Abstract
Free full text

Neuroinflammation as a cause of differential Müller cell regenerative responses to retinal injury
Abstract
Unlike mammals, some nonmammalian species recruit Müller glia for retinal regeneration after injury. Identifying the underlying mechanisms may help to foresee regenerative medicine strategies. Using a Xenopus model of retinitis pigmentosa, we found that Müller cells actively proliferate upon photoreceptor degeneration in old tadpoles but not in younger ones. Differences in the inflammatory microenvironment emerged as an explanation for such stage dependency. Functional analyses revealed that enhancing neuroinflammation is sufficient to trigger Müller cell proliferation, not only in young tadpoles but also in mice. In addition, we showed that microglia are absolutely required for the response of mouse Müller cells to mitogenic factors while negatively affecting their neurogenic potential. However, both cell cycle reentry and neurogenic gene expression are allowed when applying sequential pro- and anti-inflammatory treatments. This reveals that inflammation benefits Müller glia proliferation in both regenerative and nonregenerative vertebrates and highlights the importance of sequential inflammatory modulation to create a regenerative permissive microenvironment.
INTRODUCTION
Retinal neurodegenerative diseases in mammals ultimately result in permanent vision loss. In contrast, some nonmammalian vertebrate species can regenerate their retina post-injury through the reprogramming of Müller glia into retinal stem cells. Under physiological conditions, Müller cells play multiple and crucial roles that ensure the maintenance of retinal homeostasis and visual function (1). In response to neuronal damage, they undergo reactive gliosis and can reenter the cell cycle. In zebrafish, their efficient proliferation is accompanied by neurogenic engagement and subsequent production of retinal neurons (2). In Xenopus, we uncovered that, following mechanical injury, conditional chemogenetic rod cell ablation, or CRISPR-Cas9–mediated rhodopsin (rho) gene knockout (a model of retinitis pigmentosa), Müller glia get recruited as well. They reprogram toward a stem/progenitor state, proliferate, and eventually generate new neurons (3, 4). Numerous studies have started to identify the intrinsic regulatory network underlying the regenerative response of Müller cells to injury (2, 5–13). Recently, attention has shifted toward extrinsic factors emanating from the cellular microenvironment where Müller glia reside. Particular emphasis has been placed on the immune microenvironment, with microglia emerging as central regulators. These resident mononuclear phagocytes behave as sensors of the retinal extracellular milieu, and their activation is a hallmark of the retinal reaction to injury. Once activated, they migrate to the injured site and actively engage into the engulfment and clearance of cell debris (14). Their role extends beyond this function, and they have been identified in both zebrafish and chick as critical positive regulators of Müller glia recruitment and proliferation in degenerative contexts (14–19).
In mammals, the proliferative potential of Müller glia is extremely limited and does not result in efficient neuronal replacement (8, 13). Among the limiting factors that have been proposed to account for this weak regenerative potential is the innate immune reaction. Accordingly, inflammatory signals were shown to have opposite outcomes in the damaged brain of zebrafish and mammals, stimulating reparative neurogenesis in the former (20) while reducing it in the latter (21, 22). Similar interspecies differences were also observed within the retina of fish and birds versus mouse (9). This has led to the proposition that the nature of the neuroinflammatory response accounts for the differential regenerative capacities of nonmammalian vertebrates and mammals.
In this study, we aimed at further investigating the role of inflammation on the response of Müller cells to injury by comparing species with different regenerative capacities, the mouse and Xenopus. Despite the efficient regenerative capacity of the Xenopus laevis retina, we unexpectedly found in our models of mechanical injury and conditional chemogenetic rod ablation that the behavior of Müller cells is stage-dependent, with a very limited proliferative response observed in young tadpoles compared to old ones (3). In the present study, we further exemplify this phenomenon in our rho knockout experimental paradigm and provide data suggesting that Müller cell refractoriness at early stages is not due to an intrinsic inability to respond to injury but rather to an inadequate/insufficient inflammatory reaction. We further show through comparative analyses in Xenopus and mice that pro-mitogenic effects of inflammatory signaling on Müller glia are shared by both regenerative and nonregenerative vertebrates. Last, we highlight the importance of sequential pro- and anti-inflammatory treatments in mammals to promote both the proliferation of Müller cells and their neurogenic potential.
RESULTS
The proliferative response of Xenopus Müller cells to injury tremendously increases as tadpoles age
Taking advantage of our X. laevis model of retinitis pigmentosa based on CRISPR-Cas9–mediated rhodopsin knockout (rho crispants), we investigated the ability of Müller cells to exit quiescence at different tadpole stages. In this model, photoreceptor degeneration starts by the end of embryogenesis (from stage 40) and triggers the proliferation of cells mainly located in the inner and outer nuclear layers. In old tadpoles (stages >55), around 40% of them were identified as Müller cells (4). However, we found here that Müller glia proliferative response to degeneration differs notably with the tadpole age (Fig. 1, A and B). As previously shown after a mechanical injury or conditional chemogenetic rod cell ablation (3), only very few cells reentered the cell cycle in young rho crispant tadpoles (stage 45 to 48, <10 cells per section), despite an already extensive degeneration of photoreceptors (4). In contrast, a substantial number of 5′-bromo-2′-deoxyuridine (BrdU)–positive cells were found at older stages (stage 59, close to 100 cells per section). We ruled out a potential retinal size bias by confirming that a difference between young and late stages persisted even when the number of BrdU-labeled cells per section was normalized to the entire surface of the retinal section (fig. S1). These young (stage 45/48) and old (stages >55) stages will henceforth be referred to as refractory and permissive, respectively. In tadpoles at refractory stages, it thus appears that either Müller cells are intrinsically unable to reenter the cell cycle or the microenvironment is not conducive for their proliferative response to occur.
