Abstract
Free full text

Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response
Associated Data
Abstract
Ethylene plays its essential roles in plant development, growth, and defense responses by controlling the transcriptional reprograming, in which EIN2-C–directed regulation of histone acetylation is the first key step for chromatin to perceive ethylene signaling. But how the nuclear acetyl coenzyme A (acetyl CoA) is produced to ensure the ethylene-mediated histone acetylation is unknown. Here we report that ethylene triggers the accumulation of the pyruvate dehydrogenase complex (PDC) in the nucleus to synthesize nuclear acetyl CoA to regulate ethylene response. PDC is identified as an EIN2-C nuclear partner, and ethylene triggers its nuclear accumulation. Mutations in PDC lead to an ethylene hyposensitivity that results from the reduction of histone acetylation and transcription activation. Enzymatically active nuclear PDC synthesizes nuclear acetyl CoA for EIN2-C–directed histone acetylation and transcription regulation. These findings uncover a mechanism by which PDC-EIN2 converges the mitochondrial enzyme-mediated nuclear acetyl CoA synthesis with epigenetic and transcriptional regulation for plant hormone response.
INTRODUCTION
Ethylene is a pivotal plant hormone that plays pleiotropic roles in plant growth, development, stress response, and defense response to pathogens by controlling downstream gene transcription, protein translation and posttranslational modifications, mitochondrial retrograde signaling, chromatin remodeling, and epigenetic regulation of gene expression (1–14). Ethylene signal is perceived by ethylene receptors embedded on the endoplasmic reticulum (ER) membrane (15, 16). In the absence of ethylene, both the ethylene receptors and CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), an ER membrane–associated kinase that directly interacts with one of ethylene receptors ETHYLENE RESPONSE1 (ETR1), are activated (17, 18). CTR1 phosphorylates ETHYLENE INSENSTIVE 2 (EIN2), the essential positive regulator of ethylene signaling, at its C-terminal end (EIN2-C), leading to the repression of the EIN2 activity (19, 20). Without ethylene signaling activation, EIN2 is localized to the ER membrane at which it interacts with two F-box proteins EIN2 TARGETING PROTEIN 1/2 that mediate its protein degradation via the ubiquitin-proteasome pathway (21). Upon the perception of ethylene, both ETR1 and CTR1 are inactivated; the EIN2-C is dephosphorylated through an unknown mechanism (20). The dephosphorylated EIN2-C is cleaved and translocated into both the nucleus and the P-body (20, 22, 23). In the P-body, EIN2-C mediates the translational repression of two F-box proteins EIN3-BINDING F BOX PROTEIN 1/2 to promote the protein accumulation of ETHYLENE INSENSITIVE 3 (EIN3), the key transcription activator that is sufficient and necessary for activation of all ethylene-response genes (6, 23, 24). In the nucleus, EIN2-C mediates the direct regulation of histone acetylation of histone H3 lysine 14 (H3K14) and lysine 23 (H3K23) via histone binding protein EIN2 NUCLEAR ASSOCIATED PROTEIN 1/2, leading to an EIN3-dependent transcriptional regulation for ethylene response (25–27).
Epigenetic regulation of gene expression plays critical roles in various plant developmental processes and stress response (28–32). Histone acetylation promotes the relaxation of nucleosome between wrapped DNA and histone octamer by neutralizing the positive charges of lysine residues in histones, which is crucial for all DNA-based processes, including DNA replication and gene transcription (33, 34). Histone acetyltransferases (HATs) transfer acetyl group from acetyl coenzyme A (acetyl CoA) to conserved lysine residues on histone N-terminal tails to acetylate histones. In eukaryotes, the biosynthesis of acetyl CoA is thought to occur in the subcellular compartment where it is required because of its membrane impermeability and high instability due to the high-energy thioester bond that joins the acetyl and CoA groups (35). The availability and abundance of acetyl CoA are therefore crucial for HAT enzymatic activity and histone acetylation. For most HATs, their Michaelis constants (Kms), the concentration of substrate which permits the enzyme to achieve half Vmax, lie in the range within or greater than the cellular acetyl CoA concentrations to sense and respond to the fluctuation of acetyl CoA production (36). Therefore, the levels of histone acetylation are often linked with the availability of acetyl CoA. Such metabolic regulation of histone acetylation by nuclear synthesis of acetyl CoA has been reported to govern many major biological events including cell proliferation, cell fate determination, and environmental response (37–43). For instance, the nuclear-localized pyruvate dehydrogenases (PDHs) contribute to epigenetic remodeling in cell cycle G1 to S phase progression and embryonic genome activation by nuclear acetyl CoA synthesis in mammal (40, 41). Yet, the biological significance of nuclear localization of PDC remains a puzzle waiting to be solved. Likewise, the relationship between the nuclear acetyl CoA and histone acetylation in plant hormone response is completely unknown.
In this study, we report that functional pyruvate dehydrogenase complex (PDC) can translocate from the mitochondria to the nucleus in the presence of ethylene, generating a nuclear pool of acetyl CoA from pyruvate for the EIN2-directed acetylation of core histones for transcriptional regulation in the ethylene response. In the presence of ethylene, the PDC complex translocates to the nucleus where it interacts with EIN2-C. Mutations in PDC lead to a decreased ethylene response both at genetic level and transcriptional level. Chromatin immunoprecipitation sequencing (ChIP-seq) assay reveals that ethylene-induced elevation of histone acetylation is reduced in the PDC mutants. Biochemical assay combined with liquid chromatography–mass spectrometry (LC-MS) demonstrates that PDC regulates the nuclear acetyl CoA concentration, and ethylene-induced nuclear translocated PDC is enzymatically active to generate acetyl CoA in the nucleus for histone acetylation, which is necessary for EIN2-C’s function in the histone acetylation regulation. Both genetics and molecular evidence show that EIN2-C is required for the nuclear translocation of PDC in the ethylene response. Together, this study reveals a previously unidentified mechanism by which the PDC complex is translocated to the nucleus where it interacts with EIN2-C to provide acetyl CoA for the elevation of histone acetylation at H3K14 and H3K23 to modulate the transcriptional regulation in the ethylene response.
RESULTS
Nuclear-localized PDC interacts with EIN2-C
By tandem coimmunoprecipitation coupled with mass spectrometry (co-IP/MS) using anti–EIN2-C native antibody and anti–hemagglutinin (HA) antibody sequentially in the nuclear extracts from EIN2-YFP-HA plants treated with 4 hours of ethylene gas, three subunits from PDC, E1-2, E2-2, and E3-2, were pulled down among the top hits (fig. S1, A to C). The direct interactions between EIN2-C and these three PDC subunits identified in co-IP/MS were confirmed by in vitro pull-down assays (Fig. 1A and fig. S1, D to F). In prokaryotes and eukaryotes, PDC consists of three catalytic subunits: a PDH (E1), a dihydrolipoamide transacetylase (E2), and a dihydrolipoamide dehydrogenase (E3), and this complex is canonically localized in the mitochondrial matrix where it catalyzes the conversion from pyruvate to acetyl CoA (44). The Arabidopsis genome encodes three E1 subunits (two E1α and one E1β), three E2 subunits, and two E3 subunits localized in mitochondria (45). The interaction between EIN2-C and E1-2, E2-2, and E3-2 (henceforth referred as E1, E2, and E3) led us to examine whether it is possible that EIN2 is localized to the mitochondria. Colocalization assay of EIN2 with MitoTracker Red, a cell-permeant mitochondria-selective dye, showed that EIN2 and mitochondria were not colocalized either with or without the presence of ethylene (Fig. 1B). Given that PDC complex was pulled down from the nuclear fraction by EIN2-C co-IP/MS, we then decided to confirm whether the interactions between EIN2-C and E1, E2, and E3 occur in the nucleus in vivo. We generated EIN2-YFP-HA/E1-FLAG-BFP, EIN2-YFP-HA/E2-FLAG-BFP, and EIN2-YFP-HA/E3-FLAG-BFP transgenic plants. By using EIN2–yellow fluorescent protein (YFP)–HA or PDC-FLAG-blue fluorescent protein (BFP) as bait, we performed reciprocal in vivo coimmunoprecipitation (co-IP) assays both in the nuclear fractions and in the cytosolic fractions from those 3-day-old etiolated transgenic seedlings treated with or without 4 hours of ethylene gas. The efficiency of nuclear-cytoplasmic fractionation procedure and the purity of cytosolic and nuclear fractions were assessed by multiple marker proteins (fig. S1G). The interactions between EIN2-C and all three PDC subunits were detected only in the nuclear fractions with ethylene treatment, and no interactions were detected from cytosolic fractions regardless of the ethylene treatment (Fig. 1, C to E). Similar results were obtained by using the native EIN2-C antibody for the in vivo co-IP assay in the E1-YFP-HA, E2-FLAG-GFP, and E3-FLAG-GFP transgenic plants (fig. S1, H to J). To further evaluate the interaction between EIN2 and PDC in response to ethylene in the subcellular context, we conducted bimolecular fluorescence complementation (BiFC) assays by transient coexpression of EIN2-nYFP and E1-cYFP, E2-cYFP, and E3-cYFP, respectively, in tobacco epidermal cells. The result showed that EIN2-C physically interacts with PDC subunits only in the nucleus after ethylene treatment (Fig. 1, F to H). Furthermore, we examined the subcellular localization of EIN2-C and PDC subunits in response to ethylene in 3-day-old etiolated seedlings of EIN2-YFP-HA/E1-FLAG-BFP, EIN2-YFP-HA/E2-FLAG-BFP, and EIN2-YFP-HA/E3-FLAG-BFP plants. EIN2-C was colocalized with E1, E2, and E3 in the nucleus, respectively, with the ethylene treatment (fig. S1, K to P). Together, these results provide compelling evidence that PDC can accumulate in the nucleus to interact with EIN2-C in response to ethylene.

