Abstract
Free full text

BCR::ABL1 PROTEOLYSIS-TARGETING CHIMERAS (PROTACs): THE NEW FRONTIER IN THE TREATMENT OF Ph+ LEUKEMIAS?
Abstract
BCR::ABL1 tyrosine kinase inhibitors (TKIs) have turned chronic myeloid leukemia (CML) from a lethal condition into a chronic ailment. With optimal management, survival of CML patients diagnosed in the chronic phase is approaching that of age-matched controls. However, only one-third of patients can discontinue TKIs and enter a state of functional cure termed treatment free remission (TFR), while the remainder require life-long TKI therapy to avoid recurrence of active leukemia. Approximately 10% of patients exhibit primary or acquired TKI resistance and eventually progress to blast phase. It is thought that recurrence after attempted TFR originates from CML stem cells (LSCs) surviving despite continued suppression of BCR::ABL1 kinase. Although kinase activity is indispensable for induction of overt CML, kinase-independent scaffold functions of BCR::ABL1 are known to contribute to leukemogenesis, raising the intriguing but as yet hypothetical possibility, that degradation of BCR::ABL1 protein may accomplish what TKIs fail to achieve – eliminate residual LSCs to turn functional into real cures. The advent of BCR::ABL1 proteolysis targeting chimeras (PROTACs), heterobifunctional molecules linking a TKI-based warhead to an E3 ligase recruiter, has moved clinical protein degradation into the realm of the possible. Here we examine the molecular rationale as well as pros and con of degrading BCR::ABL1 protein. We review reported BCR::ABL1 PROTACs, point out limitations of available data and suggest directions for future research. Ultimately, clinical testing of a potent and specific BCR::ABL1 degrader will be required to determine the efficacy and tolerability of this approach.
INTRODUCTION
Chronic myeloid leukemia (CML) and one-third of acute lymphoblastic leukemia (ALL) cases are caused by BCR::ABL1, a constitutively active tyrosine kinase derived from the Philadelphia chromosome (Ph)(1). In the chronic phase of CML (CP-CML), the myeloid progenitor cell compartment is expanded, but terminal differentiation preserved. Without effective therapy, CP-CML inexorably progresses to the blast phase (BP-CML), an acute leukemia of myeloid or lymphoid phenotype with poor prognosis. Mouse models have demonstrated that BCR::ABL1 tyrosine kinase activity is required for induction of a myeloproliferative neoplasm resembling human CML(2, 3). Starting with imatinib, successive generations of BCR::ABL1 tyrosine kinase inhibitors (TKIs) have transformed CML from a lethal disease into a manageable chronic condition compatible with an almost normal life span(4–6). This remarkable success reflects the exquisite TKI sensitivity of CML progenitor cells, which are responsible for the clinical disease. In contrast, quiescent CML stem cells (LSCs) survive despite BCR::ABL 1 inhibition, which explains why only one-third of patients can discontinue TKIs and reach a state of functional cure referred to as treatment free remission (TFR)(7–9). In the remainder, active CML recurs after TKI discontinuation, indicating that fully leukemogenic LSCs have survived TKI exposure. There is evidence that BCR::ABL kinase activity is also less important at the other end of the clinical CML spectrum: TKI resistant BP-CML(10). At least half of these patients lack ‘explanatory’ BCR::ABL1 mutations, suggesting that the disease may be driven by mechanisms other than BCR:ABL1 kinase, although it remains possible that BCR::ABL1 amplification or drug efflux is involved(11). Altogether, these observations have led to the term ‘BCR::ABL1 independence’. Without effective strategies to selectively eliminate BCR::ABL protein in primary CML cells, the distinction between BCR::ABL1 kinase independence and BCR::ABL1 protein independence was of mostly semantic interest(12). However, the emergence of proteolysis-targeting chimeras (PROTACs) that degrade BCR::ABL1 has started to reframe the discussion(13). Although the notion of ‘no kinase activity, no CML’ is undisputed, there is convincing evidence that the large BCR::ABL protein chimera has scaffold functions in addition to kinase activity that contribute to leukemogenesis, suggesting that elimination of BCR::ABL1 protein might be superior to even complete inhibition of the kinase. Here we examine the data supporting scaffold functions of BCR::ABL1, review the BCR::ABL1 degraders reported to-date, and discuss future research directions.
BCR::ABL1 kinase independent cellular phenotypes and pathways
The structural features of BCR::ABL1 required to realize its full transformation potential are well described(14). Here we focus on those BCR::ABL1 functions that impact cell behavior and/or signaling even in the complete absence of kinase activity. Importantly, all available data are based cell lines or primary hematopoietic stem and progenitor cells (HSPCs) transduced with BCR::ABL1. In some cases, evidence is indirect, i.e., aberrancies persist despite inhibition of BCR::ABL1, but whether they originate from the silenced kinase itself or another intrinsic or extrinsic source is unknown. It is important to be aware of the limitations of these models. As an example, LSCs from mice with retrovirally induced CML express Alox5, which promotes leukemogenesis by enhancing leukotriene biosynthesis. Increased Alox5 expression persists upon ex vivo imatinib treatment of bone marrow (BM) cells from leukemic mice, consistent with a BCR::ABL1 kinase independent mechanism(15). However, subsequent work in human LSPCs found no evidence for increased ALOX5 expression(16). We have reported that the tetraspanin Ms4a3 is downregulated in 32D cl3 cells following transduction with p210BCR::ABL1, but not the kinase inactive p210BCR::ABL1 K1172R mutant, indicating a kinase-dependent process(10). However, after a few cell passages, imatinib treatment failed to restore MS4A3 expression, suggesting Ms4a3 expression had become uncoupled from kinase activity. These examples illustrate the challenges of distinguishing between BCR::ABL kinase-dependent and BCR::ABL1 protein-dependent functions and emphasize that readouts removed from the initial transformation event must be interpreted with caution.
