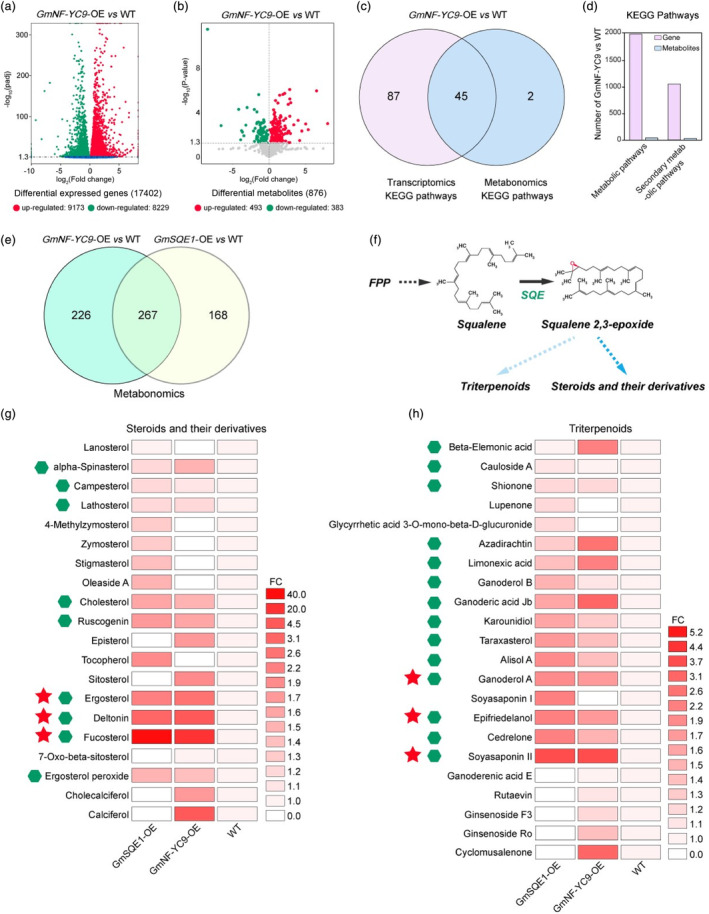
Differentially accumulated sterol metabolite analysis in WT, GmNF‐YC9‐OE, and GmSQE1‐OE soybean plants under drought stress conditions using transcriptome and metabolome. (a) Analysis of differentially expressed genes (DEGs) between GmNF‐YC9‐OE and WT plants under drought stress conditions using transcriptome technology. (b) Analysis of differentially accumulated metabolites (DAMs) between GmNF‐YC9‐OE and WT plants under drought stress conditions using metabolome technology. (c) Correlation analysis of drought stress‐induced transcriptome and metabolome data of GmNF‐YC9‐OE vs WT. (d) KEGG pathway correlation analysis of transcriptome and metabolome in GmNF‐YC9‐OE vs WT plants under drought stress conditions. (e) Correlation analysis of metabolome data among GmNF‐YC9‐OE, GmSQE1‐OE, and WT plants under drought stress conditions. (f) Analysis of plant SQE‐mediated catalysed pathways. As SQE catalysis is the rate‐limiting step in triterpenoid, steroid and steroid derivative biosynthesis, we focused on these compound classes. (g) Differentially expressed steroid and their derivative analysis among GmNF‐YC9‐OE, GmSQE1‐OE, and WT plants under drought stress conditions. (h) Differentially expressed triterpenoid analysis among GmNF‐YC9‐OE, GmSQE1‐OE, and WT plants under drought stress conditions. The green hexagon in figure g and h indicates the accumulations of 9 steroid‐related compounds and 14 triterpenoids in both GmNF‐YC9‐OE and GmSQE1‐OE plants under drought stress conditions. The red pentacle in figure g and h indicates the significant accumulations of steroid‐related compounds and triterpenoids in both GmNF‐YC9‐OE and GmSQE1‐OE plants under drought stress conditions.
Subsequently, we investigated the effects of exogenously applied fucosterol and soyasaponin II on plant stress tolerance. Interestingly, while higher concentrations of these compounds inhibited soybean growth under normal conditions, and lower concentrations promoted the growth (Figure 6a–d). Furthermore, the application of moderate concentrations of fucosterol and soyasaponin II alleviated stress‐induced growth inhibition in soybeans (Figure 6e–j). In vitro experiments conducted various crops including soybean, wheat, foxtail millet, and maize demonstrated that the spray application of moderate concentrations of these compounds enhanced drought and osmotic stress tolerance (Figure 6k–m, Figure S11). Additionally, we investigated the impact of beta‐amyrin and cycloartenol‐key products of the plant sterol synthesis pathway‐on crop stress tolerance. Consistent with previous findings, the exogenous application of beta‐amyrin and cycloartenol yielded similar results, higher concentrations inhibited soybean growth while lower concentrations not only promoted growth but also enhanced crop stress tolerance (Figure S12). Collectively, these results underscored the potential of these metabolites in enhancing stress tolerance across a variety of crucial crops.
