Abstract
Backgruound
This study aimed to develop a diabetic kidney disease (DKD) prediction model using long short term memory (LSTM) neural network and evaluate its performance using accuracy, precision, recall, and area under the curve (AUC) of the receiver operating characteristic (ROC) curve.Methods
The study identified DKD risk factors through literature review and physician focus group, and collected 7 years of data from 6,040 type 2 diabetes mellitus patients based on the risk factors. Pytorch was used to build the LSTM neural network, with 70% of the data used for training and the other 30% for testing. Three models were established to examine the impact of glycosylated hemoglobin (HbA1c), systolic blood pressure (SBP), and pulse pressure (PP) variabilities on the model's performance.Results
The developed model achieved an accuracy of 83% and an AUC of 0.83. When the risk factor of HbA1c variability, SBP variability, or PP variability was removed one by one, the accuracy of each model was significantly lower than that of the optimal model, with an accuracy of 78% (P<0.001), 79% (P<0.001), and 81% (P<0.001), respectively. The AUC of ROC was also significantly lower for each model, with values of 0.72 (P<0.001), 0.75 (P<0.001), and 0.77 (P<0.05).Conclusion
The developed DKD risk predictive model using LSTM neural networks demonstrated high accuracy and AUC value. When HbA1c, SBP, and PP variabilities were added to the model as featured characteristics, the model's performance was greatly improved.Free full text

Construction of Risk Prediction Model of Type 2 Diabetic Kidney Disease Based on Deep Learning
Abstract
Background
This study aimed to develop a diabetic kidney disease (DKD) prediction model using long short term memory (LSTM) neural network and evaluate its performance using accuracy, precision, recall, and area under the curve (AUC) of the receiver operating characteristic (ROC) curve.
Methods
The study identified DKD risk factors through literature review and physician focus group, and collected 7 years of data from 6,040 type 2 diabetes mellitus patients based on the risk factors. Pytorch was used to build the LSTM neural network, with 70% of the data used for training and the other 30% for testing. Three models were established to examine the impact of glycosylated hemoglobin (HbA1c), systolic blood pressure (SBP), and pulse pressure (PP) variabilities on the model’s performance.
Results
The developed model achieved an accuracy of 83% and an AUC of 0.83. When the risk factor of HbA1c variability, SBP variability, or PP variability was removed one by one, the accuracy of each model was significantly lower than that of the optimal model, with an accuracy of 78% (P<0.001), 79% (P<0.001), and 81% (P<0.001), respectively. The AUC of ROC was also significantly lower for each model, with values of 0.72 (P<0.001), 0.75 (P<0.001), and 0.77 (P<0.05).
Conclusion
The developed DKD risk predictive model using LSTM neural networks demonstrated high accuracy and AUC value. When HbA1c, SBP, and PP variabilities were added to the model as featured characteristics, the model’s performance was greatly improved.
INTRODUCTION
Diabetic kidney disease (DKD) is one of the most common complications of diabetes. The prevalence of DKD in China is about 20% to 40%, and it has become the second cause of endstage renal disease in China, which seriously affects the quality of life in patients with diabetes [1]. At the same time, the treatment of DKD brings a heavy economic burden to patients, their families and the whole society [2]. Clinically, the early diagnosis of DKD is usually based on the level of microalbuminuria. However, once the diagnosis of DKD is made, the kidney has been irreversibly damaged. About one-third of the patients’ condition will progress and deteriorate even if active intervention is carried out [3]. If preventive intervention was carried out for all diabetes patients, it would consume huge human resource and medical expenses. Considering that the risk of DKD in some diabetes patients is not high, to save medical expenses, it is very important to screen out the diabetes patients with high risk of DKD for future preventive intervention, therefore, a smart screen tool is needed.
Scholars used statistical methods such as meta-analysis and regressions to construct risk predictive models of DKDs [4,5]. However, the scoring systems of these models were roughly developed by researchers, and furthermore, they predict the risk of DKD based on the baseline data, without considering the variability through years, resulting in lower accuracy. We used data from Taiwan externally validated a predictive formula published on diabetes care [4], and found that it’s accuracy, precision, and recall were all low (60.9%, 43.2%%, and 53.0%, respectively) [6], indicating that the performance of the formula was not ideal.