BCR::ABL1 scaffold functions
There is substantial evidence that kinase-independent BCR::ABL1 functions contribute to leukemogenesis (Figure 1). Human CML progenitor cells demonstrate increased growth, reduced apoptosis, and aberrant migration and adhesion to BM stroma(17). Cord blood CD34+ cells transduced with p210BCR::ABL1 recapitulate this phenotype, exhibiting increased proliferation, reduced adhesion to fibronectin, and reduced chemotaxis to stroma-derived factor-1α(18). Imatinib treatment of these cells abrogates abnormal growth but does not completely restore normal adhesion and migration. In line with the latter, expression of p210BCR::ABL1K1172R has no effect on proliferation, but in part recapitulates the migration and adhesion phenotype. Adhesion and migration effects are abrogated by deletion of the BCR::ABL1 C-terminus, possibly reflecting the loss of F-actin binding(18, 19). We have shown that BCR::ABL1 induces p27 cytoplasmic mislocalization of the cyclin dependent kinase (Cdk) inhibitor p27 in a BCR::ABL1 kinase-independent fashion and that cytoplasmic p27 promotes leukemogenesis(20). Studies from the Perrotti lab showed that BCR::ABL1 expression, but not kinase activity, is required for activation of a JAK2/β-catenin survival/self-renewal pathway that includes inhibition of the tumor suppressor phosphatase PP2A(21). Importantly, this study demonstrated that p210BCR::ABL1K1172R suppressed PP2A activity in mouse Lin−Sca1+Kit+ (LSK) cells, indicating that BCR::ABL1 protein rather than kinase activity is critical for promoting LSC survival. Data from several labs have implicated tyrosine 177 of BCR as a key mediator of BCR::ABL1 scaffold functions (Figure 2). Phosphorylation of tyrosine 177 activates signaling through the RAS/RAF/mitogen activated kinase (MAPK) and phosphatidyl inositol 3’ kinase (PI3K) pathways(22, 23). Y177 is not only a BCR::ABL1 autophosphorylation site, but the target of upstream kinases, including LYN and JAK2 in CML cell lines and primary cells from TKI-resistant patients(24–27). CB CD34+ cells transduced with p210BCR::ABL1 Y77F are partially defective for expansion, proliferation, and survival compared to controls transduced with native BCR::ABL1(28). Increased generation of reactive oxygen species (ROS) in CML LSCs is thought to confer genetic instability, leading to BP. ROS remain elevated in LSCs treated with imatinib compared to HSCs, and part of the ROS production is dependent on Y177, suggesting that persistent Y177 phosphorylation may maintain ROS-induced genetic instability(29–31). Phosphorylation of the adaptor SHC is induced by serum, promoting its binding to the BCR::ABL1 SH2 domain and activation of RAS/MAPK signaling in a BCR::ABL1 kinase independent manner(32). In contrast to RAS/MAPK, the activation of STAT5 is highly dependent on BCR-ABL1 kinase, making pSTAT5Y705 a reliable biomarker of TKI activity(33). Mice transplanted with Stat5−/− bone marrow expressing GFP-p210BCR::ABL1 do not develop leukocytosis, although blood and bone marrow cells are BCR::ABL1+, suggesting that Stat5 drives the excess proliferation required for the manifestation of overt leukemia, but not LSC survival, while RAS/MAPK and PI3’K may be sufficient to support survival but insufficient to induce overt leukemia1(34). Scaffold functions of BCR::ABL1 might explain another puzzling observation. Kinase inactive p210BCR::ABL1 splice forms are present in some CML patients and may be enriched in patients with TKI resistance(35, 36). Although a direct contribution of these variants to TKI resistance is unlikely(37, 38), their persistence suggests they confer a net functional gain.

Cellular mechanisms impacted by BCR::ABL1 kinase-independent functions and pathways implicated in their regulation.
NON-ENZYMATIC ACTIVITIES IN CANCER-ASSOCIATED KINASES
Several cancer-associated kinases exhibit non-enzymatic functions that contribute to oncogenesis, serving as allosteric activators, scaffolds, or transcriptional regulators (reviewed in(39)). While mutated epidermal growth factor receptor (EGFR) is a strong driver of cancer cell proliferation, the effects of EGFR inhibitors are cytostatic(40–42). In line with this, prostate cancer cells remain viable upon inhibition of EGFR kinase. However, cell death ensues upon EGFR knockdown and is rescued by kinase inactive EGFR(43). Several Bruton’s tyrosine kinase (BTK) mutants isolated from patients with resistance to the BTK inhibitor ibrutinib are kinase inactive. Cells engineered to express these variants are resistant to ibrutinib, but sensitive to BTK degradation(44). The most common types of B-RAF mutations in lung adenocarcinoma reduce or abolish kinase activity. One of these mutants, BRAFD594G, was shown to function as an allosteric activator of wild type C-RAF(45). These observations are evidence that non-enzymatic kinase functions can contribute to TKI resistance. It will be interesting to see how wide-spread this phenomenon is.