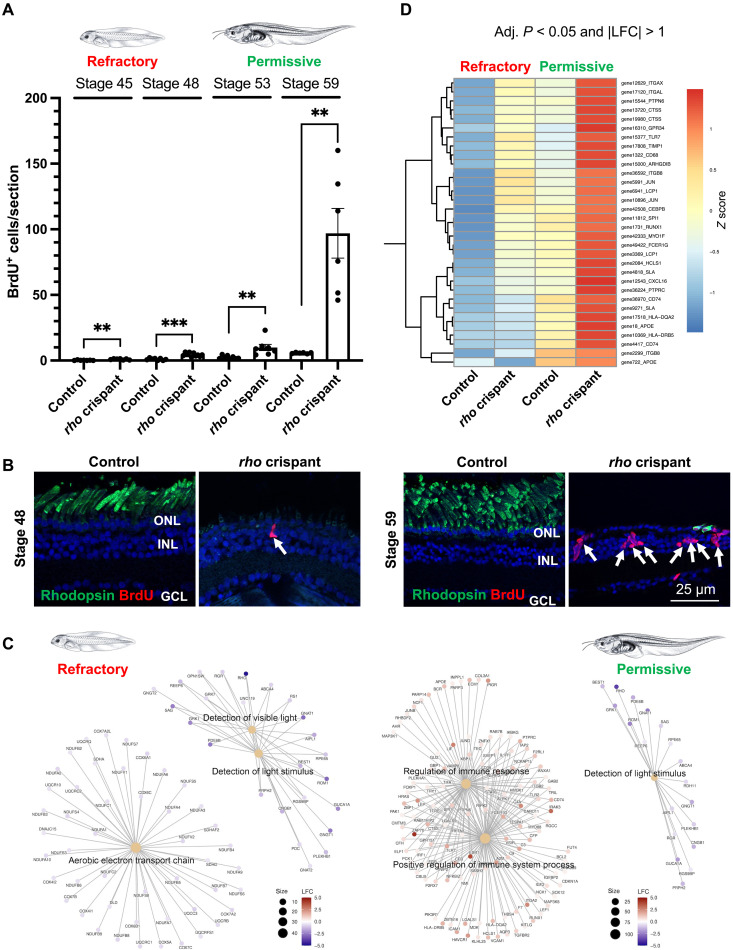
(A and B) Quantification of BrdU-positive cells in control and rho crispant tadpole retinal sections at different stages, following exposure to BrdU solution for 3 days. Data are represented as means ± SEM, and each point represents one retina. **P < 0.01; ***P < 0.001 (Mann-Whitney test). Shown in (B) are representative images of retinal sections colabeled for BrdU and rhodopsin at a young (refractory) and old (permissive) tadpole stage. Rhodopsin labeling shows the efficacy of rho knockout, and the absence of outer segments indicates degeneration of rods in rho crispants at both stages. Nuclei are counterstained with Hoechst. Arrows point to BrdU-positive cells. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (C) Gene-Concept network (cnetplot) based on the enriched biological processes at refractory and permissive stages (rho crispant tadpoles relative to controls), retrieved from GSEA results of LFC ranked gene list (see table S1). (D) Extract from the clustered heatmap of DEGs belonging to the microglial molecular signature. All genes regrouped here show a higher expression at the permissive stage in rho crispant tadpoles compared to all other conditions (refer to fig. S4 for the complete heatmap).
Inflammatory gene expression is enriched at permissive stages in response to injury
To gain insights into the molecular mechanisms underlying the variability of Müller cell response to injury, we undertook a bulk transcriptomic analysis on rho crispant and control retinas, at both refractory (stage 47 to 48) and permissive (stage 59 to 62) stages. The clustering of top expressed genes clearly discriminated rho crispant samples from controls and confirmed the reduced expression of rho from its three loci (fig. S2, A and B). Principal components analysis on all expressed genes highlighted the influence of the knockout and developmental stage on gene expression in the samples (fig. S2C). Differential expression analysis was then conducted by comparing rho crispant to control retinas at refractory or permissive stages. Log2 fold change (LFC) and adjusted P values were calculated for each gene. Using all expressed genes, we first conducted a gene set enrichment analysis (GSEA) (23, 24) on genes ranked by their LFC at refractory or permissive stages. Enriched biological processes were then identified (Fig. 1C and table S1 for the top 10 biological processes). At both refractory and permissive stages, genes associated with the detection of light stimulus were down-regulated in rho crispants compared to controls, in agreement with their expected degenerative phenotype. At the refractory stage, this was accompanied by the down-regulation of genes associated with aerobic electron transport chain, as previously described upon photoreceptor injury (25). Noticeably, damaged retinas at the permissive stage exhibited a notable enrichment, among differentially expressed genes (DEGs), of genes associated with several biological processes related to the immune response (Fig. 1C, right panel, and table S1). We then focused our transcriptomic analysis on genes that were statistically differentially expressed (|LFC| > 1, adjusted P < 0.05; fig. S3A), either at refractory stages (456 DEGs, 182 up-regulated and 274 down-regulated) or at permissive stages (272 DEGs, 136 up-regulated and 136 down-regulated). Identification of molecular cell-type signatures through an overrepresentation analysis highlighted microglia and photoreceptors as the most affected cell types at the permissive and refractory stages, respectively (fig. S3B). To assess how the expression of microglia-associated genes was altered in the different samples, we generated an expression heatmap regrouping the 83 DEGs (|LFC| > 1, adjusted P < 0.05) belonging to all microglia categories (fig. S4). Various expression profiles were observed, with some genes being down-regulated upon degeneration while others being up-regulated. Thirty-two microglial genes shown in Fig. 1D were up-regulated in rho crispant retinas at both stages, but their expression reached a much higher level at the permissive stage compared to the refractory one. Among them, we found genes encoding markers of activated microglia such as cd68 (26), cd74 (27), and cathepsin S (ctss) (28) or key inflammatory genes, including itgal (encoding Integrin Subunit Alpha L; also known as CD11a) (29, 30), itgax (encoding CD11c) (31), TIMP metallopeptidase inhibitor 1 (timp1) (32), ptprc (encoding CD45) (33), cxcl16 (34), and ptpn6 (encoding SHP-1) (35) (Fig. 1D). Taken collectively, these findings suggest that the stage-dependent regenerative potential of the Xenopus retina may stem from divergences in the nature/intensity of inflammatory reactions in response to injury.