(A) In vitro pull-down experiment to validate the interaction between EIN2-C and PDC E1, E2, and E3 subunits. IB, immunoblotting. (B) Subcellular localization of EIN2-YFP fusion protein in Arabidopsis leaf epidermal cells with MitoTracker Red staining without (top) and with (bottom) the presence of 10 μM ACC. Red arrowhead indicates mitochondria signal by MitoTracker staining; yellow arrowhead indicates EIN2-YFP fluorescence signal. Scale bars, 20 μm. (C to E) In vivo co-IP assay to examine the interaction between EIN2-C and E1 (C), E2 (D), and E3 (E) in the indicated transgenic plants. Three-day-old etiolated seedlings carrying both PDC and EIN2 fusion proteins treated with air or 4 hours of ethylene gas were fractionated to isolate cytosol and nuclei for the immunoprecipitation with either Flag-Trap magnetic agarose (DYKDDDDK Fab-Trap) or anti-HA magnetic beads, respectively. The immunoprecipitation with immunoglobulin G (IgG) beads serves as a negative control. Phosphoenolpyruvate carboxylase (PEPC) was used as cytosolic marker protein; cytochrome C (Cyt C) is a mitochondrial marker protein, and BiP is an ER marker to assess nuclear extraction purity. Histone H3 is a loading control for nuclear fractions. (F to H) Confocal microscopy images of BiFC assay showing the interaction between EIN2 and PDC E1, E2, and E3 subunits in the nucleus after ethylene treatment. Agrobacteria containing indicated paired constructs was co-infiltrated into tobacco leaves, and the YFP fluorescence was observed 2 days after infiltration with or without 4 hours of ACC treatment. NLS-BFP was used as nuclear marker. Scale bars, 50 μm.
Ethylene treatment induces the nuclear accumulation of PDC
We have observed the nuclear localization of PDC complex with the presence of ethylene (Fig. 1). To further confirm that the PDC nuclear translocation is induced by ethylene, we examined the nuclear appearance of E1, E2, and E3 proteins over a time series of ethylene gas treatments. E1 had a basal nuclear distribution in the absence of ethylene, but its nuclear levels were notably elevated by ethylene treatment and were positively correlated with the duration of ethylene treatment while E1 cytoplasmic levels showed gradual decrease as the ethylene treatment prolongs (Fig. 2A and fig. S2, A and B). Neither E2 nor E3 proteins were detected in the nucleus in the absence of ethylene, but these two proteins were accumulated in the nuclear fraction after 4 hours of ethylene treatment, and their levels were also positively correlated with the duration of ethylene treatments (Fig. 2, B and C, and fig. S2, C to F). Similar to cytoplasmic E1, we also observed that cytoplasmic E2 and E3 protein levels decrease in response to ethylene (Fig. 2, B and C, and fig. S2, C to F). Notably, the total E1 and E2 subunit protein levels were not altered by the ethylene treatments, but E3 total protein level showed slight elevation after 12-hour ethylene treatment. We then monitored the nuclear localization of PDC over a time series of ethylene treatments in living cells. In the absence of ethylene, a basal level of E1 was observed in the nucleus, but no nuclear E2 and E3 were observed (Fig. 2, D to F, and fig. S2, G to L). Upon ethylene treatment, E1, E2, and E3 accumulated in the nucleus, and their accumulation was positively associated with the duration of ethylene treatment, the cytoplasmic fraction of PDC remains in the mitochondria even with the ethylene treatment (Fig. 2, D to F, and fig. S2, G to R). This provides an additional piece of cellular evidence that ethylene induces PDC nuclear accumulation.
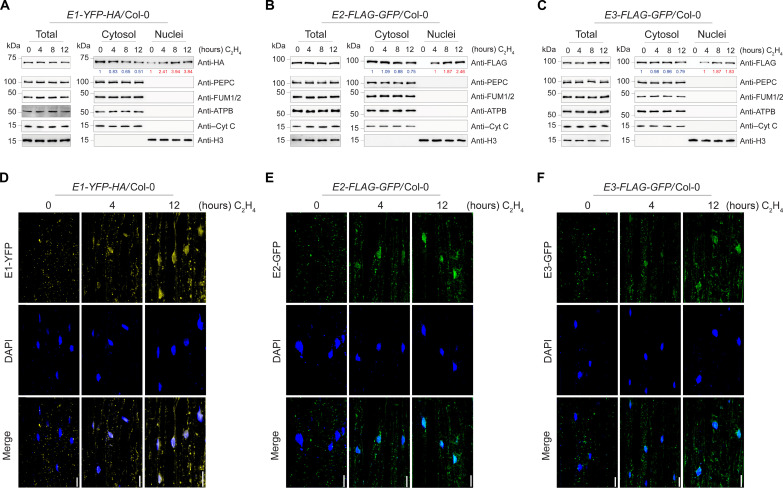
(A to C) Fractionation Western blot to examine the subcellular localization of PDC in E1-YFP-HA (A), E2-FLAG-GFP (B), and E3-FLAG-GFP (C) transgenic plants with time series of ethylene gas treatments. PDC E1 was probed with anti-HA, and E2 and E3 were probed with anti-FLAG in total protein extracts, cytoplasmic fractions, and nuclear fractions. PEPC, ATP synthase beta (ATPB), FUM1/2, Cyt C, and histone H3 were used to assess purities of nuclear and cytosolic fractionations and loading controls. Blue number indicates PDC band intensity that normalized to cytoplasmic PEPC signal. Red number indicates PDC band intensity that normalized to histone H3 signal. (D to F) Confocal microscopy images showing the subcellular localization of E1-YFP-HA (D), E2-FLAG-GFP (E), and E3-FLAG-GFP (F) with time series of ethylene gas treatments. 4′,6-Diamidino-2-phenylindole (DAPI) staining labels nuclei. Scale bars, 20 μm.
PDC subunits are required for ethylene response
Given that the interaction between PDC and EIN2-C occurs in the nucleus in response to ethylene, we decided to investigate the functions of PDC in the ethylene response. We first obtained two transferred DNA (T-DNA) insertion lines for E1 (e1-2-2 and e1-2-4) and two for E3 (e3-2-1 and e3-2-2). Reverse transcription–quantitative polymerase chain reaction (RT-qPCR) assays showed that the expression levels of these two genes were drastically reduced in their T-DNA insertion mutants (fig. S3, A to D). No E2 T-DNA homozygous plants were obtained because of homozygous lethality (46). Using CRISPR-Cas9 mutagenesis, we generated two weak alleles of e2 mutants (e2-2 #17 and e2-2 #215) that were variable (fig. S3, E to G). Phenotypical assay showed that the e2 single mutants, but not e1 nor e3 single mutant, displayed a mild ethylene insensitivity both in roots and in hypocotyls (Fig. 3, A to C). We then generated a variety of their higher-order mutants and examined their ethylene responses (fig. S3, E to G). The mild ethylene-insensitive phenotype of the e2 single mutant was significantly enhanced in e1e2 (e1-2-2 e2-2 #17 and e1-2-4 e2-2 #215) and e2e3 (e2-2 e3-2-1 #99 and e2-2 e3-2-2 #67) double mutants and in e1e2e3 triple mutants (e1-2-2 e2-2 e3-2-1 #167 and e1-2-2 e2-2 e3-2-2 #67) (Fig. 3, D to F). No significant difference was observed between the e1e3 (e1-2-2 e3-2-1 and e1-2-2 e3-2-2) double mutants and e1 or e3 single mutants (Fig. 3, A to F). Notably, e1e2e3 triple mutants exhibit mild growth defects including shortened hypocotyls and roots compared to the wild-type Columbia (Col-0) and other pdc mutants when grown in the dark in the absence of 1-aminocyclopropane-1-carboxylic acid (ACC) (Fig. 3, D to F). The triple mutant also displayed reduced root and vegetative growth under light condition (fig. S3, H and I), suggesting that PDC complex potentially influences plant growth and development. The e1e2, e2e3, and e1e2e3 ethylene-insensitive phenotype and the growth defect of e1e2e3 etiolated seedlings were complemented by introducing proE2:gE2-FLAG-GFP into each mutant background, respectively (fig. S3, J and K). These genetic results demonstrated that PDC is involved in the ethylene response, E2 is the most important subunit genetically, and E1 and E3 enhance the function of E2 in the ethylene response.