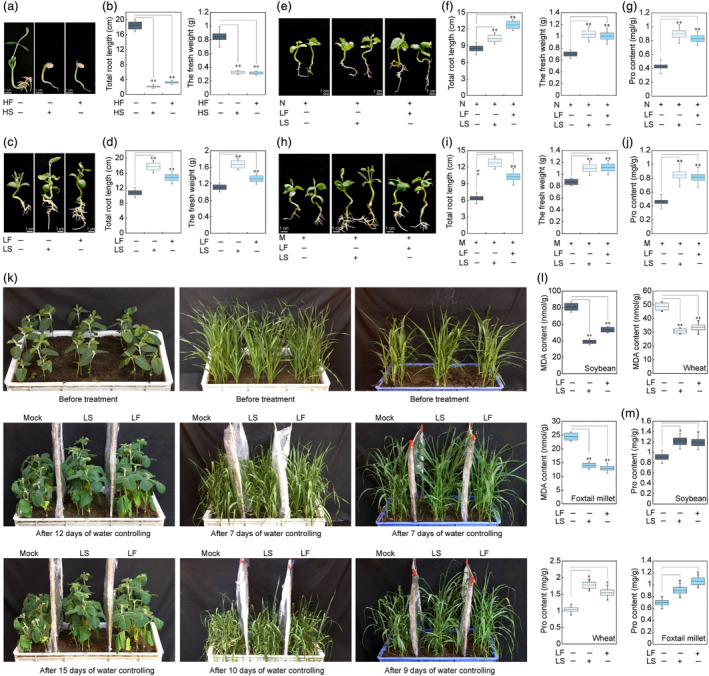
Function analysis of fucosterol and soyasaponin II in plant. (a, b) Phenotype (a) and biomass (b) analysis of soybean seedling plants under high concentrations of fucosterol and soyasaponin II treatment conditions. In figure a and b, HF indicates a high concentration of fucosterol (5 mg/L), while HS indicates a high concentration of soyasaponin II (5
mg/L), while HS indicates a high concentration of soyasaponin II (5 mg/L). (c, d) Phenotype (c) and biomass (d) analysis of soybean seedling plants under moderate concentrations of fucosterol and soyasaponin II treatment conditions. (e–g) Phenotype (e), biomass (f), and proline (Pro) content (g) analysis of soybean seedling plants under fucosterol, soyasaponin II, and 100
mg/L). (c, d) Phenotype (c) and biomass (d) analysis of soybean seedling plants under moderate concentrations of fucosterol and soyasaponin II treatment conditions. (e–g) Phenotype (e), biomass (f), and proline (Pro) content (g) analysis of soybean seedling plants under fucosterol, soyasaponin II, and 100 mM NaCl treatment conditions. In figure e–g, N indicates 100
mM NaCl treatment conditions. In figure e–g, N indicates 100 mM NaCl. (h–j) Phenotype (h), biomass (i), and proline (Pro) content (j) analysis of soybean seedling plants under fucosterol, soyasaponin II, and mannitol treatment conditions. In figure h–j, M indicates 100
mM NaCl. (h–j) Phenotype (h), biomass (i), and proline (Pro) content (j) analysis of soybean seedling plants under fucosterol, soyasaponin II, and mannitol treatment conditions. In figure h–j, M indicates 100 mM mannitol. (k) Phenotype analysis of soybean, wheat, and foxtail millet plants by spraying moderate concentration of fucosterol and soyasaponin II under drought conditions. (l, m) MDA (l) and proline (Pro) content (m) analysis of soybean, wheat, and foxtail millet plants by spraying moderate concentration of fucosterol and soyasaponin II under drought conditions. In figure c–k, LF indicates a moderate concentration of fucosterol (300
mM mannitol. (k) Phenotype analysis of soybean, wheat, and foxtail millet plants by spraying moderate concentration of fucosterol and soyasaponin II under drought conditions. (l, m) MDA (l) and proline (Pro) content (m) analysis of soybean, wheat, and foxtail millet plants by spraying moderate concentration of fucosterol and soyasaponin II under drought conditions. In figure c–k, LF indicates a moderate concentration of fucosterol (300 μg/L), while LS indicates a moderate concentration of soyasaponin II (300
μg/L), while LS indicates a moderate concentration of soyasaponin II (300 μg/L).
μg/L).
Discussion
Several studies have established a correlation between CCAAT‐box transcription factors and drought as well as heat stress (Nelson et al., 2007; Sato et al., 2019). However, the underlying mechanism remains elusive, and no direct gene targets associated with abiotic stress have been identified yet. In this study, we demonstrated that GmNF‐YC9, a member of the NF‐Y transcription factor C subunit family protein in soybean, exhibited responsiveness to both drought and salt stresses (Figure 1). Furthermore, our findings revealed that GmNF‐YC9 could form a NF‐Y transcriptional complex with GmNF‐YA2 and GmNF‐YB24 through protein–protein interactions to directly activate expression of the key enzyme involved in phytosterol synthesis pathway (GmSQE1), thereby promoting sterol synthesis to improve soybean stress tolerance (Figures 2, ,3,3, Figure S4). This discovery revealed an unrecognised role of the NF‐Y transcription factors in plants. Additionally, several subunits of NF‐Y transcription factors have been implicated in flowering regulation. For instance, the trimeric complex comprising NF‐YC and NF‐YB subunits can recruit CONSTANS‐Like transcription factors to regulate their DNA targets, thus influencing flowering time in Arabidopsis (Ben‐Naim et al., 2006; Wenkel et al., 2006). Moreover, NF‐Y has been demonstrated to modulate flowering time by mediating the control of H3K27me3 demethylation at the SOC1 locus (Hou et al., 2014). Our previous study revealed that GmNF‐YC9 showed homology with OsHAP5H, OsHAP5O, and AtNF‐YC11 (Yu et al., 2021). Interestingly, research conducted on rice indicates that OsHAP5H (OsNF‐YC9) has an association with the heading period, and overexpression of OsHAP5H delays heading under long day conditions (Li et al., 2016). However, in our experimental setup, no differences related to flowering time were observed between the GmNF‐YC9 overexpressing lines and WT plants in the growing area. Nevertheless, the role of GmNF‐YC9 in regulating flowering time required further validation.
Adaptation to oxidative stress is a fundamental aspect of plant and animal cell physiology, with implications for both crop production and human disease in the face of environmental stress (Devireddy et al., 2021; Dixon et al., 2012; Forman and Zhang, 2021; Kerchev and Van Breusegem, 2022; Zandalinas et al., 2021). Reactive oxygen species (ROS) are the primary source of cell oxidisation, leading to the oxidation of proteins, nucleic acids, and lipids (Mittler et al., 2022; Oikawa et al., 2022). Abiotic stress can cause excessive ROS accumulation in plant cells, compromising their stress tolerance (Castro et al., 2021; Kerchev and Van Breusegem, 2022). NF‐Y transcription factors have a crucial function in inhibiting the excessive ROS accumulation and enhancing the antioxidant capacity of plant cells. Studies have shown that TaNF‐YB1 regulates early wheat grain development by scavenging the excessive ROS to promote grain filling (Liu et al., 2023). In rice, OsNF‐YC5 alleviates the inhibitory effect of salt stress on seed germination by reducing ROS levels (Jin et al., 2023). Our findings demonstrated that NF‐Y transcription factors improved plant oxidation stress tolerance through the regulation of SQE activity (Figure S4, Figure 4m–r). SQE is a rate‐limiting enzyme of the IPP pathway and oxidises squalene to produce 2,3‐OSQ, which is an important precursor for steroid and triterpenoid synthesis both in plants (Prashant et al., 2016; Thimmappa et al., 2014). Research has indicated that SQE mutation alters sterol composition in Arabidopsis roots, which affects the localisation of NADPH oxidases and subsequent regulation of ROS production (Posé et al., 2009). Additionally, in gastric cancer cells, SQE mutation results in an increased squalene level, thereby conferring high antioxidant ability upon cancer cells (Garcia‐Bermudez et al., 2019). Our study demonstrated that overexpression of GmSQE1 in soybean improved significantly soybean's tolerance to oxidative stress while reducing oxidative stress‐induced cell injury (Figure 4m,n,q). These findings suggested that SQE‐mediated sterol synthesis pathway played a crucial role in modulating ROS and cell antioxidants. Furthermore, our results demonstrated that NF‐Y transcription factors could also upregulate the expression of genes associated with oxidative stress response, including heat shock proteins (HSPs) (Ul Haq et al., 2019), antioxidant enzymes (Mittler et al., 2022), and ferritins (Reyt et al., 2014) (Figure S13). This highlighted the complexity of NF‐Y transcription factors regulation on plant cell antioxidants.