Conventional machine learning methods, such as random forest, decision tree, and support vector machine (SVM), were used to develop DKD predictive models for type 2 diabetes mellitus (T2DM) patients, found that the performance of the models based on machine learning technology is obviously superior to the statistical regression based formulas [7,8]. However, these shallow artificial neural networks have limited speed and room for data training. It has to manually select and adjust the features, and are prone to over-fitting phenomenon [9], which show a high level of accuracy/precision in the process of model training, but the accuracy/precision is much lower during the testing process.
In recent years, the development of deep learning technology brings new ideas for accurate prediction of DKD. It can explore the deeper features of data, which has attracted the interest of scholars in research and clinical fields [10]. In the medical field, deep learning techniques can be used to predict the risk of disease, including breast cancer risk prediction [11-13], cardiovascular disease risk [14-16], and risk of acute respiratory disease events and death in smoking patients [17]. In imaging diagnosis, it has been used in the diagnosis and grading of diabetes retinopathy [18-21] and the diagnosis of pulmonary nodules [22]. A large number of studies have confirmed that the prediction and diagnosis tools based on deep learning technology has high accuracy, fast speed, and greatly saves labor cost, showing a strong application prospect [23]. As a type of deep learning technique, long short term memory (LSTM) networks with memory and timing, has unique advantages in fully considering the impact of previous data on subsequent outcomes [24-26].
At present, there is only one study from Japan that uses deep learning technology to build a DKD predictive model for T2DM. They extracted raw features from the previous 6 months as the reference period and selected 24 factors to find time series patterns relating to 6-month DKD aggravation, with predictive accuracy of 71% [27]. A 6-month was rather too short, and to improve the performance of predictive model, data with longer duration of follow-up is needed.
Previous studies have confirmed that the variability of glycosylated hemoglobin (HbA1c), systolic blood pressure (SBP) and pulse pressure (PP) is the key influencing factor of the development and development of DKD [28-30]. However, most current studies did not consider the influence of variability parameters on the outcome when predicting the risk of DKD.
This study intends to develop a deep learning based model to predict risks of DKD in patients with diabetes, and evaluate it’s accuracy, precision and area under the curve (AUC) in predicting the DKD risk. We would also analyze the impact of the variability of HbA1c, SBP and PP on the overall performance of the model.
METHODS
Screening of predictive risk factors
Literature review
A literature review group was formed to identify the risk factors for DKD. We searched the databases of PubMed, EBSCO, Web of Science, and Chinese database (CNKI, VIP, and Wanfang), using key words “diabetic kidney disease”/“diabetic nephropathy” and “risk factor”/“predictive factor”/“associated factors”/ “predictor/”. Then a systematical review were produced.
Focus group of physicians
The risk factors identified from the literature were reviewed by a focus group of five endocrinologists and four nephrologists. The session lasted 95 minutes. The PI facilitated the session using a semi-structured discussion guide. A research assistant took notes and record responses. The physicians were presented with a handout containing the risk factors and asked to (1) rate the importance of each risk factor on a five-point scale, from 1 (very unimportant) to 5 (very important); (2) suggest additional risk factors considered important for DKD. We then develop a list of risk factors for DKD, which was reviewed by a senior endocrinologist and a senior nephrologist.
Data source for developing the DKD risk model
The data were from Lee’s United Clinic Taiwan. Lee’s United Clinic comprised of six clinics providing multidisciplinary care for patients with diabetes. Taiwan Health Insurance Plan supports four annual follow-up visits along with access to medications, diabetes supplies, diabetes self-management education clinician visitation and primary/secondary prevention screening to patients living with diabetes.
The T2DM patients, aged ≥18 years, with urinary albumin creatinine ratio (UACR) <30 mg/g and estimated glomerular filtration rate (eGFR) ≥60 mL/min/1.73 m2, and who regularly followed up at clinics at least for 7 years were eligible for inclusion. The patients who were diagnosed with DKD at baseline and during the initial 2-year follow-up, with important data missed, with unclear pre-post relationship between nephropathy and diabetes, and fewer than two visits per year were excluded.
The Diagnostic criteria for DKD, following Chinese Clinical Guideline for Prevention and Treatment of Diabetes Nephropathy with eGFR <60 mL/min/1.73 m2 and/or UACR ≥30 mg/g (for more than 3 months or longer) [31].
Data collection
Ideally, data should include the risk factors identified from literature review and focus group, but cystatin C were not checked during the follow-up, so it was not included.