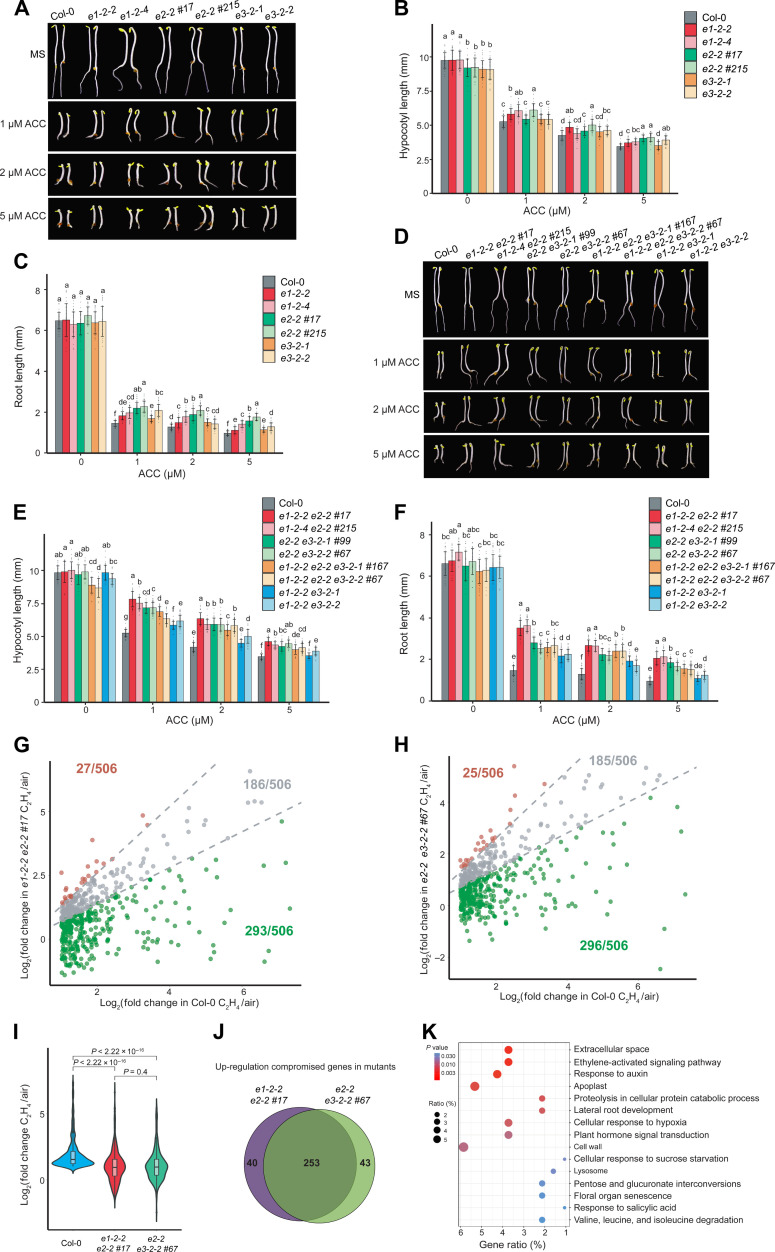
(A) Photographs of pdc single mutants and Col-0 seedlings grown on MS medium containing 0, 1, 2, and 5 μM ACC. (B and C) Measurements of hypocotyl lengths (B) and root lengths (C) of indicated mutants. (D) Representative higher-order PDC mutants and Col-0 grown on MS medium containing 0, 1, 2, and 5 μM ACC. (E and F) Measurements of hypocotyl lengths and root lengths from indicated plants. In (B), (C), (E), and (F), each value is means ± SD of at least 30 seedlings, and different letters indicate significant differences between genotypes [P ≤ 0.05, calculated by a one-way analysis of variance (ANOVA) test followed by Tukey’s post hoc test for multiple comparisons]. (G and H) Scatterplots comparing the log2FC of ethylene up-regulated genes in Col-0 with that in e1-2-2 e2-2 #17 (G) or in e2-2 e3-2-2 #67 (H) in mRNA-seq. Each dot represents a DEG that its transcription level is significantly elevated by ethylene in Col-0 (log2FC > 1, P adjusted < 0.05). In (G) and (H), red dots correspond to DEGs in the elevated group, gray dots to DEGs in the unchanged group, and green dots to DEGs in the compromised group as described in Results. (I) Violin plot of log2FC distributions of ethylene up-regulated genes in the indicated plants. P values were determined by a two-tailed t test. (J) Venn diagram showing the ethylene up-regulation compromised genes in e1-2-2 e2-2 #17 and in e2-2 e3-2-2 #67. (K) Gene ontology (GO) analysis of transcriptionally co-compromised genes in both e1-2-2 e2-2 #17 and in e2-2 e3-2-2 #67. Top 15 GO categories ranked by P values were plotted. Dot size indicates the gene percentage from each GO category of all co-compromised genes (gene ratio %); dot colors indicate the P value of the GO categories.
To further understand the functions of E1, E2, and E3 in the ethylene response, we generated their gain-of-function mutants and selected two individual transgenic lines of each with similar protein expression levels for further ethylene response analysis (fig. S4, A to C). Compared to Col-0, all the plants that overexpressed PDC E1, E2, and E3 subunits had enhanced ethylene-responsive phenotype in the presence of ACC, but the phenotypes of E2ox and E3ox did not differ from Col-0 in the absence of ACC (fig. S4D). This showed that the overexpression of PDC E2 and E3 alone was not sufficient to trigger ethylene response in the absence of the hormone when they were not accumulated in the nucleus. PDC E1ox displayed a mild ethylene hypersensitivity in the absence of ACC compared to Col-0, which is in line with the observation that there was a basal E1 in the nucleus without ethylene treatment (fig. S4D). Next, we expressed E1 and E2 fused with nuclear localization signal (NLS) peptide derived from EIN2-C (E1-NLS-GFP and E2-NLS-GFP) in Col-0 (fig. S4E). E1–NLS–green fluorescent protein (GFP) and E2-NLS-GFP fusion proteins could be localized to the nucleus in the absence of ethylene; the E1-NLS-GFP and E2-NLS-GFP plants displayed a clear ethylene hypersensitivity phenotype even in the absence of ethylene (fig. S4, F and G). Together, these data demonstrate that the nuclear localization of PDC can trigger the ethylene response.
To evaluate the function of PDC in ethylene response at a molecular level, we performed RNA sequencing (RNA-seq) using e1-2-2 e2-2 #17 and e2-2 e3-2-2 #67 double mutants treated with or without 4 hours of ethylene gas (fig. S5). We found that about 50% genes up-regulated by ethylene in Col-0 were not differentially expressed in either of the two mutants in response to ethylene (fig. S6, A and B). For further statistical quantification, we plotted the log2 fold change (log2FC) of each gene up-regulated by ethylene in Col-0 against that in each double mutant. These genes were then divided into three groups: elevated group (log2FC in mutants is 30% greater than in Col-0, plotted as red dots), unchanged group (log2FC in mutants is within 30% of that in Col-0, plotted as gray dots), and compromised group (log2FC in mutants is 30% less than that in Col-0, plotted as green dots). We found that the elevation of more than half of ethylene up-regulated genes in Col-0 was significantly compromised in both double mutants (Fig. 3, G to I). Most of the compromised genes identified from e1-2-2 e2-2 #17 seedlings were also compromised in the e2-2 e3-2-2 #67 seedlings (Fig. 3J), and the log2FC profile showed a high similarity (Fig. 3I and fig. S6C). Further gene ontology analysis using those up-regulation compromised genes showed that the ethylene signaling genes were overrepresented (Fig. 3K and fig. S6D). Together, these data provide genetic and molecular evidence that E1, E2, and E3 function coordinately to regulate ethylene response.
PDC regulates ethylene-dependent elevation of histone acetylation at H3K14 and H3K23
PDC catalyzes the synthesis of acetyl CoA, the substrate of histone acetylation. Given that the acetylation of histone at H3K14 and H3K23 is induced by ethylene, we compared their global levels in Col-0 and in different pdc mutants. The ethylene-induced global elevation of H3K14ac and H3K23ac in Col-0 was still detected in e1-2-2 e2-2 #17 but with lower levels; whereas ethylene-induced global elevation was not detected in e2-2 e3-2-2 #67 nor in e1-2-2 e2-2 e3-2-2 #67 (Fig. 4A and fig. S7, A to C). In contrast, the H3K14ac and H3K23ac levels were elevated in the plants that overexpress E2 with ethylene treatment and in the E2-NLS-GFP transgenic plants even without ethylene treatment (Fig. 4B and fig. S7, D to F). We then conducted H3K14ac and H3K23ac ChIP-seq in e1-2-2 e2-2 #17 (fig. S8, A to D). H3K14ac and H3K23ac levels were reduced in e1-2-2 e2-2 #17 compared to that in Col-0 over genes that were transcriptionally compromised in both e1-2-2 e2-2 #17 and e2-2 e3-2-2 #67 plants, and the ethylene-induced elevation was notably impaired (Fig. 4, C and D, and fig. S8, E to H). When we evaluated the ChIP signals within the first 500 bp downstream of transcription start sites as histone acetylation is associated with active transcription, an even more significant reduction in H3K14ac and H3K23ac enrichment was detected in e1-2-2 e2-2 #17 both with and without ethylene treatments when compared to those in Col-0 (Fig. 4, E and F). To further investigate the regulation of PDC on histone acetylation in ethylene-activated genes, we examined the H3K14ac and H3K23ac levels in 250 randomly selected, non-ethylene–regulated genes. These genes were chosen to match the number of co-compromised differentially expressed genes (DEGs) analyzed in Fig. 4 (C to F) and had expression levels within the 25th to 75th percentiles of the expression range of co-compromised DEGs in Col-0 without ethylene treatment (fig. S8I). No association between changes in H3K14ac or H3K23ac levels and the expression of these random genes was detected (fig. S8, J to M). This further confirms that PDC regulates H3K14ac and H3K23ac specifically in ethylene-activated genes in response to ethylene. We then conducted H3K14ac and H3K23ac ChIP-qPCR assays in the selected targets which show compromised transcriptional response to ethylene and reduced histone acetylation at H3K14 and H3K23 in pdc mutants (figs. S6D and S8, E to H), and the results further validated the ChIP-seq data (fig. S8, N and O). Given that e2-2 e3-2-2 #67 and e1-2-2 e2-2 e3-2-2 #67 have a similar ethylene-responsive phenotype as e1-2-2 e2-2 #17, we also conducted ChIP-qPCR assays in those two mutants, and we observed that ethylene-induced elevations of H3K14ac and H3K23ac enrichments were also impaired in e2-2 e3-2-2 #67 and e1-2-2 e2-2 e3-2-2 #67 as in e1-2-2 e2-2 #17 (fig. S8, N and O). In contrast, we found that in the absence of ethylene, the basal levels of H3K14ac and H3K23ac in E2-NLS are elevated when compared to that in Col-0 and also higher than that in E2ox, indicating that the nuclear-localized E2 is able to induce histone acetylation at ethylene-responsive target genes (fig. S8, P and Q). Together, these results suggest that PDC regulates the histone acetylation H3K14ac and H3K23ac in response to ethylene.