PROTEOLYSIS-TARGETING CHIMERAS (PROTACs)
General considerations
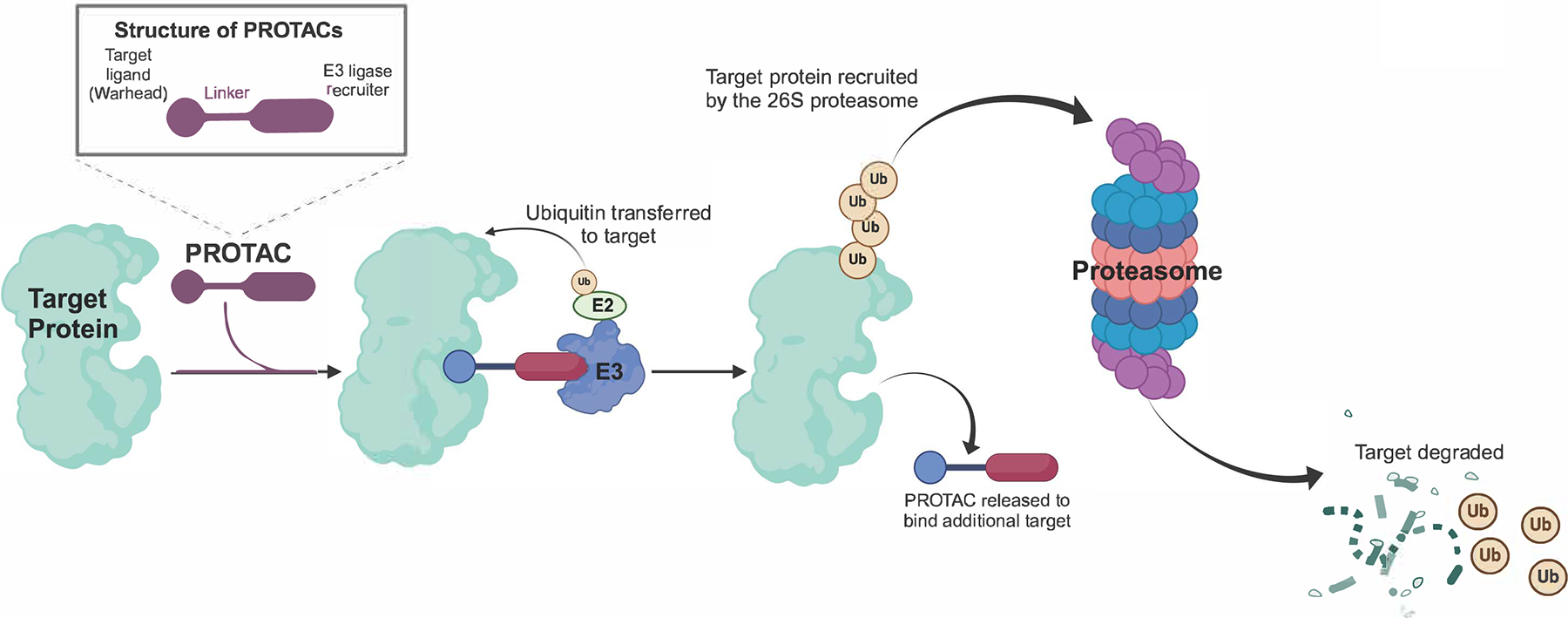
PROTACs are heterobifunctional molecules consisting of a warhead binding to the target protein connected by a linker to an E3 ligase recruiter. Physical proximity of target, E3 ligase and PROTAC in the resulting ternary complex promotes target ubiquitination and subsequent proteasomal degradation (Figure 3). Unlike TKIs, where the degree of target inhibition is determined by affinity and intracellular drug concentrations, PROTACs are recycled following each round of degradation, theoretically resulting in complete target elimination through iterative degradation cycles. PROTAC activity is not necessarily proportional to affinity, likely reflecting a requirement for flexibility to enable ubiquitin transfer to exposed lysines in the target. Degradation often reaches a concentration optimum, after which activity decreases, as formation of binary complexes is favored. This is referred to as the hook effect, a term derived from the paradoxical decrease in the dose response curve of antibody-based immune assays at increasing concentrations(46). Many PROTACs exhibit off-target protein degradation. As an example, cereblon (CRBN)-based PROTACs can cause degradation of translation termination factor GSPT1, an essential survival factor. Therefore, it is important to exclude GSPT1 degradation before attributing biological activity to degradation of the intended target protein(47). Although PROTACs derived from small molecule inhibitors often have reduced target affinity, they still retain inhibitory activity. Therefore, demonstrating that target degradation is biologically superior to inhibition requires inclusion of a degradation inactive control with identical affinity and cellular uptake.