The abundance of microglia in the retina positively correlates with the extent of Müller cell proliferation
On the basis of these transcriptomic results, we examined if the abundance of microglial cells in the retina might differ according to the tadpole stage. Labeling with fluorescent Isolectin B4 (IB4), a marker of microglia/macrophages (36), unexpectedly revealed very few microglial cells in undamaged retinas from young tadpoles (stage 45 to 48; <1 cell per section). Their number then progressively increased at later stages, reaching around 10 cells per section by stage 60 (Fig. 2, A and B). These cells were mainly located in the inner plexiform layer and had the characteristics of resting microglia, as evidenced by their ramified morphology. In the rho crispant degenerative context, the number of IB4-positive cells was significantly increased in both young and old tadpoles compared to controls (Fig. 2, C and D). However, it was here again considerably higher at the permissive stage. These results highlight that the number of microglia (and/or infiltrating macrophages) aligns with the age-dependent ability of Müller cells to proliferate: very low in the refractory period and higher at permissive tadpole stages.
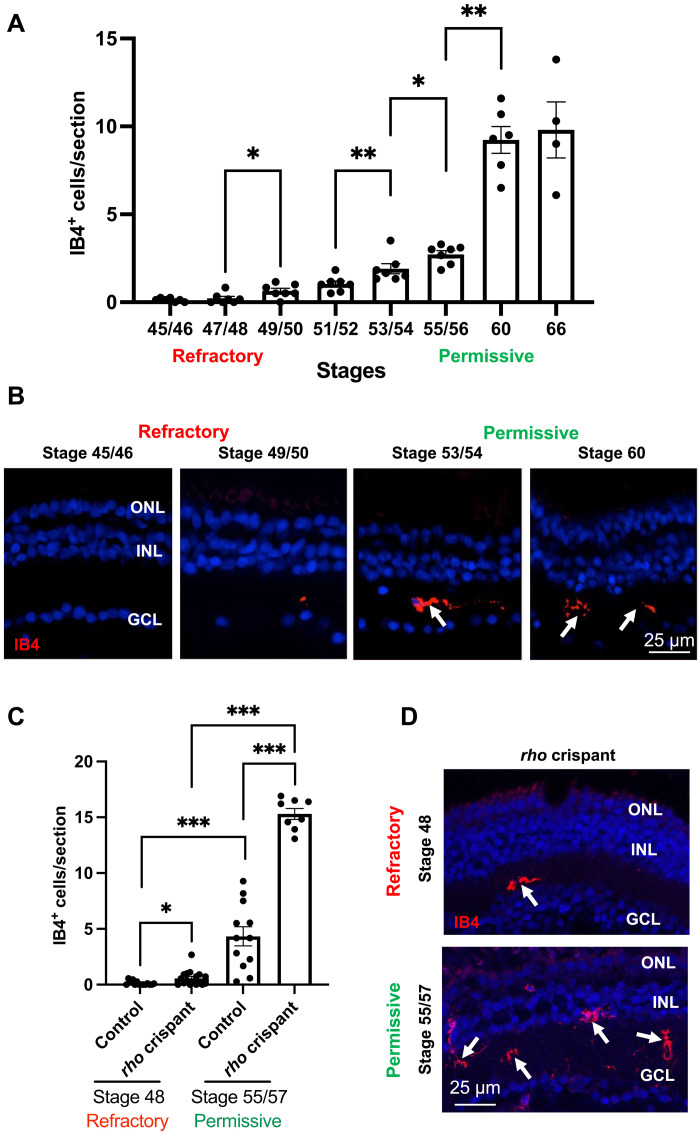
(A and B) Quantification of IB4-positive cells (microglia) on retinal sections from wild-type tadpoles at different stages. Shown in (B) are representative images of retinal sections at two refractory and two permissive stages. (C and D) Quantification of IB4-positive cells on retinal sections from control and rho crispant tadpoles at a refractory and a permissive stage. Shown in (D) are representative images of retinal sections. Nuclei are counterstained with Hoechst. Arrows point to IB4-positive cells. Data are represented as means ± SEM, and each point represents one retina. *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
Neuroinflammation enhances the proliferative response of Xenopus Müller cells to injury at permissive stages
To functionally assess the link between the intensity/effectiveness of the neuroinflammatory response and Xenopus Müller glia proliferative potential, we first asked if a stronger pro-inflammatory context might increase the proportion of Müller cells that reenter the cell cycle at permissive stages. To address this question, we turned to a degenerative model we recently developed, based on cobalt chloride (CoCl2) intraocular injection (37). In the zebrafish retina, CoCl2-mediated neurotoxic damage leads to a strong enhancement of microglial cell number (38). In Xenopus, we observed as well that the retinal degeneration caused by 25 mM CoCl2 (Fig. 3, A and B) was accompanied by a massive increase in IB4-positive cell number (Fig. 3, C to E). On the basis of their ameboid morphology, these cells presumably correspond to activated microglia and/or infiltrating macrophages. They were found about four times more numerous than in age-matched rho crispant individuals, suggesting that CoCl2 causes a particularly severe neuroinflammation reaction. In line with this, we observed a significant up-regulation of the general marker of activated phagocytic microglia/macrophages, cd68 (Fig. 3, F and G). Expression of genes encoding several other pro-inflammatory cytokines, such as tumor necrosis factor–α (TNF-α) or interleukin-1β (IL-1β) was increased as well, together with an up-regulation of anti-inflammatory ones, such as IL-10. Noticeably, this inflammatory reaction was accompanied by nearly 100% of Müller glia reentering the cell cycle, as inferred from Yes1 associated transcriptional regulator (Yap) and Proliferating cell nuclear antigen (PCNA) colabeling [Fig. 3, H to J; compared with no more than 40% in rho crispant individuals (4)].