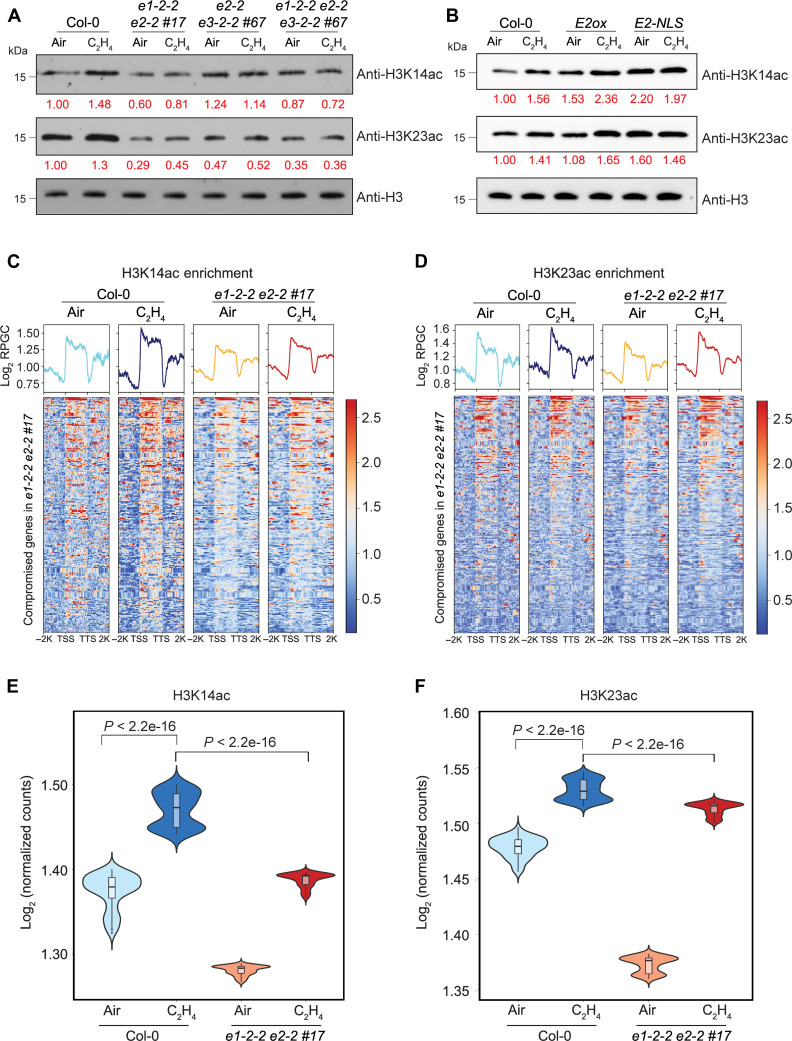
(A) Western blot analysis of total histone acetylation of H3K14 and H3K23 in response to ethylene in different genetic backgrounds as indicated in the figure. Anti-H3 Western blot served as a loading control. (B) H3K14ac and H3K23ac levels in Col-0, E2ox, and E2-NLS-GFP treated with air or 4 hours of ethylene gas. Histone H3 served as a loading control. Red number indicates the quantification of H3K14ac and H3K23ac Western blot signal intensity normalized with that of histone H3. (C and D) Heatmaps of H3K14ac (C) and H3K23ac (D) ChIP-seq signal (log2 ChIP signal) from the genes that their ethylene-induced expressions are co-compromised in e1-2-2 e2-2 #17 and e2-2 e3-2-2 #67. TSS, transcription start site; TTS, transcription termination site. (E and F) Violin plots illustrating log2 normalized H3K14ac ChIP signal (E) and H3K23ac ChIP signal (F) per bin (bin size = 1) from 500 bp downstream of TSS in the genes that were co-compromised in two pdc double mutants (Fig. 3J) in the indicated genotypes and conditions. P values were calculated by a two-tailed t test.
Enzymatically active nuclear PDC regulates acetyl CoA production in the nucleus
Because the biological function of PDC is to generate acetyl CoA, we hypothesized that the reduced histone acetylation level in the plants that lack PDC subunits results from the decreased acetyl CoA in the nucleus. We first measured the total acetyl CoA and nuclear acetyl CoA concentrations in Col-0 and in the e1-2-2 e2-2 e3-2-2 #67 triple mutant treated with 4 hours of ethylene gas by LC-MS, and the levels of total acetyl CoA and nuclear acetyl CoA were significantly decreased in the pdc triple mutant compared to levels in Col-0 while the concentrations of PDC-irrelevant CoA species malonyl CoA remain unchanged (Fig. 5, A and B, and fig. S9A). We then decided to examine whether the nuclear PDC was functional to synthesize acetyl CoA. It has been shown that dephosphorylated E1 is required for the PDC activity (44, 47); therefore, we first examined the phosphorylation status of E1 in the nucleus in response to ethylene. Western blot assay by anti-pSer antibody showed that most E1 was in a dephosphorylated state in the nucleus after ethylene treatment (Fig. 5C). Phos-tag electrophoresis assay confirmed this result (Fig. 5D). Next, we conducted LC–tandem MS (MS/MS) analysis of E1 phosphorylation. We detected phosphorylation on Ser292 (fig. S9B), a residue that is evolutionary conserved with the reported inhibitory phosphorylation site in human E1 (fig. S9C) (47). Further quantification of E1 phosphopeptides by MS showed that the ratio of phosphorylated E1 to nonphosphorylated E1 in the nucleus was significantly lower than that in the cytosol after 4 hours of ethylene treatment (Fig. 5E), suggesting that the dephosphorylated E1 is the main species in the nucleus to function in the presence of ethylene. Next, we assessed E1 activities in the nucleus using a PDH activity assay. E1 activity was detected in the nucleus in Col-0, and its activity was elevated by the ethylene treatment (Fig. 5F). However, the ethylene-induced E1 activity was not detected in e1-2-2 e2-2 #17 or in e1-2-2 e2-2 e3-2-2 #67 (Fig. 5F and fig. S9D). To further directly monitor PDC acetyl CoA biosynthesis enzyme activity in the nucleus in response to ethylene, we used a 13C isotopic tracing experiment. By feeding 2,3-13C2 pyruvate to the nuclear extracts and followed by the LC-MS/MS, we are able to measure 1,2-13C2–labeled acetyl CoA (1,2-13C2 acetyl CoA) that will be converted from 2,3-13C2 pyruvate by PDC complex (Fig. 5G and fig. S9E). This experiment assesses the nuclear PDC activity since PDC is the only enzyme that synthesizes acetyl CoA from pyruvate. As shown in Fig. 5H, we detected 1,2-13C2 acetyl CoA in the nuclear extracts of Col-0 treated with 4 hours of ethylene gas, and the 1,2-13C2 acetyl CoA levels were significantly increased in the presence of ethylene (Fig. 5H and fig. S9F). This suggests that functional PDC complex is accumulated in the nucleus in response to ethylene in Col-0. Further comparison showed no significant differences in the levels of 1,2-13C2 acetyl CoA in the nuclei of Col-0 compared to that of e1-2-2 e2-2 #17 or of e1-2-2 e2-2 e3-2-2 #67 in the absence of ethylene (Fig. 5H and fig. S9F). However, the ethylene-induced elevation of 1,2-13C2 acetyl CoA detected from Col-0 nucleus was significantly reduced from the nucleus of e1-2-2 e2-2 #17 or e1-2-2 e2-2 e3-2-2 #67 (Fig. 5H), providing further biochemical evidence that functional PDC is translocated into the nucleus in response to ethylene to synthesize acetyl CoA.

(A and B) LC-MS detection of acetyl CoA (A) and malonyl CoA (B) in purified nuclei from Col-0 and e1-2-2 e2-2 e3-2-2 #67 etiolated seedlings treated with 4 hours of ethylene gas. Total protein mass was used to normalize metabolite concentration. P values were calculated by a two-tailed t test, and each data point was plotted as a dot in the bar graph. (C) Western blot analysis of the phosphorylation status of E1 using anti-pSer antibody in the cytosolic fraction and nuclear fractions from E1ox seedlings treated with 4 hours of ethylene gas. (D) Phos-tag phosphoprotein mobility shift gel of E1 phosphorylation status in the cytosolic fraction and nuclear fractions of E1ox etiolated seedlings treated with 4 hours of ethylene gas. Black arrow shows nonphosphorylated E1; red arrow indicates phosphorylated E1 protein. PEPC, CytC, and H3 were used to assess purities of nuclear and cytosolic fractionations. (E) The ratios of phosphorylated to nonphosphorylated YHGHpSMSDPGSTYR E1 peptides in the cytosolic versus in the nuclear fractions from E1ox with 4 hours of ethylene gas treatment. Peak area values were collected from three replicates. P value was calculated by t test. (F) Pyruvate dehydrogenase activity assays in the nuclear fraction from Col-0 and indicated pdc mutants treated with 4 hours of ethylene gas. (G) Schematic diagrams to show the conversion of 2,3-13C2 pyruvate to 1,2-13C2 acetyl CoA. (H) Normalized LC-MS/MS measured the concentration of 1,2-13C2 acetyl CoA that converted from 2,3-13C2 pyruvate by using the nuclear extracts from indicated plants with or without 4 hours of ethylene gas treatment. Quantification from four replicates was normalized by total protein input. Different letters represent significant differences between each group calculated by a one-way ANOVA test followed by Tukey’s post hoc test.
EIN2 is required for PDC nuclear translocation and function in response to ethylene
Given the fact that EIN2-C interacts with PDC in the nucleus, we next explored the genetic relationship between EIN2-C and PDC subunits. By introducing E1ox, E2ox, and E3ox into the ein2-5 null mutant, we obtained E1ox/ein2-5, E2ox/ein2-5, and E3ox/ein2-5 plants with comparable protein expression levels in E1ox/Col-0, E2ox/ Col-0, and E3ox/ Col-0 separately (fig. S10, A to C). We then compared their ethylene response with their parental plants and Col-0. The ethylene hypersensitivity induced by PDC overexpression was entirely abolished in all E1ox/ein2-5, E2ox/ein2-5, and E3ox/ein2-5 plants; these plants displayed the same complete ethylene insensitivity as ein2-5 mutant (Fig. 6A). Similarly, the ethylene hypersensitivity induced by PDC overexpression was entirely abolished in all E1ox/ein3-1 eil1-1, E2ox/ein3-1 eil1-1, and E3ox/ein3-1 eil1-1 plants (fig. S10, A to D). These two genetic results suggest that both EIN2 and EIN3/EIL1 are required for the function of PDC in the ethylene response, and the involvement of PDC in ethylene response depends on EIN2- and EIN3/EIL1-directed ethylene signaling transduction.