BCR::ABL1 Degraders
Starting in 2016, a variety of BCR::ABL1 PROTACs has been reported. Dasatinib was most frequently used as warhead, but all currently approved TKIs were employed except nilotinib. CRBN and Von Hippel-Lindau (VHL) were the two most commonly used E3 ligases. The DC50 (concentration inducing 50% degradation) of published compounds ranges between 0.85 and 30,000 nM and IC50 (the concentration inducing 50% growth inhibition in K562 cells) between 0.35 and 30,000 nM. As uniform efficacy parameters are lacking, comparisons between PROTACs reported in different publications are challenging. Although the design variables are limited to warhead, linker, and E3 ligase recruiter, a multitude of compounds has been reported that defies easy classification. To provide a level of organization, we decided to discuss compounds mostly chronologically and by the labs that generated successive BCR::ABL1 PROTACs. Compounds with DC50 < 10μM, typically the best from a series, are listed in Table 1, while more extensive information is provided in Supplemental Table 1.
Table 1.
BCR::ABL1 Degraders
| Publication | Warhead and E3 Ligase |
|---|---|
| Lai AC, Toure M, Hellerschmied D, et al. Modular PROTAC Design for the | BOS-CRN |
| DAS-CRBN | |
| Burslem GM, Schultz AR, Bondeson DP, et al. Targeting BCR-ABL1 in Chronic Myeloid Leukemia by PROTAC-Mediated Targeted Protein Degradation. Cancer Res. 2019;79(18):4744–4753. | GNF5-VHL |
| Burslem GM, Bondeson DP, Crews CM. Scaffold hopping enables direct access to more potent PROTACs with in vivo activity. Chem Commun (Camb). 2020;56(50):6890–6892. | ASC-VHL |
| Shibata N, Miyamoto N, Nagai K, et al. Development of protein degradation inducers of oncogenic BCR-ABL protein by conjugation of ABL kinase inhibitors and IAP ligands. Cancer Sci. 2017;108(8):1657–1666. | HG-7-85-01-IAP |
| Zhao Q, Ren C, Liu L, et al. Discovery of SIAIS178 as an Effective BCR-ABL Degrader by Recruiting Von Hippel-Lindau (VHL) E3 Ubiquitin Ligase. J Med Chem. 2019;62(20):9281–9298. | DAS-VHL |
| Yang Y, Gao H, Sun X, et al. Global PROTAC Toolbox for Degrading BCR-ABL Overcomes Drug-Resistant Mutants and Adverse Effects. J Med Chem. 2020;63(15):8567–8583. | DAS-CRBN |
| ASC-CRBN | |
| PON-CRBN | |
| Jiang L, Wang Y, Li Q, et al. Design, synthesis, and biological evaluation of Bcr-Abl PROTACs to overcome T315I mutation. Acta Pharm Sin B. 2021;11(5):1315–1328. | PON-CRBN |
| Liu H, Mi Q, Ding X, et al. Discovery and characterization of novel potent BCR-ABL degraders by conjugating allosteric inhibitor. Eur J Med Chem. 2022;244:114810. | ASC-CRBN |
| Liu H, Ding X, Liu L, et al. Discovery of novel BCR-ABL PROTACs based on the cereblon E3 ligase design, synthesis, and biological evaluation. Eur J Med Chem. 2021;223:113645. | DAS-CRBN |
| Zhang J, Ma C, Yu Y, Liu C, Fang L, Rao H. Single amino acid-based PROTACs trigger degradation of the oncogenic kinase BCR-ABL in chronic myeloid leukemia (CML). J Biol Chem. 2023;299(8):104994. | DAS-UBR |
| E3 Recruiter | Best Compound | Structure |
|---|---|---|
| POM | BOS-6-2-2-6-CRBN |
 |
| POM | DAS-6-2-2-6-CRBN |
 |
| (S,R,S)-AHPC | GMB-475 |
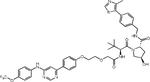 |
| (S,R,S)-AHPC | GBM-805 |
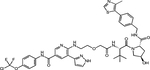 |
| MV-1 | SNIPER(ABL)-15 |
 |
| LCL161 | SNIPER(ABL)-24 |
 |
| Bestatin | SNIPER(ABL)-44 |
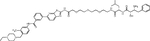 |
| MV-1 | SNIPER(ABL)-47 |
 |
| LCL161 | SNIPER(ABL)-33 |
 |
| MV-1 | SNIPER(ABL)-19 |
 |
| LCL161 | SNIPER(ABL)-39 |
 |
| (S,R,S)-AHPC | SIAIS178 |
 |
| P22D(N=4) |
 | |
| P19As(N=3) |
 | |
| P19P (N=3) |
 | |
| POM | 7o |
 |
| LEN, POM | SIAIS100 (30) |
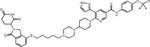 |
| LEN, THA | SAIS056 (17) |
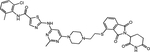 |
| ARG | ARG-PEG1-DAS |
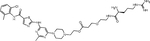 |
| LYS | LYS-PEG1-DAS |
 |
| LEU | LEU-PEG1-DAS |
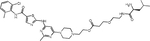 |
| PHE | PHE-PEG1-DAS |
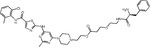 |
| Linker type (best compound) | BCR::ABL1 Degradation (best compound) | IC50 (K562 cells proliferation) | Activity in vivo |
|---|---|---|---|
 | 80% at 2.5 μM | NR | NR |
 | 60% at 1 μM | 4 nM (EC50) | NR |
 | Dmax >95%; DC50=340 nM | 1,000 nM | NR |
 | DC50=30 nM | 169 nM | K562 cells xenograft, s.c. |
 | 5 μM | NR | NR |
| 5 μM | NR | NR | |
| 10 μM | NR | NR | |
| 2 μM | NR | NR | |
| 300 nM | NR | NR | |
| 300 nM | NR | NR | |
 | 100 nM | 9 nM | NR |
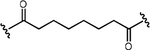 | 8.5 nM | 24 nM | K562 cells xenograft, s.c. |
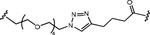 | ~10 nM | 9 nM | NR |
| 200 nM | 121 nM | NR | |