Demographic data: Age, gender, height, weight, smoking, alcohol consumption, marital status, diabetes duration, family history, education level, yearly income, medication history, and comorbidities (coronary heart disease, stroke, diabetes retinopathy, diabetes foot, and non-alcoholic fatty liver disease, etc.).
Clinical data: Following data were collected at least twice a year: body mass index (BMI), waist-hip-ratio, SBP, diastolic blood pressure, eGFR, UACR, urine protein, serum creatinine (Scr), fasting plasma glucose, fasting insulin, HbA1c, 2-hour postprandial glucose after meals, homeostasis model assessment of β-cell function, homeostasis model assessment of insulin resistance, triglyceride, total cholesterol, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol (HDLC), and variability of HbA1c, SBP, and PP during follow-up. Variability of HbA1c, SBP, and PP was defined as standard deviation (SD) from the average levels of HbA1c, SBP, and PP by calculating average values of HbA1c, SBP, and PP measurements and their SDs for each patient during the entire study period [32].
Data processing and dividing
We design the LSTM neural networks using 2 years data to predict the risk in the next 5 years, so we used 7 consecutive years data to develop the model. We process the data as following steps: (1) screen out the patients who did not develop DKD in 7 years follow-up, and marked as 0; (2) screen out the patients without DKD in the first 2 years, but developed DKD in the next 5 years, and marked as 1. Then the data were divided into two sets: 70% in the training set, and 30% in the testing set.
LSTM neural networks building
Pytorch language was used to build the networks. After the LSTM neural networks was set, the data of training set was inputted to the networks, training the LSTM networks, enabling the networks learning the features of the data, and comparing the output results with marked result, as previously described (0 represents did not developed DKD and 1 represents developed DKD). The difference values were propagated back to achieve continuous corrections and adjustments of the neural network parameters, and finally the network model achieves the optimum status. After the model is trained to the optimal state by using the training set data, the data in testing set were used to test the model’s performance.
Dropout was used to prevent the over-fitting phenomenon (the model appears to have a high accuracy in the training process, but have a much lower predictive accuracy in the test process when use the testing data set). We used dropout to solve the problem, and 20% of the neural nerves were randomly removed. The construction process of the structural design of the LSTM neural network prediction model constructed in this study is illustrated in Table 1.
Table 1.
Process of the LSTM prediction model
| The construction process is based on the structural design of the LSTM neural network prediction model. |
|---|
| class lstm (nn.Module): #define the network structure |
 def_init_ (self,input_size,hidden_size,output_size,num_layer): def_init_ (self,input_size,hidden_size,output_size,num_layer): |
  super (lstm,self)._init_() super (lstm,self)._init_() |
  self.layer1=nn.LSTM (input_size,hidden_size,num_layer) self.layer1=nn.LSTM (input_size,hidden_size,num_layer) |
  self.layer2=nn.Linear (hidden_size,output_size) self.layer2=nn.Linear (hidden_size,output_size) |
  self.dropout=nn.Dropout (p=0) self.dropout=nn.Dropout (p=0) |
 def forward (self,x): #define network output information def forward (self,x): #define network output information |
  x,_=self.layer1(x) x,_=self.layer1(x) |
  s,b,h=x.size() s,b,h=x.size() |
  x=x.view(s*b,h) #convert lstm’s 3D output to 2D sequences x=x.view(s*b,h) #convert lstm’s 3D output to 2D sequences |
  x=self.layer2(x) x=self.layer2(x) |
  x=self.dropout(x) x=self.dropout(x) |
  x=x.view(s,b,-1) x=x.view(s,b,-1) |
  return F.sigmoid(x) return F.sigmoid(x) |
| model=lstm(38,16,1,1) #define of network parameter structure |
| criterion=nn.MSELoss() #define of loss function |
| optimizer=torch.optim.Adam(model.parameters(),lr=1e-3) #define optimizer |
LSTM, long short term memory.
For disease risk prediction models, the smaller the difference between the predicted risk value and the actual disease risk, the more the more prediction accuracy. The loss function is used to measure the gap between the predicted value and the actual value. The most commonly used loss function, mean squared error (MSE), was in this study to calculate the absolute error between the real value and the estimated value. The smaller the value of MSE, the better the prediction value of the model. The functional expression is as follows (1):
Network training and output results
In this study, the network was trained with a learning rate of 10-3 epoch, 5,000; loss function, MSE; mini-batch size, 50; optimizer, adam optimization algorithm. Given that this model is used to predict the future risk of DKD in T2DM patients, its prediction result should be the probability value distributed in the (0–1) interval. We then designed the output of the neural network, which needs to be constrained. In this study, a sigmoid function is employed, which is expressed as follows (2):
Through this function, any real number of the output of the network in this study would be mapped to the interval of 0–1, that is, the output result can express the probability value of the network for a certain classification.