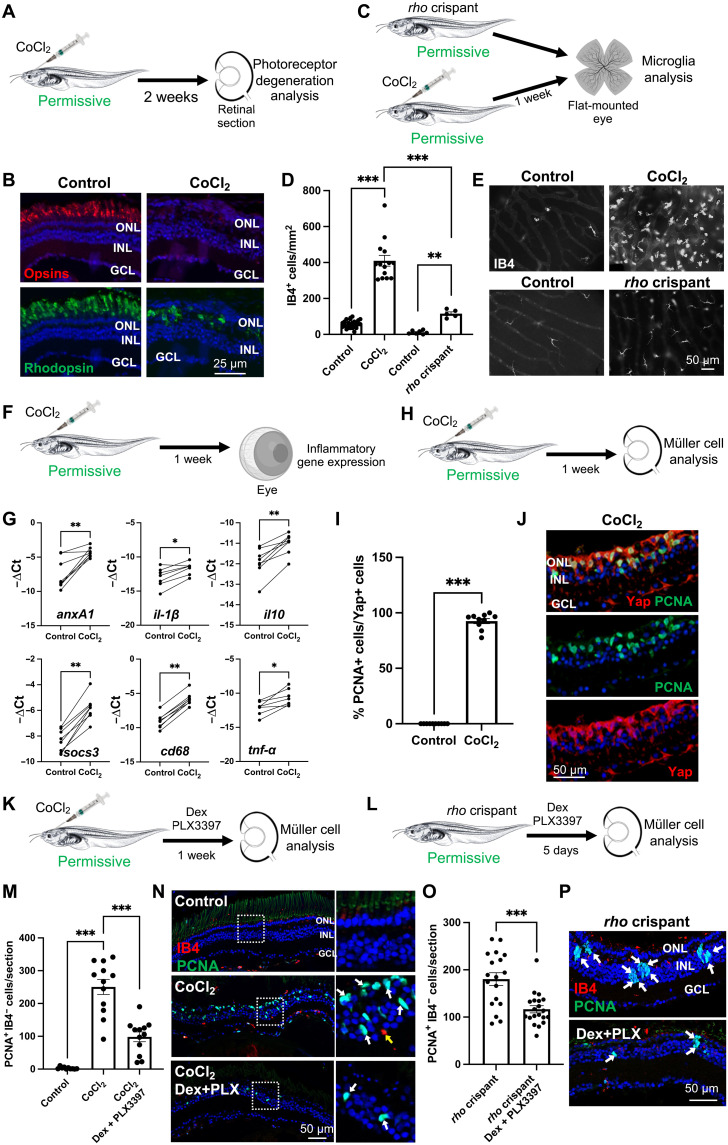
(A) Procedure used in (B). Tadpoles at a permissive stage were intraocularly injected with 25 mM CoCl2 and processed 2 weeks later for photoreceptors immunolabeling. (B) Retinal sections labeled for opsins (cones) or rhodopsin (rods). (C) Procedure used in (D) and (E). CoCl2-injected or rho crispant tadpoles and their respective controls were processed at a permissive stage for immunolabeling of microglia on flat-mounted retinas. (D and E) Quantification of IB4-positive and representative images. (F) Procedure used in (G). Tadpoles at a permissive stage were injected with CoCl2, and their retinas were dissected for qPCR analysis. (G) Expression of inflammatory genes. Shown are −ΔCt values of paired samples. Each dot represents one biological replicate. *P < 0.05; **P < 0.01 (Wilcoxon matched-paired test). (H) Procedure used in (I) and (J). Tadpoles at a permissive stage were injected with CoCl2 and processed 1 week later for immunolabeling. (I and J) Proportion of PCNA-positive cells (proliferative cells) among Yap-expressing ones (Müller cells). Shown in (J) are representative images of retinal sections. (K and L) Procedures used in (M) and (N) and (O) and (P), respectively. Tadpoles injected with CoCl2 (K) or rho crispant tadpoles (L) were soaked in PLX3397 plus Dex solution for 1 week starting 1 day before the CoCl2 injection or for 5 days as indicated and processed for immunolabeling. (M to P) Quantification of PCNA-positive, IB4-negative cells (proliferative Müller glia). Shown in (N) and (P) are representative images of sections. White boxes in (N) are enlarged on the right. White and yellow arrows point to PCNA-labeled and IB4-labeled cells, respectively. Nuclei are counterstained with Hoechst. Except in (G), data are represented as means ± SEM, and each point represents one retina. **P < 0.01; ***P < 0.001 (Mann-Whitney test). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (C) and (F) created using BioRender.com.
To question the cause-and-effect relationship between CoCl2-dependent inflammation and the intense proliferative response of Müller cells, we then treated CoCl2-injected tadpoles with a mix of pexidartinib (PLX3397) and dexamethasone (Dex). PLX3397 is a potent inhibitor of the colony-stimulating factor 1 receptor (CSF1R) and is commonly used to deplete microglia/macrophage populations (15, 39). Dex is a glucocorticoid with anti-inflammatory properties (40), which has been successfully used in the zebrafish retina to reduce the number of microglial cells (19, 41). Individually, these compounds had modest effects in Xenopus (not shown), so we used them in combination. Such a treatment in Xenopus likely diminished inflammation, as assessed by the decreased number of IB4-positive cells, while not altering the extent of CoCl2-induced cone and rod photoreceptor degeneration (fig. S5, A to C). Compared with CoCl2 injection alone, addition of these compounds significantly reduced the number of proliferative Müller glia identified as PCNA-positive/IB4-negative cells (Fig. 3, K, M, and N). Strengthening this result, the same effects were observed on both IB4-positive and Müller glia when PLX3397 and Dex were applied on rho crispant individuals (Fig. 3, L, O, and P, and fig. S5, A and D). Here again, no interference with retinal degeneration was observed (fig. S5E). Together, these data reveal that an inflammatory microenvironment is required for efficient Müller glia cell cycle reentry in the degenerative Xenopus retina at permissive stages.