| 20 nM | 8 nM | NR | |
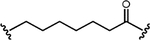 | 70% at 100 nM | 8 nM | BaF3 BCR::ABL1 cells, xenograft s.c. |
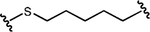 | 81.78% at 5 nM; 91.2% at 100 nM | 12 nM | NR |
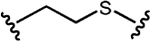 | 0.18 nM | 0.49 nM | K562 cells xenograft, s.c. |
 | 0.85 nM | 0.36 nM | NR |
| 0.99 nM | 0.46 nM | NR | |
| 0.48 nM | 0.22 nM | NR | |
| 1.56 nM | 0.30 nM | NR |
| Activity in primary CML Cells | Activity against BCR::ABL1 mutants | Off target degradation | Degradation inactive control |
|---|---|---|---|
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| YES | T315I, G250E | NR | Diastereomer |
| NR | NR | NR | Diastereomer |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | T315I; F317L; G250E, V299L | Unbiased proteomics in comparison with DAS: EPHB4, RIPK2, SRC, PDGFRß reduced | Epimer |
| NR | NR | NR | NR |
| NR | T315I | NR | NR |
| NR | E255K; T315I; H396R; V468F | FLT3 | NR |
| NR | T315I | NR | NR |
| NR | P223S; K294E; T315 M; F359I; F359V; V468F; I502L; G671R; T315I; E255V; G250E/T315I; Y253H/T315I; Y253H/255V | IKZF3,IKZF1, ZFP9 | Derivative with N - methylated glutarimide |
| NR | G250E; E255K; F317L; F317V; T315A; T315I | NR | NR |
| NR | NR | NR | NH2-PEG1-Dasa |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
| NR | NR | NR | NR |
First generation Degraders.
The Crews lab was the first to report on BCR::ABL PROTACs(13). They employed imatinib, dasatinib and bosutinib as warheads, linkers of variable length and hydrophobicity, and pomalidomide or (S,R,S)-AHPC to recruit CRBN and VHL, respectively. Although they used structural data to select the optimal point for linker attachment in each TKI, binding to ABL1 was consistently reduced compared to the parent TKI, suggesting that the increased size of PROTACs imposes additional binding constraints. TKI type and linker strongly influenced degradation activity. For example, imatinib-based PROTACs were degradation inactive, although BCR::ABL1 kinase was still inhibited, evidenced by reduction of pSTAT5 and pCRKL. While bosutinib/VHL-based degraders were inactive, dasatinib/VHL PROTACs effectively degraded ABL1, but not BCR::ABL1. In contrast, dasatinib/CRBN and bosutinib/CRBN-based compounds were consistently active against BCR::ABL1 and ABL1. These data suggest that effective BCR::ABL degradation depends on appropriate positioning within the ternary complex. DAS-6-2-2-6-CRBN, the best compound in this series, reduced viable K562 cells with a half-maximal response concentration (EC50) of 4.4 nM following 48 hours of culture.
The same group subsequently combined the allosteric TKI GNF5 with (S,R,S)-AHPC as VHL recruiter. Optimization of linker attachment and length led to the lead compound GMB-475. A diastereomeric control compound (GMB-651) with equal cell permeability but defective for recruiting VHL was prepared as a control(13). Treatment of K562 cells with GBM-475 for 18 hours reduced BCR::ABL protein with a DC50 of ~0.5μM, which translated into an IC50 of 1μM for K562 cell growth inhibition. GMB-651 was ~0.6-log less active for growth inhibition, suggesting degradation contributes to activity. Similar differences were seen for proliferation of primary CML CD34+ cells. In one sample from a blast phase CML patient, CD34+38+ and CD34+38− cells were sorted and treated with GMB-475. There was no difference in apoptosis in CD34+38+ or CD34+38− cells. Given that only a single sample was tested in this way, these data need to be considered preliminary. Of interest in the context of BCR::ABL scaffold functions, the adaptors GAB2 and SHC were highly phosphorylated in K562 cells cultured in serum containing but not in serum free media. GMB-475, but not GMB-651 reduced pSHP-2Y542 and pGAB2Y452 and abolished pSHCY239/240 in the presence of serum, implicating a BCR::ABL1 scaffold function in phosphorylation. However, no data was provided to show that this was responsible for the differences in growth inhibition. GMB-475 was also active against BaF3 cells expressing BCR::ABLT315I or BCR::ABL1G250E. The Crews lab subsequently replaced GNF5 with asciminib, yielding GMB-805, which has >10-fold higher degradation potency compared to GMB-475 and inhibited K562 cell growth with an IC50 of 169 nM, while the diastereomer was inactive at 1μM(48). GMB-805 reduced tumor growth in mice injected subcutaneously with K562 cells. As animals were treated only for three days, the diastereomer control was not included, and BCR::ABL1 degradation was not shown, it is unclear whether degradation contributed to in vivo activity.