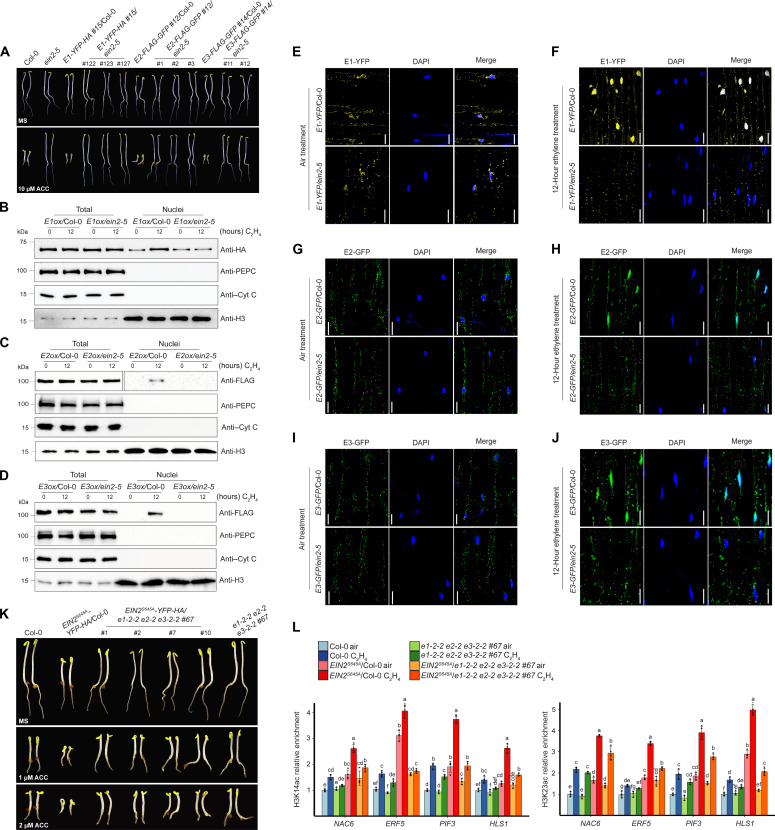
(A) Epistasis analysis of PDC E1ox, E2ox, and E3ox in the ein2-5 mutant. The seedlings were grown on MS medium containing with or without 10 μM ACC in the dark for 3 days before being photographed. (B to D) Examination of the protein levels of E1 in E1ox/Col-0 and E1ox/ein2-5 (B), E2 in E2ox/Col-0 and E2ox/ein2-5 (C), and E3 in E3ox/Col-0 and E3ox/ein2-5 (D) with 0- or 12-hour ethylene gas treatment. PEPC is cytoplasmic marker; Cyt C is a mitochondrial marker; histone H3 is a nuclear marker for a loading control. (E to J) Subcellular localization of E1-YFP-HA [(E) and (F)], E2-FLAG-GFP [(G) and (H)], and E3-FLAG-GFP [(I) and (J)] in Col-0 or ein2-5 mutant without or with 12 hours of ethylene gas treatment, respectively. Scale bars, 20 μm. (K) Photographs of 3-day-old etiolated EIN2S645A/Col-0 and EIN2S645A/e1e2e3 seedlings grown on MS medium containing indicated ACC in the dark for 3 days before being photographed. (L) ChIP-qPCR to evaluate H3K14ac (top) and H3K23ac (bottom) enrichment over selected genes in the indicated etiolated seedlings treated with air or 4 hours of ethylene gas. Different letters represent significant differences between each genotype and treatment condition that are calculated by a one-way ANOVA test followed by Tukey’s post hoc test with P ≤ 0.05.
To further confirm the EIN2 dependency of PDC in ethylene response, we examined the cellular localization of PDC in response to ethylene in the ein2-5 mutant. We found that the ethylene-induced nuclear accumulation of E1, E2, and E3 was abolished in E1ox/ein2-5, E2ox/ein2-5, and E3ox/ein2-5 (Fig. 6, B to J). Similarly, no obvious ethylene-induced nuclear accumulation of PDC proteins was observed in E1ox/ein3-1 eil1-1, E2ox/ein3-1 eil1-1, and E3ox/ein3-1 eil1-1 (fig. S10, E to J). ChIP-qPCR analyses in selected ethylene-responsive target genes (fig. S8, E to H) showed that the elevation of H3K14ac and H3K23ac levels detected in E2ox was not detected in the E2ox/ein2-5 plants, and the levels were similar to those in the ein2-5 mutant both with and without ethylene treatments (fig. S10K). In addition, the enhanced transcriptional activation in response to ethylene in E2ox was not detected in E2ox/ein2-5 plants (fig. S10L). Thus, our genetic and molecular evidence supports the conclusion that PDC functions in the ethylene response in an EIN2-dependent manner.
Acetyl CoA is important for histone acetylation, and EIN2-C is essential for the ethylene-induced histone acetylation elevation; therefore, we further investigated whether PDC regulates the function of EIN2-C. We crossed EIN2S645A, in which a point mutation at Ser645 mimics EIN2 constitutive dephosphorylation resulting in EIN2-C cleavage and nuclear accumulation, into e1-2-2 e2-2 e3-2-2 #67 (e1e2e3) background to generate EIN2S645A/e1e2e3 plants (fig. S11A). Phenotypic analysis of EIN2S645A/Col-0 and EIN2S645A/e1e2e3 showed that the ethylene hypersensitivity caused by EIN2S645A was partially rescued in the e1e2e3 mutant (Fig. 6H). However, the nuclear accumulation of EIN2-C was comparable in the EIN2S645A/Col-0 and in EIN2S645A/e1e2e3 (fig. S11, B and C), showing that PDC participates in the downstream ethylene signaling and cellular response mediated by EIN2-C rather than the initial steps of EIN2-C cleavage and translocation into the nucleus in ethylene signaling pathway. We then compared the H3K14ac and H3K23ac levels at selected ethylene-responsive genes in EIN2S645A/Col-0 and EIN2S645A/e1e2e3 plants by ChIP-qPCR. We found that the enhanced H3K14ac and H3K23ac in EIN2S645A/Col-0 were reduced in EIN2S645A/e1e2e3, although levels were still higher than that in Col-0 (Fig. 6I). Consistently, the enhancement of target gene expression in EIN2S645A/Col-0 was also partially rescued in EIN2S645A/e1e2e3 (fig. S11D). These findings suggest that PDC in the nucleus produces acetyl CoA that contributes to EIN2-C–regulated histone acetylation to mediate the ethylene response.
DISCUSSION
This study reveals a molecular mechanism by which mitochondria-resided PDC complex translocates into the nucleus to provide acetyl CoA necessary for EIN2-C–mediated histone acetylation and subsequent transcription activation in response to ethylene (fig. S12). In the presence of ethylene, PDC subunits E1, E2, and E3 accumulate in the nucleus to provide acetyl CoA for EIN2-mediated histone acetylation elevation at H3K14 and H3K23 through their interaction with EIN2-C, initiating the downstream ethylene-induced transcriptional cascade (fig. S12). Our research establishes a direct link between cell metabolism and histone modification in the ethylene response that is mediated by the key factor EIN2, opening an avenue for the study of how plant hormone and metabolisms function to ensure the chromatin and transcription regulation.
Cellular metabolism is a series of important biochemical reactions fueling development with energy and biomass, and chromatin is a mighty consumer of cellular energy generated by metabolism. The balance between metabolism and chromatin activities ensures cell homeostasis and normal cell growth. PDC functioning as a partner of EIN2-C to regulate histone acetylation levels at H3K14 and H3K23 marks in response to ethylene treatment suggests that EIN2-C recruits translocated PDC to the ethylene-responsive gene loci to provide a sufficient level of local acetyl CoA to achieve histone acetylation at H3K14 and H3K23 by HATs. Production of acetyl CoA within the nucleus may promote its availability to HAT to facilitate a rapid ethylene response. Thus, identification of the histone acetyl transferases that function together with EIN2-C and PDC complex will be an immediate goal.
In the nucleus, acetyl CoA production machineries are finely tuned to control the local metabolite levels at certain gene loci to regulate chromatin modification by interacting with different partners. For instance, PDC E2 subunit has been reported to function in the same complex with pyruvate kinase M2, HAT p300, and the transcription factor arylhydrocarbon receptor (AhR) to regulate histone acetylation for transcription regulation to facilitate cell proliferation (48). Nuclear metabolic enzyme acetyl CoA synthetase 2 (ACSS2) associates with the HAT cAMP response element-binding (CREB)-binding protein to regulate the expression of key neuronal genes through histone acetylation in the mouse hippocampus (49). In response to glucose deprivation, ACSS2 can also constitute a complex with the transcription factor EB to activate lysosomal and autophagosomal genes after its nuclear translocation (42). In ethylene response, EIN3 is the key transcription factor that determines the target gene for transcription regulation (25, 27); thus, it will be interesting to explore the connection between EIN3 and PDC complex.
In mammal, three main cytosolic enzymes that synthesize acetyl CoA, including PDC, acyl-CoA synthetase short-chain family member (ACSS2), and adenosine triphosphate (ATP) citrate lyase (ACLY or ACL), were shown to localize to the nucleus for different biological functions (38, 40, 42, 43, 48–50). PDC complex was first reported to localize to the nucleus in cell division and synthesize acetyl CoA to regulate cell proliferation through histone acetylation (40). ACSS2 catalyzes the conversion of acetyl CoA from acetate in the nucleus for the regulation of long-term spatial memory and glucose starvation response (42, 49); the nuclear ACLY uses citrate to produce acetyl CoA to promote histone acetylation at double-strand break sites for DNA repair (38). Recently, ACL subunit A2 has been reported to function with HAT1 in rice to regulate cell division in developing endosperm, suggesting the evolutionarily conserved mechanism of metabolic regulation on epigenetic modification in both plant and mammal species (51). We have noticed that although the ethylene-induced elevation of H3K14ac and H3K23ac is compromised in the pdc mutant, some extent of ethylene response still remains. It is possible that since we could not obtain the e2 null mutant because of its lethality, the weak alleles we obtained still have partially functional E2 and therefore manifest a rather weakened ethylene response. However, we could not exclude the possibility that the other acetyl CoA–producing enzymes will be functioning in the nucleus to modulate histone acetylation in response to ethylene in the absence of the nuclear PDC in pdc mutants. Investigating the involvement of other acetyl CoA synthesis pathways, such as ACSS (42, 49) and ACLY (38), in the ethylene response will potentially address the question.