Interestingly, we observed a reversal of SQE‐mediated cell antioxidant behaviours between human cells and soybean. While the mutation of SQE in human cells led to strong cell antioxidant activity (Garcia‐Bermudez et al., 2019), our findings demonstrated that overexpression of GmSQE1 in soybean enhanced oxidative stress tolerance (Figure 4m,n,q). Further analysis reveals large differences in sterol metabolism process between mammals and plants, as almost all plants steroids are derived from lanosterol and cycloartenol, while only lanosterol serves as the precursor of all animal steroids (Figure S14) (Prashant et al., 2016; Yoshioka et al., 2020). Both lanosterol and cycloartenol are produced directly from 2,3‐OSQ (Prashant et al., 2016; Yoshioka et al., 2020). In plants, 2,3‐OSQ also serves as a substrate for the amyrin synthase (AYSE)‐mediated amyrin metabolism pathway for production of triterpenoids such as soyasaponin (Figure S14) (Mertens et al., 2016). These substantial disparities indicate that steroids in plants exhibit a greater diversity and complexity compared to those observed in mammals, highlighting the crucial roles of plant‐specific cycloartenol synthase (CYSE)‐mediated cycloartenol metabolism and amyrin synthase (AYSE)‐mediated amyrin metabolism pathways in plant stress tolerance, including oxidative stress tolerance. Indeed, our targeted metabolome analysis revealed significant accumulations of multiple steroids and triterpenoids (including their derivatives) following stress treatments (Figure 5g,h). Spraying these compounds in vitro significantly improves drought stress tolerance of wheat, maize, and foxtail millet (Figure 6, Figures S11, S12), suggesting that the effect is also relevant to monocots and the function is crucial on enhancing plant stress tolerance. These findings suggest that these compounds, through genetic modification or selection to enhance their production, or by their chemical application, can be used to improve stress tolerance and quality in crops. However, further elucidation is required regarding the molecular mechanism underlying how plant steroids affect cell viability. Together, our research uncovered an undiscovered NF‐Y‐SQE module that modulated sterol synthesis and played a crucial role in plant stress tolerance (Figure 7). Application of plant steroids in vitro showed potential for enhancing crop tolerance to abiotic stress (Figure 7). Furthermore, we hypothesised that phytosterols may exhibit significant potential in the field of medicine, particularly in enhancing the microenvironment of animal cells through augmenting their antioxidative capacity.
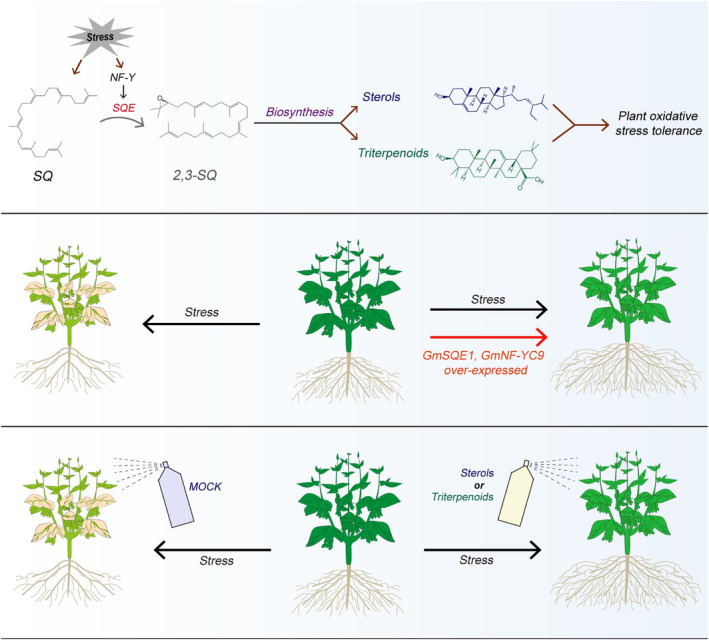
The model of NF‐Y‐SQE module and soybean steroids improving plant stress tolerance. NF‐Y‐SQE module participates in regulating plant stress tolerance by enhancing SQE‐mediated squalene metabolic pathways. Application of plant steroids or triterpenoids in crops enhanced their tolerance to drought stress.
Materials and methods
Generation and detection of transgenic soybean plants
Soybean cultivated variety (Glycine max, ‘Tiefeng 8’) was utilised to amplify GmNF‐YC9 (Glyma.13G189400) and GmSQE1 (Glyma.08G063700). The open reading fames (ORFs) of GmNF‐YC9 and GmSQE1 were cloned into the pTF101 vector to generate the recombinant vectors, which were then transformed into Agrobacterium strain EHA105. Soybean variety “Williams 82” was used as the recipient material for transformation, and stable transformation was performed following the protocol described by Chen et al. (2018). T3 homozygous generations of GmNF‐YC9 and GmSQE1 transgenic soybean plants were confirmed by PCR and qPCR (Table S2), which were used for functional analysis.
Analysis of stress tolerance on transgenic soybean plants
For 150 mM NaCl and 200
mM NaCl and 200 mM mannitol treatments, T3 homozygous GmNF‐YC9 and GmSQE1 transgenic soybean seeds were surface‐sterilised with chlorine for 6
mM mannitol treatments, T3 homozygous GmNF‐YC9 and GmSQE1 transgenic soybean seeds were surface‐sterilised with chlorine for 6 h, following the method described by Chen et al. (2018) and Cheng et al. (2021). The sterilised seeds were then transferred to a B5 vitamin solid medium for growth under normal conditions. After 7
h, following the method described by Chen et al. (2018) and Cheng et al. (2021). The sterilised seeds were then transferred to a B5 vitamin solid medium for growth under normal conditions. After 7 days of growth, the seedlings were transferred into a B5 vitamin solid medium supplemented with 150
days of growth, the seedlings were transferred into a B5 vitamin solid medium supplemented with 150 mM NaCl and 200
mM NaCl and 200 mM mannitol for stress tolerance analysis. After 7
mM mannitol for stress tolerance analysis. After 7 days of treatment, the seedlings were analysed.
days of treatment, the seedlings were analysed.