Inhibitor of apoptosis proteins (IAP)-based PROTACs.
IAPs are a family of E3 ligases that suppress apoptosis, are negatively regulated by second mitochondria-derived activator of caspases (SMACs) and have important roles in various processes, including inflammation. Upregulation of IAPs or downregulation of SMACs is common in cancer, and several SMAC mimetics are in clinical development (reviewed in(49)). The labs of Mikihiko Naito and Masaaki Kurihara reported a series of IAP-based PROTACs termed SNIPERs (for Specific and Non-genetic IAP-dependent Protein Erasers) that combine dasatinib, GNF5, HG-7-85-01 (a preclinical type II inhibitor with activity against native and T315I mutant BCR::ABL1(50)) or imatinib with various ligands recruiting XIAP, cIAP1 and/or cIAP2(51–53). In the initial study, imatinib was linked to the cIAP recruiter bestatin by alkyl linkers of different length, yielding weak degraders active only at 30 μM(51). A subsequent study took a systematic approach, connecting the same suite of TKIs through polyethylene glycol (PEG) linkers of variable length with the IAP ligands bestatin, MV-1 or an LCL-161 derivative (Table 1, Supplemental Table 1). A compound combining dasatinib with an LCL-161 derivative termed SNIPER(ABL)-39 proved to be a highly effective BCR::ABL1 degrader, with effects at 10 nM, maximal activity at 100 nM, and a hook effect at higher concentrations. SNIPER(ABL)-39 inhibited the proliferation of K562, KCL22 and KU812 CML cell lines with an IC50 of ~10 nM, while BCR::ABL1-negative cell lines and cells expressing BCR::ABL1T315I were resistant, as expected. SNIPER(ABL)-39 also induced strong degradation of IAP proteins through self-ubiquitination. The same group subsequently reported on SNIPERs using several myristate pocket binders as warheads, including GNF-2, GNF-5, and an “ABL001 derivate”(53). Bioluminescence resonance energy transfer (BRET)-based assays revealed binding affinities (IC50) in the low nanomolar range for the ABL-001 based compounds. BCR::ABL1 degradation and growth inhibition of CML cell lines was seen at mid-nanomolar concentrations, with a hook effect at higher concentrations. As with the orthosteric TKI-based PROTACs, cIAPs were degraded. Pre-treatment with E3 ligand alone reduced cIAP, but this affected BCR::ABL1 degradation only slightly, suggesting newly synthesized cIAP is sufficient for BCR::ABL1 degradation. Nonetheless, the degradation of cIAP is clearly an undesirable feature of SNIPERs.
Advanced VHL- and CRBN-based PROTACs.
The lab of Biao Jiang reported a BCR::ABL1 PROTAC termed SIAIS178 that emerged as the most potent degrader from a series of compounds based on dasatinib as warhead and (S,R,S)-AHPC as VHL recruiter(54). SIAIS178 inhibits K562 cell proliferation with an IC50 of 24 nM and degrades BCR::ABL1 with a DC50 of 8.5 nM. Ternary complex formation between SIAIS178, VHL and BCR::ABL1 was demonstrated in BRET assays. This thorough study provided the most convincing in vivo data reported thus far, showing dose-dependent efficacy in mice subcutaneously injected with K562 cells. Importantly, a degradation inactive epimer was less active, suggesting BCR::ABL1 degradation is more effective than kinase inhibition in vivo. Unbiased proteomics was used to comprehensively map proteins targeted by SIAIS178 in comparison with dasatinib. The levels of several known dasatinib targets were reduced, while others were unaffected, although binding affinity was maintained, confirming that proper positioning of the target to facilitate access to lysines is critical. Unexpectedly, levels of ABL2, but not ABL1 nor BCR expression were reduced, although BCR::ABL1 degradation was evident on immunoblots. As the degradation defective epimer was not included for comparison, the interpretation of the proteomics data is challenging. They subsequently synthesized a series of asciminib-based PROTACs, including extensive SAR studies to optimize linker length and attachment to warhead and the E3 ligase ligand lenalidomide. The most potent molecule, compound 30 (SIAIS100), has a DC50 value of 2.7 nM and Dmax of 91.2% against BCR::ABL1 and an IC50 of 12 nM in K562 cells. Proteomics identified IKZF1, IKZF3, and ZFP91 as off-targets, in line with prior publications implicating them as lenalidomide neo-substrates(55). SIAIS induced degradation of several single BCR::ABL1 mutants as well as the BCR::ABL1G250E/T315I, BCR::ABL1Y253H/T315I, and BCR::ABL1Y253H/E255V compound mutants. However, there was no activity against BCR::ABL1F359I, BCR::ABL1I502L and BCR::ABL1V468F, likely reflecting impaired asciminib binding(56).