In this research, we found that ethylene treatment triggers the translocation of PDC from the cytosol to the nucleus to facilitate H3K14ac and H3K23ac elevation for ethylene-responsive transcriptional regulation. However, the question remains how the translocation of PDC complex occurs at molecular level because PDC is a large protein complex without known NLS. Recent studies have shown that specialized contact points between mitochondria and nucleus aid in the exchanges of metabolites and proteins between these two organelles, and mammalian PDC directly enters the nucleus at those contact points across the nuclear envelop through mitofusin 2–mediated mitochondria-nucleus tethering, which is independent of the nuclear pore complex (NPC) (52, 53). However, whether it is possible that nuclear PDC proteins are transported into the nucleus from the cytosol following their protein synthesis is unknown. Investigating how Arabidopsis PDC enters the nucleus and whether its nuclear entry follows the similar NPC-independent mechanism to regulate histone acetylation in response to ethylene will provide more insights into the mitochondria and nucleus retrocommunication in the establishment of the proper ethylene response, providing a perspective of metabolic regulation in plant hormone response research.
MATERIALS AND METHODS
Plant materials, growth condition, and hormone treatments
Arabidopsis (Arabidopsis thaliana) accession Columbia (Col-0) was used as the wild type in this study, and all the mutant genotypes are in this ecotype background. The T-DNA insertion mutants of pdc e1-2-2 (or e1-2-2, AT1G59900, SALK_074384), pdc e1-2-4 (or e1-2-4, AT1G59900, SALK_047438), pdc e3-2-1 (or e3-2-1, AT3G17240, SALK_082094), and pdc e3-2-2 (or e3-2-2, AT3G17240, SALK_027039) were ordered from Arabidopsis Biological Resource Center. The T-DNA insertions of e1-2-2, e1-2-4, e3-2-1, and e3-2-2 were confirmed by genotyping PCR to identify the homozygotes. Primers used for genotyping are listed in table S1. Arabidopsis seeds were first surface-sterilized with solution containing 50% bleach and 0.01% Triton X-100 for 10 min, washed with sterile distilled water for four times, sown on Murashige and Skoog (MS) medium plates containing 1% sucrose and 1% phytoblend agar, and stratified for 3 days at 4°C in the dark. For seed propagation, 1- or 2-week-old green seedlings were transferred into soil (Promix-HP) and grown in a growth chamber setting with the long-day photoperiod (16-hour light/8-hour dark) at 22°C till maturity.
Ethylene triple response assay was performed with sterilized seeds on MS plates containing various concentrations of ACC (Sigma-Aldrich) (0, 1, 2, 5, and 10 μM ACC). Approximately 60 seeds from each genotype collected in the similar time period were sown on the same MS medium plate per concentration. Seeds sown on ACC plates were kept at 4°C in the dark for 3 days for stratification, and then they were exposed to light for 4 hours and transferred in the dark growth chamber for 3 days at 22°C. For phenotypic analysis, representative seedlings were selected for photograph. Their hypocotyl and root lengths were measured using Fiji ImageJ software.
For all imaging, RNA, ChIP, and protein-related assays, Arabidopsis seedlings were grown on MS plates in the air-tight containers with a flow of hydrocarbon-free air at 22°C for 3 days and subsequently treated with ethylene gas [10 parts per million (ppm)] or hydrocarbon-free air for indicated hours before collect samples.
Plasmid construction and generation of transgenic Arabidopsis plants
For yeast two-hybrid assays, the full-length coding regions of E1, E2, and E3 were obtained by RT-PCR from Col-0 cDNA and then cloned into pEXPAD502 [activation domain (AD)] and pDBLeu (BD) backbone. For recombinant protein expression, E1 and E2 cDNA was first cloned into pENTR vector and then cloned into pVP13, and E3 cDNA was amplified and inserted into pMAL-pX2 for maltose-binding protein (MBP) fusion. For plant transformation, full-length E1 cDNA was cloned into pENTR vector, and then E1-pENTR was cloned into pEarleyGate 101 vector using recombination reaction between attL and attR sites (LR reaction) to generate E1-YFP-HA. E2 and E3 cDNA was cloned into pCambia1300 to generate E2-FLAG-GFP and E3-FLAG-GFP. To generate PDC-BFP fusion protein, E1, E2, and E3 cDNA was inserted into modified pCambia1300 with replacement of GFP to BFP. All these genes were expressed under the control of the CaMV 35S promoter. The promoter region and genomic sequence of E2 were cloned from Col-0 genomic DNA and integrated into pCambia1300 backbone. These constructs were introduced into Agrobacterium tumefaciens strain GV3101 through electroporation and transformed into Arabidopsis Col-0 ecotype or desired genetic backgrounds using the floral dip method. Positive T1 transformants were selected on MS medium with appropriate antibiotics. Homozygous T3 transgenic seedlings with single T-DNA insertion were used for further analysis. Table S1 contains details of plasmid and primer sequence information.
Yeast two-hybrid assay
Yeast two-hybrid assay was performed using the ProQuest Two-Hybrid System (Invitrogen) by polyethylene glycol–mediated transformation following the manufacturer’s instructions. Briefly, the bait and prey plasmids (pAD-E1, pBD-E1, pAD-E2, pBD-E2, pAD-E3, and pBD-E3) were cotransformed into the yeast strain AH109 as the experiment design. The positive transformants were selected on SD/-Trp-Leu (two-dropout) medium. The growth on SD/-Trp-Leu-His (three-dropout) medium supplemented with appropriate concentrations of 3-amino-1,2,4-triazole indicates interaction between corresponding proteins.
In vitro protein purification and pull-down assays
MBP and recombinant MBP-E1, MBP-E2, MBP-E3, and 6XHis-EIN2-C proteins were expressed in the Escherichia coli strain BL21-CodonPlus(DE3)-RIPL (Agilent) with protein induction by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when OD600 (optical density at 600 nm) reached 0.5 for 4 hours at 37°C, and then they were purified using amylose resin column (New England Biolabs, for MBP-related proteins purification) or nickel-nitrilotriacetic acid (Ni-NTA) agarose column (Qiagen, for 6XHis-EIN2-C purification) according to the manufacturer’s menu followed by sonication. MBP-tagged PDC proteins or MBP proteins were added and incubated with 6XHis-EIN2-C–bound Ni-NTA resin for 1 hour at 4°C with gentle rocking. After five times washing with pull-down buffer to remove nonspecific binding, Ni-NTA resin beads were collected by brief centrifugation and then resuspended and denatured in 2× protein sample buffer by boiling. Proteins were then resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and detected with corresponding antibodies.
Confocal imaging and MitoTracker Red staining
Confocal images were acquired by a Zeiss LSM710 confocal microscope with a ×40 (water immersion) or ×63 lens (water immersion). For subcellular localization analysis of EIN2 with MitoTracker staining, true leaves of 3-week-old green seedlings were submerged in ½ MS solution (mock), and 10 μM ACC was dissolved in ½ MS solution (ACC treatment) for 4 hours and then vacuum-infiltrated for 30 min with the addition of 200 nM MitoTracker Red (MitoTracker Red CMXRos, Invitrogen, M7512) before imaging. Images of EIN2-YFP/MitoTracker localization analysis were taken from leaf epidermal cells in central areas in the adaxial side of stained true leaves of Arabidopsis seedlings. For other confocal imaging experiments such as the colocalization of EIN2 and PDC as well as the subcellular localization of PDC under time series of ethylene treatment, 3-day-old etiolated seedlings were used after each indicated treatment, and the images were taken from the similar middle regions of hypocotyls. The excitation/emission spectra for fluorescent proteins and fluorophores used in this study are as follows: YFP, 514 nm/520 to 540 nm; GFP, 488 nm/501 to 528 nm; 4′,6-diamidino-2-phenylindole, 405 nm/437 to 476 nm; BFP, 405 nm/410 to 500 nm, and MitoTracker Red, 579 nm/585 to 615 nm.
Nuclear-cytoplasmic fractionation
Three-day-old etiolated seedlings treated with air or ethylene were first snap-frozen and homogenized in liquid nitrogen (LN2) to fine powders. Approximately 2 g of frozen powders was resuspended in 10 ml of lysis buffer [20 mM tris-HCl (pH 7.4), 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM sucrose, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail] and then filtered through a 40-μm cell strainer (nylon mesh, Fisher brand). A total of 10% of the flow-through was saved as total extract for the following immunoblot analysis, and the rest of the flow-through was then centrifuged at 1500g for 10 min at 4°C. The supernatant was then transferred to a new tube and centrifuged for 10 min at 10,000g at 4°C, and this supernatant was saved as the cytoplasmic fraction. The pellet was washed for five times with 5 ml of nuclear washing buffer/nuclear resuspension buffer 1 [20 mM tris-HCl (pH 7.4), 2.5 mM MgCl2, 25% glycerol, 0.2% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail]. Then, the pellet was resuspended in 500 μl of the nuclear resuspension buffer 2 [20 mM tris-HCl (pH 7.4), 10 mM MgCl2, 250 mM sucrose, 0.2% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail] and then carefully overlaid on top of the 500 μl of the nuclear resuspension buffer 3 [20 mM tris-HCl (pH 7.4), 10 mM MgCl2, 1.7 M sucrose, 0.2% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail] followed by centrifuge at 16,000g for 45 min at 4°C. The supernatant was then carefully removed, and the final nuclear pellet was saved as the nuclear fraction for the following biochemical assays. The efficiency of the cell fractionation was confirmed by anti–phosphoenolpyruvate carboxylase (anti-PEPC; Abcam, ab347793) for the cytoplasmic fraction, anti–cytochrome C (anti-Cyt C; Agrisera, AS08 343A), anti-fumarase 1 + 2 (anti-FUM1/2; Agrisera, AS16 3966), and anti-ATPB (Agrisera, AS16 3976) for mitochondria contamination examination, anti–luminal-binding protein (anti-BiP; Agrisera, AS09 481) for ER contamination examination, and anti–histone 3 (anti-H3; Abcam, ab1791) for the nuclear fraction in the immunoblots.