Evaluation of the model performance
The predictive accuracy, precision, recall and the AUC of the receiver operating characteristic (ROC) curve were used to evaluate the performance of the model. AUC ≤0.5 means the model does not have the predictive value; AUC ≤0.7, AUC >0.8, AUC >0.9; represent the performance of the model is low, good, and great, respectively. The overall performance of the pre-trained models is evaluated using four criteria (TP, true positive; TN, true negative; FP, false positive; and FN, false negative). The system’s performance is assessed by using the Equations 3 to 5:
Statistical analysis
After all data were collected and processed, the data were finally entered into Excel 2016 software by double check and the related database was established. Using SPSS version 22.0 software (IBM Co., Armonk, NY, USA) to determine the correlation coefficient of each parameter, MATLAB simulation software and pytorch language were used for data extraction, data set processing, LSTM neural network design, network training and result output. ROC curve was used to calculate the AUC, indicating the discrimination of the model. The chi-squared test was used for inter-group comparisons of accuracy, precision, and recall; and the z-test was used for AUC comparisons. There is a statistical difference when the P value is less than 0.05.
Institutional Review Board statement
This study involves human participants and was approved by Human Trial Review Committee of Taiwan Lee’s United Clinic (No.14-055-B2). Participants gave informed consent to participate in the study before taking part.
RESULTS
DKD risk predictors
This study analyzed and summarized the risk factors of DKD based on literature review and physician focus group. After the focus group interviews, we removed some attributes such as “tea drinking,” “high fat diet,” “sweet food” reported from single Chinese paper, and “genes.” Of course, genetic factors are very important for DKD development, but it is expensive and not commonly checked. The attributes must included, but not limited in data collection were as follows: older age, longer duration of diabetes, family history of DKD, smoking, diabetes retinopathy, mean HbA1c, hypertension, without regular exercise, high blood uric acid, insulin resistance, high BMI, SBP, HDL-C, triacylglycerol (TG), UACR, and cystatin C, meat intake, number of oral hypoglycemic drugs. The variability of HbA1c, SBP, and PP were also the predictors of DKD (Table 2). The important predictors the machine selected were (from higher weight size to lower weight size): Scr, age, HDL-C, HbA1c, PP, SBP, TG, variabilities of HbA1c, SBP, and PP.
Table 2.
Major attributes for patients
| Variable | Training set (n=4,228) | Testing set (n=1,812) | P value |
|---|---|---|---|
| Age, yr | 54.31±10.98 | 54.90±11.49 | 0.636 |
| Smoking | 1,289 (30.5) | 545 (30.1) | 0.831 |
| Hypertension | 875 (20.7) | 168 (20.0) | 0.753 |
| Duration of diabetes, yr | 9.65±6.59 | 8.71±5.92 | 0.411 |
| Family history of DKD | 393 (9.3) | 185 (10.2) | 0.475 |
| Diabetes retinopathy | 799 (18.9) | 368 (20.3) | 0.082 |
| Exercise | 3,412 (80.7) | 1,493 (82.4) | 0.476 |
| Meat intake, g/day | 190.13±24.31 | 188.79±30.42 | 0.357 |
| No. of oral hypoglycemic drugs | 2 (0–3) | 2 (0–3) | 0.821 |
| BMI, kg/m2 | 23.88±3.43 | 24.51±3.32 | 0.423 |
| HbA1c, % | 9.38±1.87 | 8.97±1.82 | 0.114 |
| Blood uric acid, μmol/L | 341.82±99.93 | 349.55±104.66 | 0.370 |
| SBP, mm Hg | 129.43±15.16 | 132.00±10.56 | 0.401 |
| HOMA2-IR | 2.55 (1.80–3.65) | 2.61 (1.83–3.46) | 0.454 |
| TG, mmol/L | 2.09 (1.95–2.22) | 1.96 (1.82–2.22) | 0.094 |
| HDL-C, mmol/L | 1.34±0.34 | 1.42±0.33 | 0.463 |
| UACR, mg/g | 13.86±7.35 | 14.08±9.95 | 0.097 |
| HbA1c variability | 1.36±0.83 | 1.25±0.59 | 0.379 |
| SBP variability | 11.36±5.65 | 11.03±6.41 | 0.081 |
| PP variability | 6.81±2.21 | 7.63±2.10 | 0.122 |
Values are presented as mean±standard deviation, number (%), or median (interquartile range). HOMA2 Calculator software (https://www.dtu.ox.ac.uk/homacalculator/) was used to calculate homeostatic model assessment of β-cell function and insulin resistance (HOMA2-β and HOMA2-IR, respectively).