Neuroinflammation triggers quiescence exit of Xenopus Müller cells upon degeneration at refractory stages
We next questioned if the failure of Müller cells to respond to injury in young tadpoles might be due to insufficient inflammatory reaction. To test this hypothesis, we assayed if CoCl2-induced neuroinflammation might activate the proliferation of Müller cells at refractory stages (Fig. 4, A to D). As expected, CoCl2 intraocular injection in young tadpoles increased the number of IB4-positive cells, and this increase was mitigated upon Dex and PLX3397 cotreatment (Fig. 4B). We next analyzed the extent of Müller cell proliferation and found an average of 82 cells per section that were 5-ethynyl-2’-deoxyuridine (EdU)-positive/IB4-negative (Fig. 4C), compared to less than 5 in aged-matched rho crispant individuals (Fig. 1A). This demonstrates that Müller cells are not intrinsically unable to reenter the cell cycle at refractory stages. Furthermore, the increase in proliferative Müller cells was abolished when CoCl2-injected tadpoles were raised in a medium containing Dex and PLX3397 (Fig. 4C), thus highlighting the pivotal role of inflammation in Müller cell reactivation. To strengthen these results, we turned to alternative approaches and stimulated neuroinflammation in young rho crispants, using either zymosan (Fig. 4, E and F to H) or heat-killed bacteria (42) injection (Fig. 4, E and I to K). Zymosan, a polysaccharide from the cell wall of Saccharomyces cerevisiae (43), was previously successfully used in the zebrafish retina to trigger inflammation (41). Both regimens in Xenopus resulted in a significant increase in IB4-positive cell number within the retina (Fig. 4, F and I). Compared to controls, this was associated with enhanced proliferation of Müller cells, as assessed by quantification of BrdU-positive/IB4-negative cells (Fig. 4, G and J) and BrdU/Yap colabeling (fig. S6). Together, these data reveal that an inflammatory environment is sufficient to push Xenopus Müller cells out of quiescence at a refractory stage.
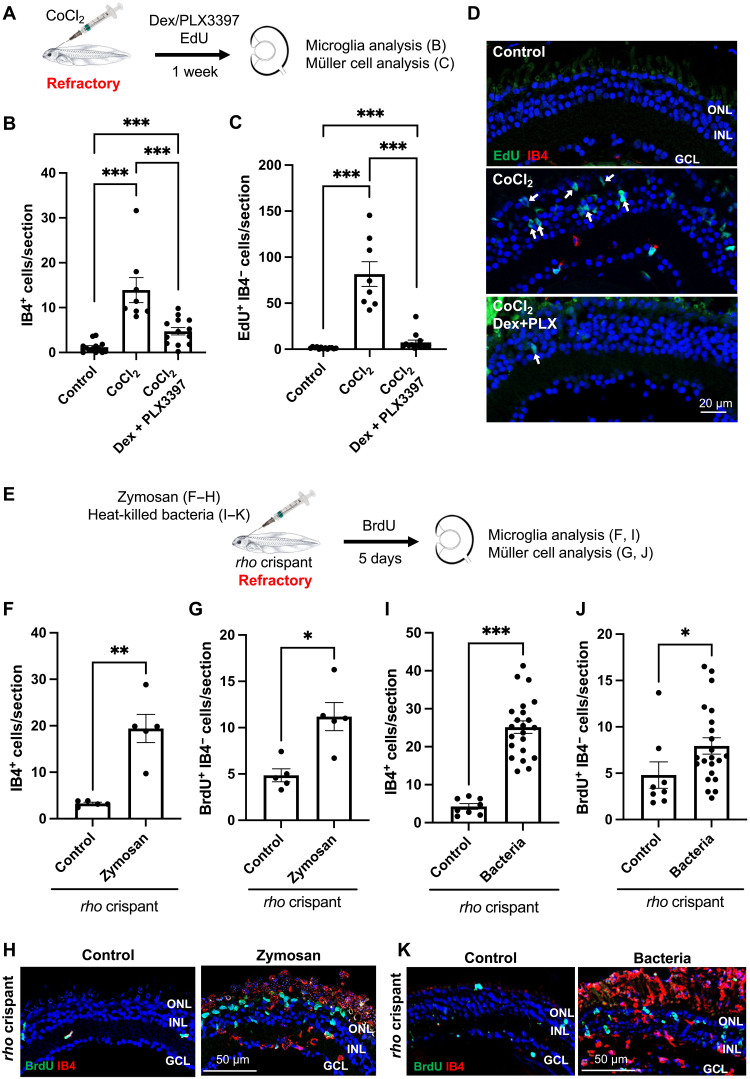
(A) Procedure used in (B) to (D). Tadpoles at a refractory stage were intraocularly injected with 25 mM CoCl2, soaked for 1 week in PLX3397 plus Dex solution, exposed to EdU for the last 3 days and then processed for immunolabeling on retinal sections. (B to D) Quantification of IB4-positive cells [microglia (B)] and EdU-positive, IB4-negative cells [proliferative Müller glia (C)] on retinal sections. Shown in (D) are representative images of retinal sections. Arrows point to EdU-positive/IB4-negative cells in the INL and ONL. (E) Procedure used in (F) to (K). rho crispant tadpoles at a refractory stage were intraocularly injected with zymosan [(F) to (H)] or heat-killed bacteria [(I) to (K)], exposed to BrdU solution for 5 days, and then processed for immunolabeling on retinal sections. (F to K) Quantification of IB4-positive cells [microglia (F) and (I)] and BrdU-positive, IB4-negative cells [proliferative Müller glia (G) and (J)]. Shown in (H) and (K) are representative images of retinal sections. Nuclei are counterstained with Hoechst. Data are represented as means ± SEM, and each point represents one retina. *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
LPS-induced inflammatory response stimulates the proliferation of mouse Müller cells
Given the ability of inflammatory signaling to activate Xenopus Müller cells, at a stage where they typically remain quiescent upon injury, we then logically wondered if such pro-proliferative activity could be leveraged to stimulate mammalian Müller cells. To address this question, we cultured adult mouse retinal explants, a well-established model of retinal degeneration (44, 45), in the presence of lipopolysaccharide (LPS). This bacterial endotoxin is commonly used to stimulate the pro-inflammatory response of brain microglia both in vitro and in vivo (46, 47). Of note, in this organotypic model, microglia are the only present inflammatory cells because infiltration of circulating monocytes/macrophages cannot occur. We determined that the proliferation of Müller cells in control explants starts at 5 days ex vivo (DEV), as assessed by colabeling of EdU and SOX9, a marker of Müller cells (Fig. 5, A to C) (48). Yet, the extent of Müller glia cell cycle reentry remained modest, with an average of 17 proliferative cells per field. From DEV5 onward, LPS treatment significantly increased this number to almost 100 cells per field. Prolonged microglial activation in mammals has been associated with deleterious effects in neural tissues (21, 22). We thus investigated if LPS challenge could have exacerbated retinal damage or stress, which could account for the amplified proliferative response of Müller cells. We first examined cell death in retinal explants following LPS exposure, by calculating the expression ratio of two pro- and anti-apoptotic genes (Bax and Bcl2, respectively), a well-recognized indicator of apoptosis extent (fig. S7, A and B) (49). No significant differences in Bax/Bcl2 mRNA ratios were observed between control and LPS-treated retinal explants. We next assessed expression of the Müller cell marker glial fibrillary acidic protein (GFAP), whose up-regulation is considered a hallmark of retinal stress (1). LPS treatment did not modify Gfap mRNA levels compared to the control group (fig. S7, A and C). Together, these results suggest that the higher amount of proliferative Müller cells in LPS-treated explants does not result from enhanced neurotoxicity or from increased stress reactivity of Müller cells.