For stress tolerance analysis during the soybean seedling stage, T3 homozygous GmNF‐YC9 and GmSQE1 transgenic soybean seeds were sown in flowerpots filled with equally weighted nutrient soil for growth under normal conditions. Four‐week‐old soybean seedlings were treated with either water controlling or 250 mM NaCl. After 14
mM NaCl. After 14 days of drought or 7
days of drought or 7 days of salt treatment, the stress tolerance phenotypes of transgenic soybean seedlings were analysed. The method was expounded upon by Wang et al. (2020) and Yu et al. (2021).
days of salt treatment, the stress tolerance phenotypes of transgenic soybean seedlings were analysed. The method was expounded upon by Wang et al. (2020) and Yu et al. (2021).
For drought tolerance analysis at soybean adult stage, we utilised rain‐proof installations to systematically investigate the effects of water stress on transgenic soybean plants. Every year at the end of May (Beijing, China), we planted transgenic soybean seeds in the field for growth under natural conditions. Around the middle of August, soybean plants began to enter periods of pod filling. We thus stopped irrigation to control water at the beginning of August and utilised rain‐proof installations to prevent weather from affecting drought treatment (Figure S15). After 1 month of water controlling treatment, the soil water content was measured using the method described by Yu et al. (2021) (Table S3), and the phenotype under drought conditions was recorded. In the middle of October, soybean plants began to be harvested, and agronomic traits were recorded for analysis.
Bimolecular fluorescence complementation (BiFC) assay
The ORFs of bait protein and candidate protein were cloned into the pUC‐SPYNE and pUC‐SPYCE vectors, respectively. Subsequently, the recombinant vectors were co‐transformed into Arabidopsis protoplasts through PEG‐mediated transfection, followed by incubation in the dark at 25 °C. The detailed method was described by Yoo et al. (2007). YFP fluorescence of the protoplasts was observed using a confocal laser scanning microscope (LSM 700; Zeiss, Oberkochen, Germany).
°C. The detailed method was described by Yoo et al. (2007). YFP fluorescence of the protoplasts was observed using a confocal laser scanning microscope (LSM 700; Zeiss, Oberkochen, Germany).
Luciferase complementation imaging (LCI) assay
The bait protein and candidate protein genes were individually inserted into pCAMBIA1300‐nLUC and pCAMBIA1300‐cLUC vectors. Subsequently, Agrobacterium GV3101 harbouring the nLUC and cLUC derivative binary recombinant plasmids were separately introduced via co‐injection into leaves of 4‐week‐old Nicotiana benthamiana. The firefly luciferase complementation imaging (LCI) transient expression assay was performed following the protocol established by Fujikawa and Kato (2007).
In vitro GST pull‐down assay
The open reading frames (ORFs) of bait protein and candidate protein genes were cloned into the pCold™ TF vector (TaKaRa, Bio) and pGEX‐4T‐1 vector (GE Healthcare, Chicago, IL), respectively. Subsequently, the recombinant plasmids were introduced into the E. coli strain transetta (TransGen, Biotech). Expressions of the corresponding proteins were induced overnight using 0.5 mM isopropyl β‐D‐thiogalactoside (IPTG) (Inalco SPA, San Luis Obispo, CA) at 16
mM isopropyl β‐D‐thiogalactoside (IPTG) (Inalco SPA, San Luis Obispo, CA) at 16 °C, and then a pull‐down assay was performed. The experimental details were described by Yu et al. (2021).
°C, and then a pull‐down assay was performed. The experimental details were described by Yu et al. (2021).
In vitro EMSA binding assay
The genes encoding three subunit proteins of the NF‐Y transcription factor were cloned into expression vectors harbouring the tag proteins (His, GST, and MBP), respectively, and subsequently transferred into competent cells of the E. coli strain Rosetta. These recombinant proteins were employed for an EMSA using a biotin end‐labelled duplex DNA probe. The method was previously described by Yu et al. (2021).
Effect of fucosterol and soyasaponin II on crop growth under different treatments
Three‐week‐old of soybean, wheat, foxtail millet, and maize seedlings grown under normal conditions were subjected to controlled water treatment. During the treatment, the seedlings were sprayed with aqueous solutions of mock (40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM fatty alcohol polyoxyethylene ether (APE), which was a leaf surfactant), fucosterol (20
mM fatty alcohol polyoxyethylene ether (APE), which was a leaf surfactant), fucosterol (20 μL of fucosterol at a concentration of 600
μL of fucosterol at a concentration of 600 mg/L was added into 40
mg/L was added into 40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM APE), and soyasaponin II (20
mM APE), and soyasaponin II (20 μL of soyasaponin II at a concentration of 600
μL of soyasaponin II at a concentration of 600 mg/L was added into 40
mg/L was added into 40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM APE). The sprays were applied four times each week, with at least 1 day between each spraying. The changes in crop phenotypes were observed and recorded during controlled water treatment. The leaf samples of crops were collected at the onset of phenotypic variations in order to quantify physiological and biochemical alterations.
mM APE). The sprays were applied four times each week, with at least 1 day between each spraying. The changes in crop phenotypes were observed and recorded during controlled water treatment. The leaf samples of crops were collected at the onset of phenotypic variations in order to quantify physiological and biochemical alterations.