DKD, diabetic kidney disease; BMI, body mass index; HbA1c, glycosylated hemoglobin; SBP, systolic blood pressure; HOMA2-IR, homeostasis model assessment of insulin resistance; TG, triacylglycerol; HDL-C, high-density lipoprotein cholesterol; UACR, urinary albumin/creatinine ratio; PP, pulse pressure.
Performance comparison between LSTM and SVM models
Based on the 7-year consecutive data of Lee’s United Clinic in Taiwan, the model was trained to use the first 2 years data to predict the DKD risk in next 5 years. According to the inclusion and exclusion criteria, a total of 6,040 T2DM patients were included in the study, and 30% (1,812 cases) of which were divided into the test set. Diabetes patients (n=1,268) did not develop DKD and 544 patients developed DKD.
The prediction performance of LSTM model were: 411 TP, 120 FP, 133 FN, and 1,148 TN, while for the SVM model, 283 TP, 163 FP, 261 FN, and 1,105 TN. Therefore, in this study, the precision of the LSTM model was 411/(411+120)×100%=77%, while the precision of the prediction model constructed based on SVM algorithm was 283/(283+163)×100%=63%, and the precision of the LSTM model was higher (F=128.521, P<0.001). The accuracy of LSTM model was (411+1,148)/(411+1,148+120+133)×100%=86%, much higher than that of SVM model: (283+1,105)/(283+1,105+163+261)×100%=76% (F=71.785, P<0.001). The recall for LSTM model was 411/(411+133)×100%=76%, much better than that of SVM model: 283/(283+261)×100%=52% (F=226.598, P<0.001). Thus, the overall performance of LSTM model superior to that of SVM model (Fig. 1).
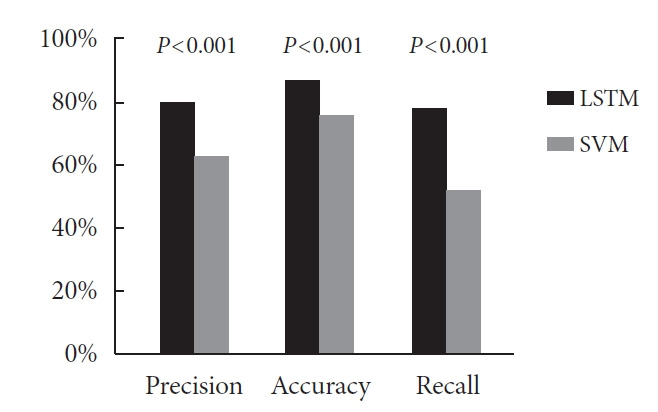
Comparison of precision, accuracy and recall of models constructed by long short term memory (LSTM) and support vector machine (SVM).
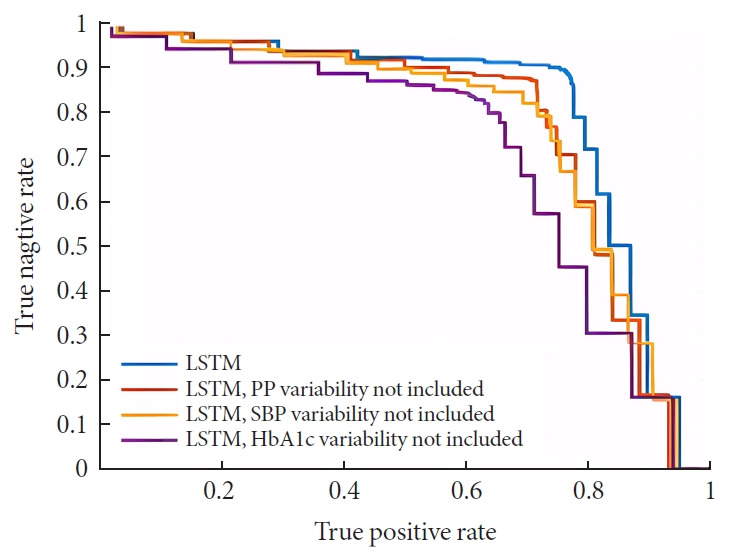
In this study, the optimal cutoff point and AUC for LSTM model were 0.55 and 0.83 and which of the SVM model were 0.75 and 0.73 indicating that the performance of the LSTM model was superior to that of SVM algorithm. The ROC curves of the two are shown in Fig. 2.