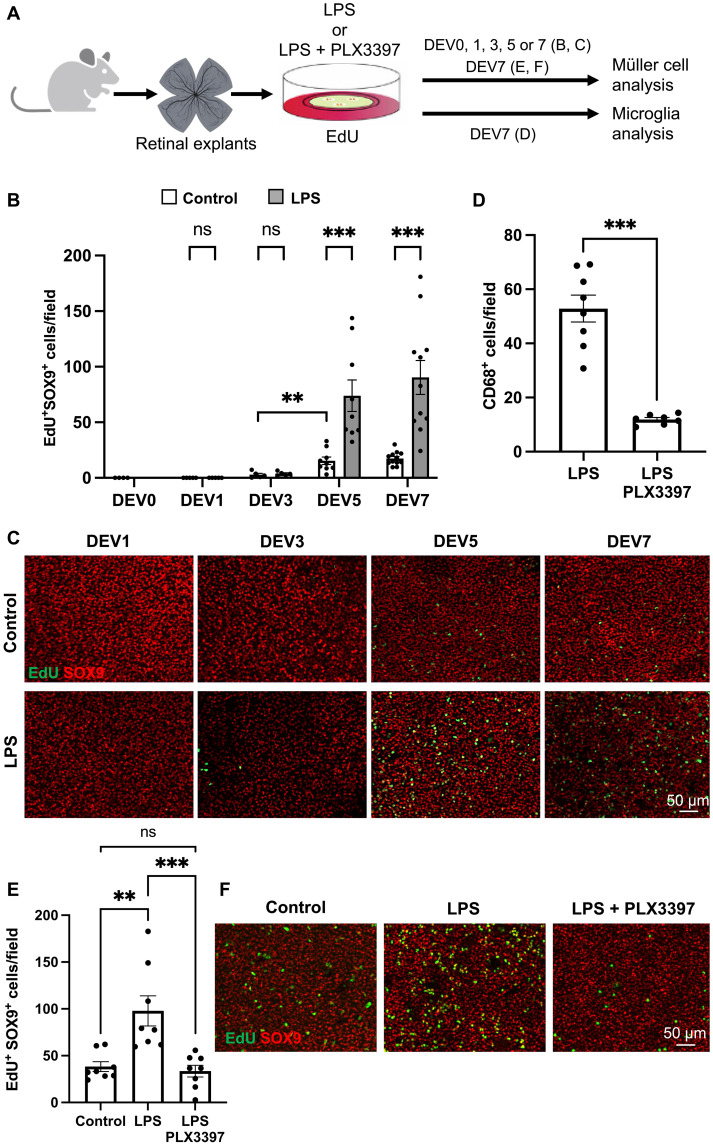
(A) Procedure used in (B) to (F). Retinas from wild-type mice were flattened and cultured for 1 to 7 days (DEV1 to DEV7) in the presence of EdU alone (control) or together with either LPS alone [(B) and (C)] or LPS plus PLX3397 [(D) to (F)]. Retinal explants were then processed for immunostaining analysis. DEV0 corresponds to an uncultured retina. Created using BioRender.com. (B and C) Quantification of double EdU-positive and SOX9-positive cells (proliferative Müller cells) at different time points in control and LPS-treated retinal explants. Shown in (B) are representative images of flat-mounted retinas. (D) Quantification of CD68-positive cells (activated microglia) in retinal explants treated with LPS or with LPS plus PLX3397. (E and F) Quantification of double EdU-positive and SOX9-positive cells in control, LPS-treated, and LPS plus PLX3397-treated retinal explants. Shown in (F) are representative images of flat-mounted retinas. Data are represented as means ± SEM, and each point represents one retinal explant. ns, not significant; **P < 0.01; ***P < 0.001 (Mann-Whitney test).
We then aimed at determining if the LPS mitogenic effect on Müller glia requires microglial cells. To do so, we analyzed the proliferative response of Müller cells to LPS in microglia-depleted explants, through a cotreatment with PLX3397. Of note, previous findings in another murine model of retinal degeneration showed that PLX3397-dependent microglia depletion does not influence the extent of retinal degeneration (50). As expected, exposure to LPS alongside PLX3397 significantly reduced the number of CD68-positive microglial cells, compared to LPS alone (Fig. 5, A and D). Such a regimen also led to a significant decrease in the number of EdU/SOX9 colabeled cells (Fig. 5, A, E, and F). This was neither due to a reduced number of Müller cells nor to an altered survival rate, as inferred by the constant number of SOX9-labeled cells (fig. S8, A and B) and the stable Bax/Bcl2 mRNA ratio (fig. S8, A and C). To exclude any potential nonspecific effects of PLX3397, we also used PLX5622, a highly selective CSF1R inhibitor (51). As observed with PLX3397, exposure to LPS plus PLX5622 significantly reduced the number of both microglial cells and proliferative Müller glia, compared to LPS alone (fig. S9). Together, these data thus suggest that activated microglia mediate the effect of LPS on Müller glia cell cycle reentry within mouse retinal explants.