Five‐week‐old of soybean, wheat, foxtail millet, and maize seedlings cultivated under normal conditions were subjected to osmotic stress treatment by irrigating them with 200 mM mannitol aqueous solutions. During osmotic stress treatment, the seedlings were sprayed with mock (mock, 40
mM mannitol aqueous solutions. During osmotic stress treatment, the seedlings were sprayed with mock (mock, 40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM APE), fucosterol (fucosterol aqueous solutions, 20
mM APE), fucosterol (fucosterol aqueous solutions, 20 μL fucosterol (600
μL fucosterol (600 mg/L) into 40
mg/L) into 40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM APE), and soyasaponin II aqueous solutions (soyasaponin II aqueous solutions, 20
mM APE), and soyasaponin II aqueous solutions (soyasaponin II aqueous solutions, 20 μL soyasaponin II (600
μL soyasaponin II (600 mg/L) into 40
mg/L) into 40 mL distilled water containing 200
mL distilled water containing 200 μL of 40
μL of 40 mM APE). The sprays were applied four times each week, with at least 1 day between each spraying. The phenotype changes of the crops were observed and recorded during controlled water treatment. Leaf samples of the crops were taken when differences in the leave phenotype began to appear in order to measure physiological and biochemical changes.
mM APE). The sprays were applied four times each week, with at least 1 day between each spraying. The phenotype changes of the crops were observed and recorded during controlled water treatment. Leaf samples of the crops were taken when differences in the leave phenotype began to appear in order to measure physiological and biochemical changes.
Metabolites extraction, UHPLC–MS–MS analysis, and data processing
The samples of stress induced WT and transgenic soybean seedlings (GmSQE1‐OE6 and GmNF‐YC9‐OE8 plants) were freeze‐dried, and then crushed with a mixer mill. A 50 mg aliquot of each individual samples were precisely weighed and transferred to Eppendorf tubes, which were used for metabolites extraction and UHPLC–MS analysis (Allwegene Technology Co., Ltd, Beijing, China). The methods of plant metabolites extraction were described in detail by Vos et al. (2007).
mg aliquot of each individual samples were precisely weighed and transferred to Eppendorf tubes, which were used for metabolites extraction and UHPLC–MS analysis (Allwegene Technology Co., Ltd, Beijing, China). The methods of plant metabolites extraction were described in detail by Vos et al. (2007).
The UHPLC separation was performed using an EXIONLC System (Sciex). The mobile phase A was 0.1% formic acid in water, while mobile phase B was acetonitrile. The column temperature was set at 40 °C, and the auto‐sampler temperature was set at 4
°C, and the auto‐sampler temperature was set at 4 °C with an injection volume was 2
°C with an injection volume was 2 μL. A Sciex QTrap 6500+ (Sciex Technologies) was applied for assay development. Typical ion source parameters were as follows: IonSpray Voltage, +5500/−4500
μL. A Sciex QTrap 6500+ (Sciex Technologies) was applied for assay development. Typical ion source parameters were as follows: IonSpray Voltage, +5500/−4500 V; Curtain Gas, 35
V; Curtain Gas, 35 psi; Temperature, 400 °C; Ion Source Gas, 1:60
psi; Temperature, 400 °C; Ion Source Gas, 1:60 psi; Ion Source Gas 2, 60
psi; Ion Source Gas 2, 60 psi; DP, ±
psi; DP, ± 100
100 V (Allwegene Technology Co., Ltd, Beijing, China).
V (Allwegene Technology Co., Ltd, Beijing, China).
SCIEX Analyst Work Station Software (Version 1.6.3) was employed for MRM data acquisition and processing. MS raw data (.wiff) files were converted to the TXT format using MSconventer. An in‐house R program and database were applied to peak detection and annotation (Allwegene Technology Co., Ltd, Beijing, China).
Measurement of squalene, cycloartenol, and beta‐amyrin by LC–MS
The samples of untreated, 25 mM terbinafine treated, and 100
mM terbinafine treated, and 100 mM mannitol induced soybean seedlings were freeze‐dried, and then crushed with a mixer mill. A 50
mM mannitol induced soybean seedlings were freeze‐dried, and then crushed with a mixer mill. A 50 mg aliquot of each sample was accurately weighed and transferred to Eppendorf tubes for metabolites extraction. The extracted metabolites were used for squalene, cycloartenol, and beta‐amyrin content measurement by UHPLC–MS analysis. The method of extracting plant metabolites was described in detail by Vos et al. (2007).
mg aliquot of each sample was accurately weighed and transferred to Eppendorf tubes for metabolites extraction. The extracted metabolites were used for squalene, cycloartenol, and beta‐amyrin content measurement by UHPLC–MS analysis. The method of extracting plant metabolites was described in detail by Vos et al. (2007).
Squalene (5 mg/mL) (Sigma, USA), cycloartenol (5
mg/mL) (Sigma, USA), cycloartenol (5 mg/mL) (Sigma, USA), and beta‐amyrin (5
mg/mL) (Sigma, USA), and beta‐amyrin (5 mg/mL) (Sigma, USA) were used in appropriate amounts to form standard solutions. These solutions were then diluted with methyl alcohol to create a suitable standard series of standards at concentrations of 5, 10, 20, 50, 100, 200, and 500
mg/mL) (Sigma, USA) were used in appropriate amounts to form standard solutions. These solutions were then diluted with methyl alcohol to create a suitable standard series of standards at concentrations of 5, 10, 20, 50, 100, 200, and 500 ng/mL. The peak areas of the standard substances at different concentrations were used to draw the standard curves that exhibited a good linear relationship with an R
2 value >0.99. The standard curves were drawn based on the peak areas of the standard substance at different concentrations and their corresponding concentrations. With an R
2 value >0.99, it indicated that the standard curves exhibit a strong linear relationship and could be utilised for calculating the concentrations of squalene, cycloartenol, and beta‐amyrin in the samples.
ng/mL. The peak areas of the standard substances at different concentrations were used to draw the standard curves that exhibited a good linear relationship with an R
2 value >0.99. The standard curves were drawn based on the peak areas of the standard substance at different concentrations and their corresponding concentrations. With an R
2 value >0.99, it indicated that the standard curves exhibit a strong linear relationship and could be utilised for calculating the concentrations of squalene, cycloartenol, and beta‐amyrin in the samples.
Author contributions
ZSX coordinated the project, conceived and designed experiments, and edited the manuscript; TFY, HZH, HLW, SYC, and XYS performed the experiments and wrote the first draft; SYC, WJZ, LZ, and JTW generated and analysed LC–MS and cell antioxidant‐related data; ZWL, LZ, JC, YBZ, MC, SLS, and LGJ contributed valuable discussions; QYJ reviewed and revised this paper; YZM coordinated the project and edited the manuscript. All authors have read and approved the final manuscript.
Supporting information
Figure S1 Generation and stress tolerance identification of GmNF‐YC9‐OE soybean plants.
Figure S2 Functional analysis of GmNF‐YB24 and GmNF‐YA2 candidate proteins.
Figure S3 Differentially expressed genes of GmNF‐YC9‐OE soybean plants by transcriptome sequencing.