Impact of variability parameters on the DKD prediction
To verify the effects of the variability of HbA1c, SBP, and PP on the performance of the model, three neural network models were constructed, with each model excluded HbA1c variability, SBP variability, or PP variability, were compared with the optimal prediction model which include all the variability parameters (Table 3).
Table 3.
Impact of variability parameters on diabetic kidney disease prediction
| HbA1c variability not included | SBP variability not included | PP variability not included | Optimal LSTM | |
|---|---|---|---|---|
| Precision | 0.64a | 0.65a | 0.70a | 0.77 |
| Accuracy | 0.78a | 0.79a | 0.81a | 0.86 |
| Recall | 0.61a | 0.65a | 0.67a | 0.76 |
HbA1c, glycosylated hemoglobin; SBP, systolic blood pressure; PP, pulse pressure; LSTM, long short term memory.
Comparison of accuracy and AUC area of DKD models
The results show that the best cutoff point of the optimal LSTM prediction model with all variability parameters is 0.55, and the AUC value is 0.83. When we remove the risk factor of HbA1c, SBP, or PP variability one by one, the accuracy of each model were 78% (P<0.001), 79% (P<0.001) and 81% (P<0.001), and the best cutoff point for the DKD risk predictive models were 0.60, 0.55, and 0.60, respectively, with AUC values of 0.72, 0.75, and 0.77, respectively (Fig. 3). The comparison between the model included all HbA1c, SBP, or PP variabilities with other models, which missed one of three variabilities, that the AUC of LSTM model with all variability parameters was significantly better than that of the other three prediction models, all P< 0.05, indicating that the inclusion of variability parameters such as HbA1c, SBP, and PP can significantly increase the overall performance of the model.
DISCUSSION
In this study, we used LSTM neural network developed DKD risk prediction model for diabetes patients, with high accuracy and AUC. At the same time, HbA1c variability, SBP variability and PP variability were added as new risk factors of DKD to develop the DKD risk prediction model and improved the model’s performance.
At present, only one study from Japan successfully constructed the DKD risk prediction model using deep learning technology, with the accuracy of 71% and AUC of 0.743 [27]. The accuracy and AUC value of our study were higher. The following reasons may explain the difference. First, the different deep learning technology made the difference. The Japanese study used convolutional neural network while our study used LSTM neural network, and the latter has unique advantages over convolutional neural network in DKD risk model building, because LSTM has memory and timing concept, fully considering the impact of previous metabolic control on the occurrence of DKD subsequent years [33]. Second, before developing the model, we not only identified the risk factors through systematical reviews physician specialists focus group, but also included HbA1c, SBP, and PP variability as important characteristic parameters in the model for training. Compared with our study, the factors included in this Japanese study were only routine indexes of serum and urine tests of patients in the previous 6 months, such as blood glucose, HbA1c, and blood lipid, and did not fully consider the important influence of fluctuation of HbA1c and blood pressure on the development of DKD. Therefore, the feature parameters included in our study are more comprehensive and have more prediction value, which can improve the overall performance of the prediction model to a certain extent. Finally, our study used 7 years consecutive data while the Japanese study use 6 months data. Since the process of developing DKD is relatively long, involving years, so 6-month of data is not long enough to let DKD happen.
It was also demonstrated that in our study, when HbA1c, SBP, and PP variability were added as risk factors to the characteristic parameters to develop the model, the overall performance was significantly improved. Some studied demonstrated that HbA1c, SBP, and PP variabilities impacted the development of DKD in patients with T2DM [34], but the mechanism is still unclear, which may closely related to oxidative stress, endothelial dysfunction and increased inflammatory factors caused by the fluctuation of HbA1c, SBP, and PP. Oxidative stress, endothelial dysfunction, and increased inflammatory factors are key pathogenic factors leading to kidney tissue damage and impaired renal function leading to the development of DKD [35-39]. Further prospective studies are needed to elucidate the mechanism on the fluctuation of HbA1c, SBP, and PP impacting the development of DKD.