TNF-α is a mediator of microglia-dependent Müller cell proliferation
We next aimed at identifying candidate cytokines that might relay the effect of LPS-activated microglia on the proliferation of Müller cells. To this aim, we first analyzed in retinal explants the expression of genes encoding typical pro-inflammatory factors, namely, IL-6, IL-1β, and TNF-α as well as the chemotactic molecule CCL2 (Fig. 6A). Compared to the controls, chronic LPS exposure up-regulated the expression of Ccl2, Il-1β, and Il-6 from DEV1 onward. A gradual increase in Tnf-α levels was observed as well, starting at DEV3. We then turned to functional experiments on retinal explants, focusing on IL-6 and TNF-α because they were previously shown to be involved in the proliferation of Müller cells (16, 52–54). Contrasting with results obtained in zebrafish (54), adding IL-6 did not modify the number of murine Müller cells exiting quiescence (Fig. 6, B to D). In contrast, TNF-α could convert quiescent Müller cells into proliferative ones, with an efficacy even higher than that of LPS. This effect was not potentiated by a cotreatment with IL-6. Last, we questioned which cells might produce TNF-α in response to LPS. We found that LPS-dependent increase in Tnf-α expression was abolished upon cotreatment with PLX3397 (Fig. 6E). LPS-dependent up-regulation of Tnf-α thus likely occurs in activated microglial cells. We, however, cannot exclude a contribution of Müller glia in the production of TNF-α because these cells are known as well to express multiple inflammatory factors (55, 56). These findings nevertheless suggest that TNF-α mediates the pro-proliferative effects of microglia on Müller cells in response to LPS activation.
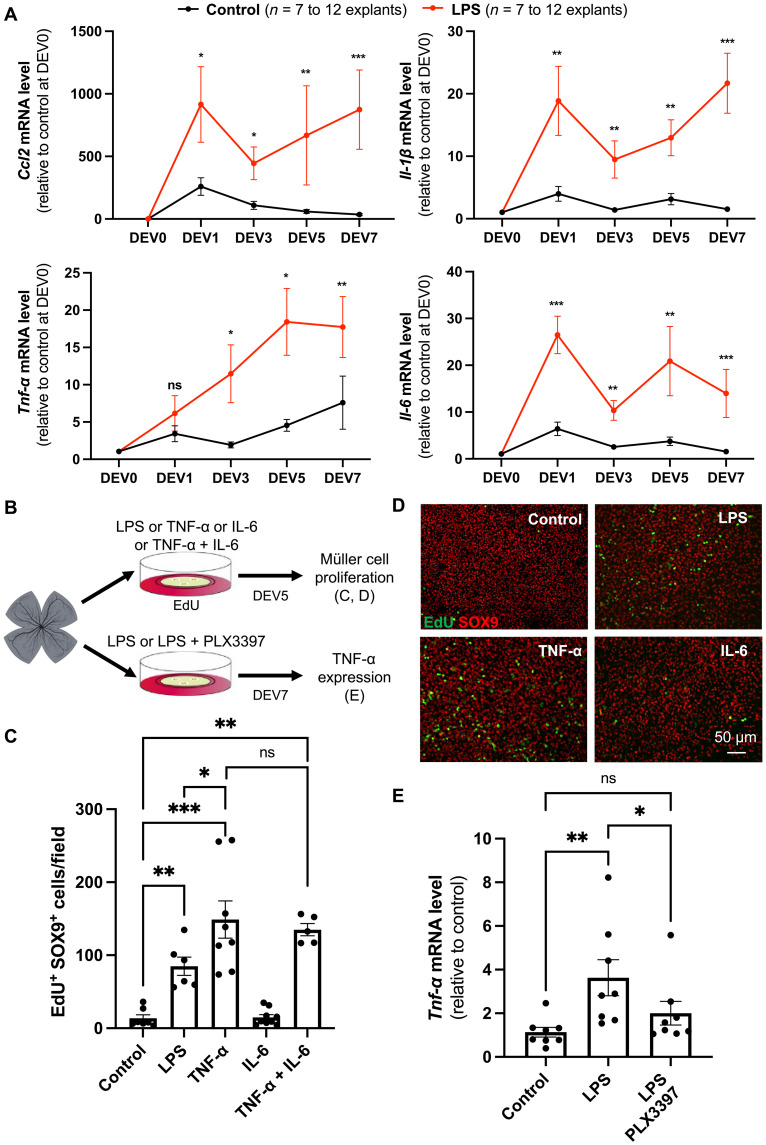
(A) qPCR analysis of Ccl2, Il-1β, Tnf-α, and Il-6 expression in control and LPS-treated retinal explants at different culture time points, relative to uncultured control condition (DEV0). (B) Procedure used in (C) to (E). Retinas from wild-type mice were flattened and cultured for 5 days in the presence of EdU alone (control) or in combination with LPS, TNF-α, IL-6, or TNF-α plus IL-6. Retinal explants were then processed for immunostaining analysis [(C) and (D)]. Other retinal explants were cultured for 7 days in the presence of LPS or LPS plus PLX3397 and subjected to qPCR analysis (E). Created using BioRender.com. (C and D) Quantification of double EdU-positive and SOX9-positive cells (proliferative Müller cells). Shown in (D) are representative images of flat-mounted retinas. (E) qPCR analysis of Tnf-α gene expression. Data are represented as means ± SEM, and each point represents one retinal explant. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test).
Inflammatory signaling is required for mitogenic factors to drive Müller cell proliferation
Diverse signaling pathways were previously identified by our team and others as able to trigger the proliferation of Müller cells in retinal explants. This includes the Hippo pathway, through its terminal effector YAP, and the epidermal growth factor (EGF) pathway (57, 58). We thus sought to identify their potential interplay with inflammatory signaling. Retinal explants were cultured in the presence of LPS for 7 days following transduction with AAV-YAP5SA (Fig. 7, A to C). This adenovirus (Shh10 variant) (59) allows the expression of a constitutively active form of YAP (YAP5SA), specifically in Müller cells (57). As expected, the number of EdU/SOX9 colabeled cells was strongly enhanced upon exposure to AAV-YAP5SA compared to the control and reached a higher level than the one observed following LPS treatment alone. This number was further increased in explants treated with both AAV-YAP5SA and LPS. We then performed similar experiments using EGF (Fig. 7, A, D, and E). Here again, we observed significantly more EdU/SOX9 colabeled cells when EGF was added together with LPS, compared to EGF alone. Collectively, these data reveal that the proliferative activity of Müller cells triggered by mitogenic factors can be further enhanced by stimulation of inflammation.