Figure S4 Interaction analysis between the NF‐Y transcription factor and the potential target gene GmSQE1.
Figure S5 Effect of terbinafine on soybean plant growth and content analysis of squalene, cycloartenol, and β‐amyrin under different treatments by LS‐MS.
Figure S6 Identification and stress tolerance identification of GmSQE1 overexpressing soybean plants.
Figure S7 Agronomic trait analysis of GmNF‐YC9‐ and GmSQE1‐OE soybean plants under normal conditions.
Figure S8 Agronomic trait analysis of GmSQE1‐ and GmNF‐YC9‐OE soybean plants under drought conditions.
Figure S9 Oxidative stress tolerance analysis of GmNF‐YC9‐ and GmSQE1‐OE soybean plants.
Figure S10 Effect of terbinafine on soybean oxidative stress tolerance.
Figure S11 Effect of fucosterol and soyasaponin II on crop stress tolerance.
Figure S12 Effect of cycloartenol and beta‐amyrin on crop stress tolerance.
Figure S13 Expression level analysis of the genes related with response to ROS in GmNF‐YC9‐OE and WT soybean plants.
Figure S14 Difference analysis of SQE‐mediated catalytic pathway in humans and higher plants.
Figure S15 Stress tolerance of soybean materials by application of rain‐proof installations in the experimental field.
Table S1 Different types of seeds divided into four groups (Field).
Table S2 GmNF‐YC9 and GmSQE1 gene‐specific primers used in this article.
Table S3 Relative soil water content analysis of the experimental field.
Table S4 Specific primers of the candidate proteins of GmNF‐YC9.
Table S5 qPCR primers for detecting DEGs associated with ROS response.
Acknowledgements
This research was financially supported by the National Key R & D Program of China (2022YFF1001600), China Postdoctoral Science Foundation (2022M720495), the National Natural Science Foundation of China (31900744 and 32372131), Hainan Seed Industry Laboratory (B21HJ0215), Beijing Natural Science Foundation (6234044) and the Key Research and Development Program of Hainan Province (ZDYF2022XDNY135). We are grateful to Drs. Li‐Juan Qiu and Shi Sun of the Institute of Crop Science, CAAS for kindly providing soybean seeds and Dr. Wen‐Sheng Hou of the Institute of Crop Science, CAAS for providing soybean transforming technology.
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- Ben‐Naim, O. , Eshed, R. , Parnis, A. , Teper‐Bamnolker, P. , Shalit, A. , Coupland, G. , Samach, A. et al. (2006) The CCAAT binding factor can mediate interactions between CONSTANS‐like proteins and DNA. Plant J. 46, 462–476. [Abstract] [Google Scholar]
- Benveniste, P. (2004) Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55, 429–457. [Abstract] [Google Scholar]
- Brodersen, P. , Sakvarelidze‐Achard, L. , Schaller, H. , Khafif, M. , Schott, G. , Bendahmane, A. and Voinnet, O. (2012) Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis . Proc. Natl. Acad. Sci. USA 109, 1778–1783. [Europe PMC free article] [Abstract] [Google Scholar]
- Castro, B. , Citterico, M. , Kimura, S. , Stevens, D.M. , Wrzaczek, M. and Coaker, G. (2021) Stress‐induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants. 7, 403–412. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen, L. , Cai, Y. , Liu, X. , Yao, W. , Guo, C. , Sun, S. , Wu, C. et al. (2018) Improvement of soybean agrobacterium‐mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int. J. Mol. Sci. 19, 3039. [Europe PMC free article] [Abstract] [Google Scholar]
- Cheng, Y. , Wang, X. , Cao, L. , Ji, J. , Liu, T. and Duan, K. (2021) Highly efficient Agrobacterium rhizogenes‐mediated hairy root transformation for gene functional and gene editing analysis in soybean. Plant Methods 17, 73. [Europe PMC free article] [Abstract] [Google Scholar]
- Devireddy, A.R. , Zandalinas, S.I. , Fichman, Y. and Mittler, R. (2021) Integration of reactive oxygen species and hormone signaling during abiotic stress. Plant J. 105, 459–476. [Abstract] [Google Scholar]
- Dixon, S.J. , Lemberg, K.M. , Lamprecht, M.R. , Skouta, R. , Zaitsev, E.M. , Gleason, C.E. , Patel, D.N. et al. (2012) Ferroptosis: an iron‐dependent form of nonapoptotic cell death. Cell 149, 1060–1072. [Europe PMC free article] [Abstract] [Google Scholar]
- Forman, H.J. and Zhang, H. (2021) Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 20, 689–709. [Europe PMC free article] [Abstract] [Google Scholar]
- Fujikawa, Y. and Kato, N. (2007) Split luciferase complementation assay to study protein‐protein interactions in Arabidopsis protoplasts. Plant J. 52, 185–195. [Abstract] [Google Scholar]
- Garcia‐Bermudez, J. , Baudrier, L. , Bayraktar, C. , Shen, Y.S. , La, K. , Guarecuco, R. , Yucel, B. et al. (2019) Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 567, 118–122. [Europe PMC free article] [Abstract] [Google Scholar]
- He, J.X. , Shozo, F. , Li, T.C. , Kang, S.G. , Seto, H. , Suguru, T. , Shigeo, Y. et al. (2018) Sterols regulate development and gene expression in Arabidopsis . Plant Physiol. 131, 2003. [Abstract] [Google Scholar]
- Hou, X. , Zhou, J. , Liu, C. , Liu, L. , Shen, L. and Yu, H. (2014) Nuclear factor Y‐mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5, 4601. [Abstract] [Google Scholar]
- Jin, X. , Li, X. , Xie, Z. , Sun, Y. , Jin, L. , Hu, T. and Huang, J. (2023) Nuclear factor OsNF‐YC5 modulates rice seed germination by regulating synergistic hormone signaling. Plant Physiol. 193, 2825–2847. [Abstract] [Google Scholar]
- Kerchev, P.I. and Van Breusegem, F. (2022) Improving oxidative stress resilience in plants. Plant J. 109, 359–372. [Abstract] [Google Scholar]