Yang et al. took a systematic click chemistry approach to discover CRBN-based PROTACs. Using PEG-based linkers of variable length, and pomalidomide derivatives as E3 ligase recruiters. While imatinib-based PROTACs were inactive, the most potent dasatinib, asciminib and ponatinib-based PROTACs had a DC50s of 10, 200 and 20 nM, respectively. Computational simulations suggested that adjacent domains such as the ABL1 SH2 and SH3 domains may interfere with ternary complex formation, explaining in part differences in potency. Linker length greatly influenced potency. PROTACs were generally less active in cell proliferation assays of BCR::ABL1 expressing cell lines than the corresponding TKIs, although it was not investigated whether this reflects reduced affinity or reduced uptake. Interestingly, the ponatinib-based PROTAC compound 19 (DC50 20 nM) had no degradation activity towards the ponatinib targets SRC, KIT, and PDGFR, confirming that degradation is subject to more stringent structural requirements and thus potentially more specific than kinase inhibition. Indeed, compound 19 had reduced toxicity against H9C2 rat cardiomyocytes and HUVECs compared to ponatinib. However, as cellular uptake data were not reported, reduced toxicity may also reflect reduced cellular uptake.
Jiang et al. synthesized a series of PROTACs based on GZD824 (olverembatinib), PEG or carbon chain linkers and VHL, CRBN, and cIAP1 recruiters(57, 58). They also included adamantyl as hydrophobic tag, which can flag proteins for proteasomal degradation by mimicking a misfolded protein(59). A compound termed 7o with 6-carbon chain and a CRBN ligand was most potent, with a DR50<100 nM and IC50 of 9.7 nM against BaF3 cells expressing BCR::ABL1T315I. This compound exhibits a half-life of 48 hours following intraperitoneal injection and showed in vivo activity in a subcutaneous tumor model of BaF3 cells expressing BCR::ABLT315I. Importantly, BCR::ABL1 degradation was demonstrated in tumor tissue.
Single amino acid-based PROTACs.
Presented as the first amino acid, a basic (e.g., Arg) or hydrophobic (e.g., Phe) residue can be directly recognized by the UBR1 family of E3 ubiquitin ligases(60). Based on this mechanism, Zhang et al. appended Arg, Lys, Leu, or Phe to dasatinib through PEG linkers(61). The resulting degraders were highly potent, with a DC50 of 0.48 – 1.56 nM, while an NH2-PEG1-dasatinib control was inactive. Degradation was correlated with growth inhibition of K562 cells at concentrations <1 nM. The Arg-PEG1-dasatinib degrader reduced tumor growth in mice injected subcutaneously with K562 cells, with BCR::ABL1 degradation in residual tumors. While this data highlights the efficacy of single amino acid–based PROTACs, their utility remains unclear, since off-target activity was not reported.
PROTACs targeting other BCR::ABL1 domains.
In addition to the orthosteric and allosteric sites, there are additional structural BCR::ABL1 motifs that may be targetable. For example, ABL interacting protein 1 (ABI-1) binds to the ABL1 SH3 domain and it would be conceivable to use an ABI-1 peptide as warhead(62). A conceptually interesting molecule termed PMI Bcr/Abl-R6 was recently reported, in which N-terminal sequences of the BCR oligomerization domain were replaced with a p53/MDM2 inhibitor peptide (PMI). PMI Bcr/Abl-R6 targets Ph+ cells by disrupting BCR::ABL1 oligomerization (inhibiting kinase activity), recruiting BCR::ABL1 to MDM2 for degradation, and promoting apoptosis by releasing TP53 from MDM2(63). Activity in cell lines, primary cells and a mouse model was shown. An obvious challenge for this approach, as for all peptide-based therapeutics will be to turn the tool compounds into an orally bioavailable molecule.