Co-IP and immunoblot assays
For co-IP between EIN2-C and PDC following nuclear-cytoplasmic fractionation, the cytoplasmic fraction was used directly for co-IP assays. The nuclear fraction was first sonicated for 30 s by Bioruptor at 4°C to release the nuclear content, and the cleared nuclear lysate after centrifuge at 16,000g for 10 min at 4°C was used for co-IP. A 10% input aliquot for each co-IP experiment was saved from the cleared supernatant before the rest of the supernatant was subject to immunoprecipitation. Cytoplasmic EIN2-YFP-HA full-length proteins and nuclear EIN2-C–YFP–HA proteins were immunoprecipitated by Pierce anti-HA magnetic beads (Thermo Fisher Scientific), PDC FLAG-BFP fusion proteins were immunoprecipitated by DYKDDDDK Fab-Trap agarose beads (ChromoTek) for 2 hours at 4°C with gentle rotating, and then beads were washed five times with ice-cold 1× phosphate-buffered saline buffer. The EIN2-C native antibody was prebound with the equilibrated protein G–coated magnetic beads (Dynabeads, Thermo Fisher Scientific) with 1% bovine serum albumin for 3 hours with gentle rotation at 4°C and incubated at 4°C after cleared cell fractionation addition with gentle rocking overnight for co-IP. Dynabeads was magnetically precipitated using a DynaMagnetic rack (Thermo Fisher Scientific) and then washed five times with 0.5 ml of ice-cold 1× PBS buffer to eliminate nonspecific binding. After IP and washing, proteins were then released from beads using 2× Laemmli sample buffer by heating at 85°C for 8 min and subject to SDS-PAGE. For EIN2-related Western blots, protein samples were denatured in 2× Laemmli sample buffer with the addition of 3 M urea at 75°C in water bath for 5 min to avoid protein aggregation and degradation.
For protein immunoblots, proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (0.2 μm; Bio-Rad) by the wet-tank transfer method and blocked with 5% nonfat milk in TBST (Tris-Buffered Saline with 0.1% Tween 20) for 1 hour at room temperature before the overnight inoculation in primary antibody at 4°C with gentle rotation. The following antibodies and dilutions were used for immunoblotting: anti-HA (BioLegend, #901503), 1:10,000; anti-FLAG (rabbit) (Cell Signaling Technology, #14793), 1:2000; anti-FLAG (mouse) (Sigma-Aldrich, F3165),1:5000; anti-H3 (Abcam, ab1791), 1:10,000; anti-PEPC (Abcam, ab34793), 1:2000; anti-Cyt C (Agrisera, AS08 343A), 1:2000; anti-FUM1/2 (Agrisera, AS16 3966), 1:2000; anti-BiP (Agrisera, AS09 481), 1:2000; and anti–EIN2-C [developed in-house (20)], 1:2000. Goat anti-mouse kappa light chain antibody (Bio-Rad, #105001G) and goat anti-rabbit [immunoglobulin G (H + L)–horseradish peroxidase (HRP) conjugate #1706515, Bio-Rad] were used as secondary antibodies at 1:10,000 dilution. HRP activity was detected by enhanced chemiluminescence (GE Healthcare) according to the manufacturer’s instructions using either a ChemiDoc Imaging System (Bio-Rad) or conventional x-ray films.
BiFC assay
The BiFC assay was performed following previously published methods with minor revisions (54). Briefly, Agrobacterium strain GV3101 carrying plasmids of interest (EIN2-nYFP, E1-cYFP, E2-cYFP, and E3-cYFP), NLS-BFP/pCambia1300 served as nuclear marker, and p19 was cultured overnight at 28°C, centrifuged, and resuspended in the fresh infiltration buffer [10 mM MES/KOH (pH 5.7), 10 mM MgCl2, and 100 μM acetosyringone]. The different combinations of BiFC pairs, NLS-BFP strain, and p19 strain were evenly mixed to yield a final OD600 of 0.4 of each strain. Then, the mixture was kept at 28°C for 1 hour and then infiltrated into Nicotiana benthamiana (tobacco) leaves. The YFP fluorescence from tobacco epidermal cells was observed under confocal microscopy (Zeiss LSM710) after 2 days of infiltration. For ACC treatment, leaf discs of infiltrated tobacco leaves were submerged in ½ MS solution (mock), and 20 μM ACC was dissolved in ½ MS solution for 4 hours before confocal microscopy.
CRISPR-Cas9 mutagenesis
The e2-2 #17 and e2-2 #215 single mutants, e2-2 e3-2-1 #99 double mutant, and e1-2-2 e2-2 e3-2-1 #167 and e1-2-2 e2-2 e3-2-2 #67 triple mutants were generated by the CRISPR-Cas9 system in Col-0, e3-2-1 single mutant, and e1-2-2 e3-2-1 and e1-2-2 e3-2-2 double mutant backgrounds, respectively, by following the previous publication (55, 56), because E2 and E3 are linked in chromosome 3 in proximity. e2-2 e3-2-2 #67 was generated by out-crossing e1-2-2 e2-2 e3-2-2 #67 to Col-0. CRISPR-Cas9 transgene was removed by out-crossing to Col-0, and Cas9-free homozygous mutants were used for genetic and molecular experiments. Briefly, pairs of two guide RNA (gRNA) sequences targeting E2 locus were designed by using CRISPR-PLANT v2 (http://omap.org/crispr2/index.html) online tool. gRNA sequences with high specificity in Arabidopsis genome and high predicted efficiency were chosen for PCR amplification using pDT1T2 vector as template and later incorporated into the binary vector pHEE401E. Agrobacterium-mediated floral dipping method was used to transform the E2 gRNA containing pHEE401E vectors into each corresponding genetic background. To genotype deletion mutations, plant genomic DNAs extracted from hygromycin-positive T1 plants were used to PCR amplify of E2 gRNA-targeted regions and followed by Sanger sequencing to identify the CRISPR-Cas9–directed mutation and to obtain homozygotes of e2 mutations.
Transcriptome analysis
RNA was extracted from e1-2-2 e2-2 #17 and e2-2 e3-2-2 #67 double mutant etiolated seedlings with or without 4 hours of ethylene gas treatment. RNA extraction was conducted according to the manufacturer’s recommendation using the RNeasy Plant Kit (Qiagen) with deoxyribonuclease I digestion and clean up [Qiagen, RNeasy Mini Kit (250)]. For mRNA-seq, libraries were generated according to the instructions from the manufacturer using the NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490) and the NEBNext Ultra II RNA Library Prep Kit for Illumina (E7770) with 1 μg of RNA as input. Indexed libraries were sequenced on the platform of HiSeq2000 (Illumina). Paired-end raw fastq data were evaluated with FastQC, and low-quality reads were removed with Trim Galore (Babraham Institute). The trimmed and filtered reads were then mapped to the Arabidopsis reference genome (TAIR10) with STAR (57). Mapped reads were counted by featureCounts (Subread 2.0.1), and differentially regulated genes were identified using R package DESeq2 (58) with a P adjusted < 0.05 and |log2(FC)| > 1. The RNA-seq data for Col-0 air and ethylene treatment were obtained from a previous study [Gene Expression Omnibus (GEO) accession code GSE77396] (26) and reanalyzed with the pipeline described above. For PDC-dependent DEG (compromised DEGs) identification, typical ethylene-responsive up-regulated DEGs were selected from Col-0 RNA-seq dataset based on P adjusted < 0.05 and log2(FC) > 1 and show P adjusted < 0.05 in pdc mutants air versus C2H4 DESeq2 comparison. The scatterplots, violin plots, and dot plots were generated by R package ggplot2, and the heatmaps of log2FC were generated by R package pheatmap. Gene ontology (GO) enrichment analysis for DEGs was performed by agriGO v2 (59).
Histone extraction
Approximately 2 g of ground fine powders of etiolated seedlings was homogenized in 20 ml of nuclear extraction buffer [250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM Pipes (pH 6.8), 0.5% Triton X-100, 1 mM PMSF, 1× protease inhibitor, and 10 mM sodium butyrate as histone deacetylase inhibitor] for 10 min. After filtering through a single layer of Miracloth twice, the flow-through was then centrifuged at 2880g for 20 min at 4°C to collect the nuclei. The crude nuclear pellet was then resuspended in 800 μl of 0.4 M H2SO4 by gentle pipetting and was incubated for 2 hours for complete lysis followed by centrifuge at 16,000g at 4°C. Total histone in the supernatant phase was then precipitated by 33% trichloroacetic acid (TCA) for at least 2 hours and then centrifuged at maximum speed for 10 min at 4°C. The histone pellet was washed twice with ice-cold acetone containing 0.1% HCl and once with pure acetone. The air-dried histone pellet was then dissolved in 2× Laemmli buffer containing 2 M urea, denatured by boiling, and subject to immunoblots.
ChIP-seq analysis
Chromatin immunoprecipitation was performed according to previous publication (24). Briefly, 3-day-old etiolated e1-2-2 e2-2 #17 seedlings treated with air or 4-hour ethylene were harvested and cross-linked in 1% formaldehyde, and the chromatin was isolated and sheared by Bioruptor. Anti-H3K14ac (Millipore; 07-353) and anti-H3K23ac (Millipore; 07-355) antibodies together with Dynabeads were added to the sonicated chromatin followed by incubation overnight to precipitate histone-bound DNA fragments. DNA eluted from chromatin-immunoprecipitated was prepared for library construction by using NEBNext Ultra II DNA Library Prep with Sample Purification Beads according to the standard operating procedures and then sequenced using HiSeq2000 (Illumina). Paired-end raw reads were first mapped to the Arabidopsis genome (TAIR10) using Bowtie2 software with default parameters. Duplicate reads were removed using SAMtools. H3K14ac and H3K23ac ChIP-seq data for Col-0 air and ethylene treatment were obtained from a previous study (GEO accession code GSE77396) (26). Two replicate ChIP-seq files were merged for the following analyses according to a previous publication (26). ChIP-seq coverage tracks shown in the genome browser IGV (Integrative Genomics Viewer) were generated using deepTools 3.5.0 (60) bamCoverage function normalized by reads per genomic content (1× RPGC, 1× normalization) and bin size = 1. The ChIP-seq signals from 2 kb upstream transcription start site to 2 kb downstream transcription end site of PDC specifically regulated gene were calculated with bamCompare and computeMatrix (deepTools) and plotted with plotHeatmap and plotProfile in deepTools or ggplot2 in R.