- Kou, S. , Chen, L. , Tu, W. , Scossa, F. , Wang, Y. , Liu, J. , Fernie, A. et al. (2018) The arginine decarboxylase gene ADC1, associated to the putrescine pathway, plays an important role in potato cold‐acclimated freezing tolerance as revealed by transcriptome and metabolome analyses. Plant J. 96, 1283–1298. [Abstract] [Google Scholar]
- Laranjeira, S. , Amorim‐Silva, V. , Esteban, A. , Arro, M. , Ferrer, A. , Tavares, R.M. , Botella, M.A. et al. (2015) Arabidopsis squalene epoxidase 3 (SQE3) complements SQE1 and is important for embryo development and bulk squalene epoxidase activity. Mol. Plant 8, 1090–1102. [Abstract] [Google Scholar]
- Li, Q. , Yan, W. , Chen, H. , Tan, C. , Han, Z. , Yao, W. , Li, G. et al. (2016) Duplication of OsHAP family genes and their association with heading date in rice. J. Exp. Bot. 67, 1759–1768. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu, G. , Zhang, R. , Li, S. , Ullah, R. , Yang, F. , Wang, Z. , Guo, W. et al. (2023) TaMADS29 interacts with TaNF‐YB1 to synergistically regulate early grain development in bread wheat. Sci. China Life Sci. 66, 1647–1664. [Abstract] [Google Scholar]
- Liu, X.L. , Wang, L. , Wang, X.W. , Yan, Y. , Yang, X.L. , Xie, M.Y. , Hu, Z. et al. (2020) Mutation of the chloroplast‐localized phosphate transporter OsPHT2;1 reduces flavonoid accumulation and UV tolerance in rice. Plant J. 102, 53–67. [Abstract] [Google Scholar]
- Man, J. , Shi, Y. , Huang, Y. , Zhang, X. , Wang, X. , Liu, S. , He, G. et al. (2023) PnMYB4 negatively modulates saponin biosynthesis in Panax notoginseng through interplay with PnMYB1. Hortic Res. 10, 134. [Europe PMC free article] [Abstract] [Google Scholar]
- Mantovani, R. (1999) The molecular biology of the CCAAT‐binding factor NF‐Y. Gene 239, 15–27. [Abstract] [Google Scholar]
- Men, S.Z. , Boutté, Y. , Ikeda, Y. , Li, X.G. , Palme, K. , Stierhof, Y.D. , Hartmann, M.A. et al. (2008) Sterol‐dependent endocytosis mediates post‐cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10, 237–244. [Abstract] [Google Scholar]
- Mertens, J. , Pollier, J. , Robin, V.B. , Irene, L.V. , José, F.Z. and Alain, G. (2016) The bHLH Transcription Factors TSAR1 and TSAR2 Regulate Triterpene Saponin Biosynthesis in Medicago truncatula. Plant Physiol. 170, 194–210. [Abstract] [Google Scholar]
- Mittler, R. , Zandalinas, S.I. , Fichman, Y. and Van Breusegem, F. (2022) Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 23, 663–679. [Abstract] [Google Scholar]
- Nelson, D.E. , Repetti, P.P. , Adams, T.R. , Creelman, R.A. , Wu, J. , Warner, D.C. , Bensen, R.J. et al. (2007) Plant nuclear factor Y (NF‐Y) B subunits confer drought tolerance and lead to improved corn yields on water‐limited acres. Proc. Natl. Acad. Sci. USA 104, 16450–16455. [Europe PMC free article] [Abstract] [Google Scholar]
- Oikawa, K. , Goto‐Yamada, S. , Hayashi, Y. , Takahashi, D. , Kimori, Y. , Shibata, M. , Yoshimoto, K. et al. (2022) Pexophagy suppresses ROS‐induced damage in leaf cells under high‐intensity light. Nat. Commun. 13, 7493. [Europe PMC free article] [Abstract] [Google Scholar]
- Phillips, D.R. , Rasbery, J.M. , Bartel, B. and Matsuda, S.P. (2006) Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314. [Abstract] [Google Scholar]
- Posé, D. , Castanedo, I. , Borsani, O. , Nieto, B. , Rosado, A. , Taconnat, L. , Ferrer, A. et al. (2009) Identification of the Arabidopsis dry2/sqe1‐5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J. 59, 63–76. [Abstract] [Google Scholar]
- Pospíšil, P. (2016) Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 7, 1950. [Europe PMC free article] [Abstract] [Google Scholar]
- Prashant, D.S. , Jacob, P. , Sayantan, P. , Jedrzej, S. , Hassan, M. , Meital, Y. , Tamar, U. et al. (2016) Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nature plant 3, 16205. [Abstract] [Google Scholar]
- Rasbery, J.M. , Shan, H. , LeClair, R.J. , Norman, M. , Matsuda, S.P.T. and Bartel, B. (2007) Arabidopsis thaliana squalene epoxidase is essential for root and seed development. J. Biol. Chem. 282, 17002–17013. [Abstract] [Google Scholar]
- Reyt, G. , Boudouf, S. , Boucherez, J. , Gaymard, F. and Briat, J.F. (2014) Iron and ferritin dependent ROS distribution impact Arabidopsis root system architecture. Mol. Plant 9, 133. [Abstract] [Google Scholar]
- Sangwan, V. , Orvar, B.L. , Beyerly, J. , Hirt, H. and Dhindsa, R.S. (2002) Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31, 629–638. [Abstract] [Google Scholar]
- Sato, H. , Suzuki, T. , Takahashi, F. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2019) NF‐YB2 and NF‐YB3 have functionally diverged and differentially induce drought and heat stress‐specific genes. Plant Physiol. 180, 1677–1690. [Abstract] [Google Scholar]
- Schaller, H. (2003) The role of sterols in plant growth and development. Prog. Lipid Res. 42, 163–175. [Abstract] [Google Scholar]
- Schaller, H. (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol. Biochem. 42, 465–476. [Abstract] [Google Scholar]
- Schrick, K. , Mayer, U. , Horrichs, A. , Kuhnt, C. , Bellini, C. , Dangl, J. , Schmidt, J. et al. (2000) FACKEL is a sterol C‐14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. [Europe PMC free article] [Abstract] [Google Scholar]
- Senthil‐Kumar, M. , Wang, K. and Mysore, K.S. (2013) AtCYP710A1 gene‐mediated stigmasterol production plays a role in imparting temperature stress tolerance in Arabidopsis thaliana . Plant Signal. Behav. 8, e23142. [Europe PMC free article] [Abstract] [Google Scholar]
- Shen, C.C. , Liu, H.Y. , Guan, Z.Y. , Yan, J.J. , Zheng, T.Z. , Yan, W.H. , Wu, C.Y. et al. (2020) Structural insight into DNA recognition by CCT/NF‐YB/YC complexes in plant photoperiodic flowering. Plant Cell 32, 3469–3484. [Abstract] [Google Scholar]