FUTURE DIRECTIONS
Whether BCR::ABL1 degraders will be the next frontier in CML therapy will depend on several factors. Based on a fairly extensive body of data, it is reasonable to believe that degrading BCR::ABL1 will be superior to inhibiting kinase activity. However, in the publications reviewed in this manuscript, only five PROTACs were compared with degradation inactive controls, and none provided data demonstrating comparable cellular uptake and target engagement of active and inactive compounds. Only two compounds were screened for off-target degradation by unbiased proteomics. As both showed off-target effects, attribution of biological activity to BCR::ABL degradation alone was not impossible and future work needs to address this deficiency. Only a minority of studies reported in vivo activity data, and those that did used exclusively subcutaneous xenografts of leukemia cell lines. These models have little in common with CML, where a hierarchy of cells with various levels of BCR::ABL1 dependence interact with their microenvironment in a complex fashion. Given these limitations, it is challenging to identify the most promising compounds amongst the published inhibitors. Two molecules with very extensive datasets and nanomolar activity (SIAIS178(54) and SIAIS100(64)) also have documented off-target degradation, raising concerns about their use in biological studies. Ongoing work in our lab is focused on complete characterization of a series of novel BCR::ABL1 degraders. Preliminary data of the lead molecule LPA81 indicate high potency and specificity, but additional work is needed to confirm this data(65). Another unknown is what maximal BCR::ABL1 kinase inhibition alone might achieve clinically. Data from the aborted EPIC trial of frontline ponatinib in CP-CML suggest that highly effective kinase inhibition early on may dramatically increase the rates of deep molecular response(66). Evidently, the caveat here is that effectiveness may in part reflect ponatinib’s off-target activity. A trial of ‘high dose’ (200 mg twice daily) of asciminib, effective and well tolerated in patients with BCR::ABL1T315I, in newly diagnosed CP-CML may give the answer(67). A fundamental limitation of all published BCR::ABL1 PROTAC studies is the dearth of data on primary CML cells. Only one study used CD34+38− cells, which are at least enriched in CML stem cells (LSCs) but tested only one sample(68). If LSCs are indeed more sensitive to BCR::ABL1 degradation than kinase inhibition, that would be a very strong impetus to develop a clinical BCR::ABL1 PROTAC. For such a compound to be clinically viable in the setting of CP-CML, it would be imperative to develop an orally bioavailable molecule. To our knowledge, no such compound has been reported. Even without highly bioavailable PROTACs, rigorously designed ex vivo experiments using established LSC readouts such as long term culture-initiating cells (LTC-ICs) and degradation deficient PROTAC controls may provide convincing evidence. Another concern is toxicity. ABL1 TKIs are well tolerated, but whether the same will be the case for ABL degraders is unknown. Abl1 null mice are born runted and at a lower rate than expected, and Abl1/Abl2 double knockout is embryonically lethal(69, 70). Are ABL TKIs well tolerated because ABL1 and ABL2 are critical only during development or because the scaffold functions remain intact? Generating mice transgenic for a kinase deficient Abl1/2 variant would be informative as would be toxicity studies with longer drug exposure. Another major knowledge gap is that we do not know whether the E3 ligases used by BCR::ABL1 PROTACs, are expressed in LSCs, especially human LSCs defined by more robust combinations of immunophenotypic markers. For instance, the CD34+CD35+CD38−KITlowCD45RA+ immunophenotype may afford much greater enrichment for LSCs than CD34+38−(71). Despite a wealth of reported pathways with potential for synthetic lethality when targeted in combination with BCR::ABL1 TKIs, few candidates have been tested clinically, in part reflecting the reluctance of patients responding to TKIs to enroll in complex, time-intense combination trials, an impediment that would not apply to a single agent BCR::ABL1 degrader(72). At the other extreme of the clinical CML spectrum is BP-CML(73, 74). Current thinking holds that this disease is mostly driven by alternative pathways, but surprisingly, despite often complex chromosomal rearrangements, loss of BCR::ABL has been reported only in a single patient(75). Is this because BCR::ABL1 scaffold functions are still required? If so, this would provide a strong rationale for developing a BCR::ABL1 degrader to treat advanced CML and Ph+ ALL. Generating data to validate or reject this concept should be a priority for the field.
ACKNOWLEDGEMENTS
MWD was supported in part by R01CA268496, R01CA257602, and R01CA254354. WT was supported in part by NIH R35GM148266 and NIH R01CA284689.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
Michael Deininger is a consultant for Blueprint Medicines, Novartis, CTIBioPharma, Incyte, Dava Oncology, Pfizer, Cogent. Weiping Tang is a cofounder and shareholder of Chimergen Therapeutics Inc.
REFERENCES
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/166141329
Article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Proteolysis-Targeting Chimeras (PROTACs) targeting the BCR-ABL for the treatment of chronic myeloid leukemia - a patent review.
Expert Opin Ther Pat, 33(6):397-420, 01 Jan 2023
Cited by: 2 articles | PMID: 37494069
Review
Targeting BCR-ABL1 in Chronic Myeloid Leukemia by PROTAC-Mediated Targeted Protein Degradation.
Cancer Res, 79(18):4744-4753, 16 Jul 2019
Cited by: 96 articles | PMID: 31311809 | PMCID: PMC6893872
Differentiation status of primary chronic myeloid leukemia cells affects sensitivity to BCR-ABL1 inhibitors.
Oncotarget, 8(14):22606-22615, 01 Apr 2017
Cited by: 13 articles | PMID: 28186983 | PMCID: PMC5410248
Integrated drug profiling and CRISPR screening identify BCR::ABL1-independent vulnerabilities in chronic myeloid leukemia.
Cell Rep Med, 5(5):101521, 22 Apr 2024
Cited by: 2 articles | PMID: 38653245 | PMCID: PMC11148568
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01 CA257602
Grant ID: R01 CA268496
Grant ID: R01 CA284689
Grant ID: R01 CA254354
NIGMS NIH HHS (1)
Grant ID: R35 GM148266
U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI) (3)
Grant ID: R01CA254354
Grant ID: R01CA257602
Grant ID: R01CA268496
U.S. Department of Health & Human Services | National Institutes of Health (NIH) (2)
Grant ID: R35GM148266
Grant ID: R01CA284689
U.S. Department of Health & Human Services | NIH | National Cancer Institute (3)
Grant ID: R01CA257602
Grant ID: R01CA254354
Grant ID: R01CA268496
U.S. Department of Health & Human Services | National Institutes of Health (2)
Grant ID: R35GM148266
Grant ID: R01CA284689