In vivo E1 phosphorylation detection
Three-day-old E1-YFP-HA transgenic etiolated seedlings after 4 hours of ethylene treatment were first subject to the nuclear-cytoplasmic fractionation procedure with the addition of phosphatase inhibitors (20 mM NaF (sodium fluoride), 1 mM Na2MoO4, 10 mM Na3VO4, 50 mM β-glycerophosphate, and 1× PhosStop cocktail) to preserve phosphorylation modifications. Nuclear pellets were sonicated briefly by Bioruptor to release nuclear proteins and cleared by centrifuge at 10,000g for 10 min at 4°C. Cytoplasmic and nuclear E1 proteins were then immunoprecipitated by GFP-Trap (ChromoTek) for 2 hours at 4°C with gentle rotating, and then GFP-trap beads were washed five times with IP buffer and boiled in 2× protein sample buffer followed by subsequential immunoblotting. Anti-HA was used to assess E1-YFP-HA loading, and anti-pSer (P3430, Sigma-Aldrich) was used to detect phosphorylated E1 in each fractionation. IPed E1 used for LC-MS/MS phosphorylation detection was also purified following the above-mentioned procedure.
Phos-tag gel electrophoresis assay
The nuclear-cytoplasmic fractionation using 3-day-old E1-YFP-HA transgenic etiolated seedlings was performed with aforementioned buffers’ recipes but without the addition of EDTA and subjected to Phos-tag electrophoresis analysis following the manufacturer’s instructions. Briefly, the E1 proteins from cytosolic fraction and nuclear fraction were resolved by using 6% SDS-PAGE containing 100 μM MnCl2 and 50 μM Phos-tag (FUJIFILM Wako). After electrophoresis, the poly-acrylamide gel was washed five times with the transfer buffer (20 mM tris, 150 mM glycine, and 20% methanol) with the addition of EDTA (1 mM as final concentration) for 10 min with gentle shaking and then washed twice in the buffer without EDTA 10 min before wet-tank transfer overnight at 50 V using the transfer buffer containing 0.01% SDS followed by immunoblotting.
LC-MS/MS analysis of E1 phosphorylation
After GFP-Trap purification using nuclear and cytoplasmic fractions of 3-day-old E1-YFP-HA transgenic etiolated seedlings, SDS-PAGE, and CBB staining/distaining, E1-YFP-HA protein bands were excised and subjected to in-gel digestion with trypsin. Peptide mixtures were desalted using C18 ZipTips (Millipore). Data were acquired in the data-dependent acquisition mode. Peptides were analyzed by LC-MS/MS on an Easy LC 1200 UPLC (ultra performance liquid chromatography) liquid chromatography system (Thermo Fisher Scientific) connected to a Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were first trapped using trapping column Acclaim PepMap 100 (75 μm by 2 cm, nanoViper 2Pk, C18, 3 μm, 100 A) and then separated using analytical column AUR2-25075C18A, 25CM Aurora Series Emitter Column (25 cm by 75 μm, 1.6 μm C18) (IonOpticks). The flow rate was 300 nl/min, and a 120-min gradient was used. Peptides were eluted by a gradient from 3 to 28% solvent B (80% (v/v) acetonitrile/0.1% (v/v) formic acid) over 100 min and from 28 to 44% solvent B over 20 min, followed by a short wash at 90% solvent B. Precursor scan was from mass/charge ratio (m/z) 375 to 1600 (resolution, 120,000; AGC 3.0E6), and top 20 most intense multiply charged precursors were selected for fragmentation. Peptides were fragmented with higher-energy collision dissociation with normalized collision energy. MS/MS data were converted to peaklist using PAVA (peaklist generator that provides centroid MS2 peaklist) (61), and data were searched using Protein Prospector against the TAIR10 database (https://arabidopsis.org/). The precursor mass tolerance was set to 5 ppm, and the MS/MS2 tolerance was set to 20 ppm. Carbamidomethylcysteine was searched as a constant modification. Variable modifications include protein N-terminal acetylation, peptide N-terminal Gln conversion to pyroglutamate, and Met oxidation. Subsequent searches were performed to find peptides modified by phosphorylation. The search parameters were as above but also allowing for phosphorylation on serine, threonine, and tyrosine. A phosphorylated peptide YHGHS(phophos)MSDPGSTYR and an unphosphorylated form YHGHSMSDPGSTYR were identified. Extracted ion chromatography on each precursor ion was extracted using Skyline and quantified from each sample (62). The phosphorylation/nonphosphorylation ratio is calculated by dividing intensity of phosphorylated peptide to that of unmodified peptide and then compared between nuclear and cytosolic fraction after ethylene treatment. Three biological replicates were performed. The mass spectrometry E1 phosphorylation data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) with the dataset identifier PXD040689.
E1 PDH activity assay
Pyruvate dehydrogenase activity was determined by using the PDH Activity Assay Kit (Sigma-Aldrich, MAK183) per the manufacturer’s protocol with adaptation. In short, clean nuclear pellets after nuclear-cytoplasmic fractionation performed on 3-day-old etiolated seedlings from indicated genetic backgrounds were resuspended in 500 μl of PDH assay buffer and later subject to brief sonication shearing and centrifuge to obtain soluble nuclear proteins. Fifty microliters of the cleared nuclear contents was then used to perform the colorimetric enzymatic assay using the Pyruvate dehydrogenase assay kit. The reaction mix was incubated at 37°C. Absorbance at 450 nm was measured by using the Flex Station 3 Multi-Mode Microplate Reader every 1 min and then converted to the amount of NADH (reduced form of nicotinamide adenine dinucleotide; nanomoles per well) generated based on NADH standard curve. Each experimental group was performed with at least three replicates.
Isotope labeling and LC-MS/MS analysis of 1,2-13C2 acetyl CoA
Purified nuclei from 3-day-old etiolated seedlings from each indicated genetic backgrounds were resuspended in lysis buffer from nuclear-cytoplasmic fractionation and subjected to sonication for 30 s in Bioruptor to release nuclear content. Cleared nuclear lysates were added with 2,3-13C2 pyruvate to 2 mM as the final concentration. Reaction mixtures were gently rotated in the dark at 22°C for 3 hours for 13C isotope labeling reaction. Then, TCA was added to the reaction mixture to reach 10% as final concentration to stop the labeling and precipitate protein. Acetyl CoA extraction was performed according to literature with adjustments (63). Briefly, Oasis HLB SPE columns was first conditioned with 1 ml of methanol and equilibrated with 1 ml of water using a vacuum manifold. Cleared reaction mixture after TCA precipitation and centrifuge was loaded onto the SPE column, and SPE column was later washed once with 1 ml of water after binding. Five hundred microliters of elution solution (25 mM ammonium acetate in methanol) was used to elute acetyl CoA from SPE column and dried under nitrogen. Four replicates were performed for each genotype under each treatment. The above dried samples were reconstituted in 100 μl of mobile phases A/B (80/20) and vortexed for 1 min. Thirty microliters of the sample was thoroughly mixed with 10 μl of internal standard (Malonyl-13C3 coenzyme A). The mixed sample was directly injected on to C18 column and analyzed on Shimadzu LC-MS 8060 using gradient program. The LC-MS/MS analysis was carried out in positive electrospray ionization and multiple reaction monitoring mode (+ESI LC-MS/MS MRM). The m/z transitions used for quantification were 812.1+/305+ for 1,2-13C2 acetyl CoA and 857.1+/405+ for internal standard. The mobile phases used were the following: (A) 100% water containing 200 mM ammonium acetate and (B) 90/10 acetonitrile/water containing 10 mM ammonium acetate and 0.05% difluoro acetic acid. The LC flow was maintained at 0.25 ml min−1, and the column was maintained at room temperature. The internal calibration method was used for the calculation of analyte concentrations based on 6-point calibration curve.
Acknowledgments
We thank D. Hernandez for lab maintenance. We thank C. Petucci and Metabolomics Core of Cardiovascular Institute at University of Pennsylvania for nuclear acetyl CoA LC-MS quantification and K. S. Browning for nuclear-cytoplasmic fractionation setup. We thank P. Oliphint, A. Webb, and Microscopy and Imaging Facility at The University of Texas at Austin for the support of confocal microscope imaging. E1 phosphorylation MS analysis was supported by the Carnegie endowment fund to the Carnegie mass spectrometry facility, and we thank A. V. Reyes for assistance in MS analysis. We thank S. Briggs and JadeBio Inc. for proteomic services. We also thank Arabidopsis Biological Resource Center for the Arabidopsis T-DNA insertion mutants.
Funding: Z.S. was supported by the Research Awards from Department of Integrative Biology and the Graduate Continuing Fellowship from The University of Texas at Austin. This work was supported by grants from the National Institute of Health to H.Q. (NIH-2R01 GM115879) and Lorene Morrow Kelley Endowed Faculty Fellowship from University of Texas at Austin to H.Q.
Author contributions: Z. Shao and H.Q. designed the study. Z.S. performed most of the experiments and analysis. L.B. performed LC-MS/MS analysis of 1,2-13C2 acetyl CoA, and Y.B. performed MS analysis of E1 phosphorylation. Z. Shen performed co-IP/MS. J.G.B. and P.K. helped genetic material generation and imaging. S.K.A., T.J.D., and M.A.B. helped with biochemical assays and high-throughput sequencing and contributed to manuscript editing. S.-L.X., Z.-Y.W., S.P.B., and H.Q. supervised the study. Z.S. and H.Q. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The high-throughput sequencing data generated in this study have been deposited in the GEO database (accession no. GSE212540). The mass spectrometry E1 phosphorylation data have been deposited to the ProteomeXchange Consortium via the PRIDE with the dataset identifier PXD040689.
REFERENCES AND NOTES
Articles from Science Advances are provided here courtesy of American Association for the Advancement of Science
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/165709971
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE77396
ProteomeXchange
- (1 citation) ProteomeXchange - PXD040689
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation.
Cell, 158(1):84-97, 01 Jul 2014
Cited by: 323 articles | PMID: 24995980
EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling.
Nat Commun, 7:13018, 03 Oct 2016
Cited by: 63 articles | PMID: 27694846 | PMCID: PMC5063967
The pyruvate dehydrogenase complex at the epigenetic crossroads of acetylation and lactylation.
Mol Genet Metab, 143(1-2):108540, 16 Jul 2024
Cited by: 0 articles | PMID: 39067348
Review

 1
,
2
,*
1
,
2
,*