This study also has several limitations. First, the study subjects in this study were from Taiwan, and the factors impact DKD development may be partially influenced by the fact that there is a degree of variation in different regions. Second, the data used to train the model and test the performance of the model were from the same source. They were all from Taiwan Lee’s United Clinic, and this may affect the generalization of the model. So in the future, external validation is needed. Third, the overall performance of the model still has room for improvement; therefore, continues adjustment is warranted.
In conclusion, our study used LSTM neural networks to develop the DKD prediction model with higher performance, as indicated by precision, accuracy, recall, and AUC value. When variability of HbA1c, SBP, and PP was added to the model as featured characteristics, the performance of the model was greatly improved, which suggests that minimizing the variability of HbA1c, SBP, and PP would help to prevent DKD in patients with diabetes. The LSTM based DKD prediction model brings a new idea to screen diabetes patients with high risk of DKD, enabling precise early preventative interventions, so as to prevent DKD economically. The results not only urge patients and health care professionals to consider the important impact of variability of metabolic parameters on DKD development, but also provide new ideas and methods for developing predicting models for other chronic diseases such as diabetic retinopathy.
Acknowledgments
The authors sincerely thank all participants for their time and effort in this study.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Conception or design: Q.L.
Acquisition, analysis, or interpretation of data: C.Y., F.T., Z.G., W.W., F.B., H.L., Y.L., Q.L.
Drafting the work or revising: C.Y., F.T., J.D.M., Q.L.
Final approval of the manuscript: all authors.
FUNDING
This study was supported by National Key R&D program of China (2021YFE0204800) and Key R&D Program of Hainan Province (ZDYF2021SHFZ236).
REFERENCES
Articles from Diabetes & Metabolism Journal are provided here courtesy of Korean Diabetes Association
Full text links
Read article at publisher's site: https://doi.org/10.4093/dmj.2023.0033
Read article for free, from open access legal sources, via Unpaywall:
https://www.e-dmj.org/upload/pdf/dmj-2023-0033.pdf
Citations & impact
Impact metrics
Article citations
Construction of Risk Prediction Model of Type 2 Diabetic Kidney Disease Based on Deep Learning (Diabetes Metab J 2024;48:771-9).
Diabetes Metab J, 48(5):1008-1011, 12 Sep 2024
Cited by: 0 articles | PMID: 39313234 | PMCID: PMC11449815
Construction of Risk Prediction Model of Type 2 Diabetic Kidney Disease Based on Deep Learning (Diabetes Metab J 2024;48:771-9).
Diabetes Metab J, 48(5):1003-1004, 12 Sep 2024
Cited by: 0 articles | PMID: 39313232 | PMCID: PMC11449817
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[Blood Lipid Indicators and Different Clinical Classifications of Dyslipidemia and Diabetic Kidney Disease: Correlation and Predictive Value].
Sichuan Da Xue Xue Bao Yi Xue Ban, 54(5):1013-1018, 01 Sep 2023
Cited by: 1 article | PMID: 37866961 | PMCID: PMC10579073
Predicting diabetic kidney disease for type 2 diabetes mellitus by machine learning in the real world: a multicenter retrospective study.
Front Endocrinol (Lausanne), 14:1184190, 04 Jul 2023
Cited by: 9 articles | PMID: 37469989
Developing screening tools to estimate the risk of diabetic kidney disease in patients with type 2 diabetes mellitus.
Technol Health Care, 32(3):1807-1818, 01 Jan 2024
Cited by: 0 articles | PMID: 37980579
[Development of a prediction model for the incidence of type 2 diabetic kidney disease and its application based on a regional health data platform].
Zhonghua Liu Xing Bing Xue Za Zhi, 45(10):1426-1432, 01 Oct 2024
Cited by: 0 articles | PMID: 39444128
Funding
Funders who supported this work.
Key R&D Program of Hainan Province (1)
Grant ID: ZDYF2021SHFZ236
Key R&D Program of Hainan Province (1)
Grant ID: ZDYF2021SHFZ236
National Key R&D program of China (1)
Grant ID: 2021YFE0204800
National Key R&D program of China (1)
Grant ID: 2021YFE0204800

 8
8