- Singh, A.K. , Kumar, S.R. , Dwivedi, V. , Rai, A. , Pal, S. , Shasany, A.K. and Nagegowda, D.A. (2017) A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215, 1115–1131. [Abstract] [Google Scholar]
- Souter, M. , Topping, J. , Pullen, M. , Friml, J. , Palme, K. , Hackett, R. , Grierson, D. et al. (2002) Hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031. [Abstract] [Google Scholar]
- Spanova, M. and Daum, G. (2011) Squalene‐biochemistry, molecular biology, process biotechnology, and applications. Eur. J. Lipid Sci. Technol. 113, 1299–1320. [Google Scholar]
- Thimmappa, R. , Geisler, K. , Louveau, T. , O'Maille, P. and Osbourn, A. (2014) Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257. [Abstract] [Google Scholar]
- Ul Haq, S. , Khan, A. , Ali, M. , Khattak, A.M. , Gai, W.X. , Zhang, H.X. , Wei, A.M. et al. (2019) Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20, 5321. [Europe PMC free article] [Abstract] [Google Scholar]
- Vos, R.C.D. , Moco, S. , Lommen, A. , Keurentjes, J.J. , Bino, R.J. and Hall, R.D. (2007) Untargeted large‐scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2, 778–791. [Abstract] [Google Scholar]
- Wang, T.T. , Yu, T.F. , Fu, J.D. , Su, H.G. , Chen, J. , Zhou, Y.B. , Chen, M. et al. (2020) Genome‐wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front. Plant Sci. 11, 604690. [Europe PMC free article] [Abstract] [Google Scholar]
- Wenkel, S. , Turck, F. , Singer, K. , Gissot, L. , Le Gourrierec, J. , Samach, A. and Coupland, G. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. [Abstract] [Google Scholar]
- Willemsen, V. , Friml, J. , Grebe, M. , van den Toorn, A. , Palme, K. and Scheres, B. (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE 1 function. Plant Cell 15, 612–625. [Abstract] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [Abstract] [Google Scholar]
- Yoshioka, H. , Coates, H.W. , Chua, N.K. , Hashimoto, Y. and Ohgane, K. (2020) A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate. Proc. Natl. Acad. Sci. USA 117, 15923–17158. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu, T.F. , Ying Liu, Y. , Fu, J.D. , Ma, J. , Fang, A.W. , Chen, J. , Zheng, L. et al. (2021) The NF‐Y‐PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 19, 1–17. [Europe PMC free article] [Abstract] [Google Scholar]
- Zandalinas, S.I. , Fritschi, F.B. and Mittler, R. (2021) Global warming, climate change, and environmental pollution: recipe for a multifactorial stress combination disaster. Trends Plant Sci. 26, 588–599. [Abstract] [Google Scholar]
- Zanetti, M.E. , Blanco, F.A. , Beker, M.P. , Battaglia, M. and Aguilar, O.M. (2010) A C subunit of the plant nuclear factor NF‐Y required for rhizobial infection and nodule development affects partner selection in the common bean‐Rhizobium etli symbiosis. Plant Cell 22, 4142–4157. [Abstract] [Google Scholar]
- Zhang, C. , Zhang, H. , Zhang, M. , Lin, C. , Wang, H. , Yao, J. , Wei, Q. et al. (2019) OSBPL2 deficiency upregulate SQLE expression increasing intracellular cholesterol and cholesteryl ester by AMPK/SP1 and SREBF2 signaling pathway. Exp. Cell Res. 383, 111512. [Abstract] [Google Scholar]
- Zhang, H. , Zhu, J. , Gong, Z. and Zhu, J.K. (2022) Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. [Abstract] [Google Scholar]
- Zhang, W.B. , Tang, Y. , Hu, Y.L. , Yang, Y.H. , Cai, J.J. , Liu, H.L. , Zhang, C.Y. et al. (2021) Arabidopsis NF‐YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light‐regulated hypocotyl elongation. Plant Cell 33, 2360–2374. [Abstract] [Google Scholar]
- Zhou, Y.Y. , Zhang, Y. , Wang, X.W. , Han, X. , An, Y. , Lin, S.W. , Shen, C. et al. (2020) Root‐specific NF‐Y family transcription factor, PdNF‐YB21, positively regulates root growth and drought resistance by abscisic acid‐mediated indoylacetic acid transport in Populus. New Phytol. 227, 407–426. [Abstract] [Google Scholar]
- Zhu, J.K. (2016) abiotic stress signaling and responses in plants. Cell 167, 313–324. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1111/pbi.14349
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/pbi.14349
Citations & impact
Impact metrics
Article citations
Association between cMIND diet and hypertension among older adults in China: a nationwide survey.
Aging Clin Exp Res, 36(1):182, 05 Sep 2024
Cited by: 0 articles | PMID: 39235675 | PMCID: PMC11377468
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
GmANKTM21 Positively Regulates Drought Tolerance and Enhanced Stomatal Response through the MAPK Signaling Pathway in Soybean.
Int J Mol Sci, 25(13):6972, 26 Jun 2024
Cited by: 0 articles | PMID: 39000082
GmWRKY17-mediated transcriptional regulation of GmDREB1D and GmABA2 controls drought tolerance in soybean.
Plant Mol Biol, 113(4-5):157-170, 16 Nov 2023
Cited by: 4 articles | PMID: 37973764
Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants.
BMC Plant Biol, 20(1):123, 20 Mar 2020
Cited by: 22 articles | PMID: 32192425 | PMCID: PMC7082914
Biological Networks Underlying Abiotic Stress Tolerance in Temperate Crops--A Proteomic Perspective.
Int J Mol Sci, 16(9):20913-20942, 01 Sep 2015
Cited by: 54 articles | PMID: 26340626 | PMCID: PMC4613235
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Beijing Natural Science Foundation (1)
Grant ID: 6234044
China Postdoctoral Science Foundation (1)
Grant ID: 2022M720495
Hainan Seed Industry Laboratory (1)
Grant ID: B21HJ0215
Key Research and Development Program of Hainan Province (1)
Grant ID: ZDYF2022XDNY135
National Key Research and Development Program of China (1)
Grant ID: 2022YFF1001600
National Natural Science Foundation of China (2)
Grant ID: 32372131
Grant ID: 31900744
Natural Science Foundation of Beijing Municipality (1)
Grant ID: 6234044

 1
,
6
and
1
,
6
and 