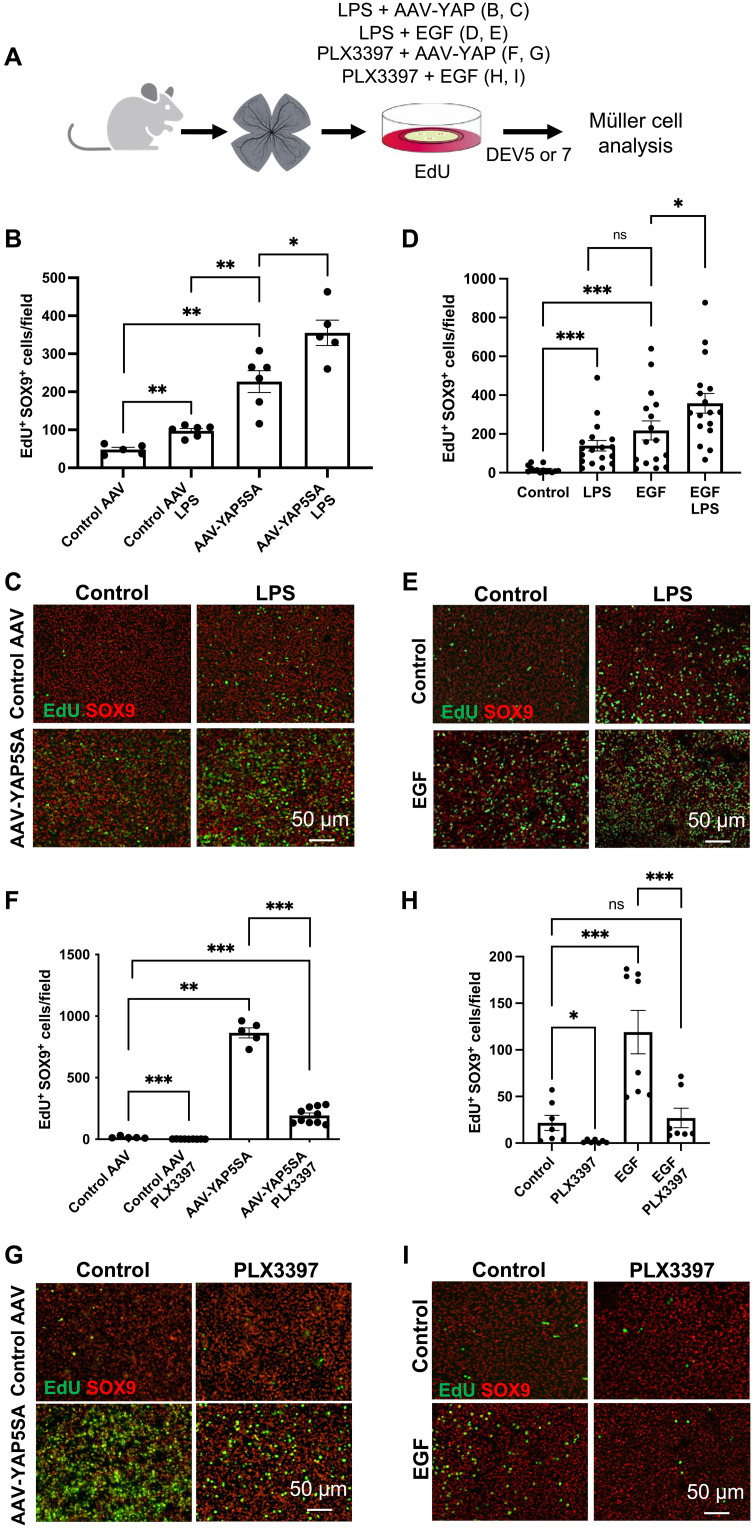
(A) Procedure used in (B) to (I). Retinas from wild-type mice were flattened and cultured for 5 days in the presence of EdU and LPS or PLX3397, together with EGF, or for 7 days if together with AAV-GFP (control AAV) or AAV-YAP5SA. Retinal explants were then processed for immunostaining analysis. Created using BioRender.com. (B to E) Quantification of double EdU-positive and SOX9-positive cells (proliferative Müller cells) in retinal explants exposed to LPS and transduced with either control AAV or AAV-YAP5SA [(B) and (C)] or treated with EGF [(D) and (E)]. Shown in (C) and (E) are representative images of flat-mounted retinas. (F to I) Quantification of double EdU-positive and SOX9-positive cells in retinal explants exposed to PLX3397 and transduced with either control AAV or AAV-YAP5SA (F) or treated with EGF (H). Shown in (G) and (I) are representative images of flat-mounted retinas. Data are represented as means ± SEM, and each point represents one retinal explant. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test).
On the basis of the beneficial effects of such combinatorial treatments on the proliferation of Müller cells, we next wondered if microglial inflammatory signaling might be required for EGF and YAP mitogenic activity. We thus induced microglia depletion through PLX3397 treatment in AAV-YAP5SA–transduced or EGF-exposed explants. We observed a potent inhibition of YAP and EGF pro-proliferative effects on Müller cells in explants treated with PLX3397 (Fig. 7, F to I). These results suggest that microglial cells are absolutely essential for Müller glia cell cycle reentry upon stimulation by mitogenic factors.
Sequential pro- and anti-inflammatory treatments promote both the proliferative and neurogenic potential of Müller cells
In regenerative species, the proliferation of Müller cells following injury is accompanied by a reprogramming process, leading to the expression of neurogenic genes (2, 8). Among critical factors sufficient to drive Müller glia reprogramming into neurons is the transcription factor ASCL1. Because antineurogenic effects of inflammation were reported in mammals (60), we questioned the neurogenic competence of proliferative Müller glia in LPS-treated retinal explants by examining Ascl1 expression. We found that LPS-driven proliferation of Müller cells was accompanied by a slight down-regulation of Ascl1 (Fig. 8, A and B). This finding suggests that, while chronic LPS immune challenge triggers quiescent Müller cells to reenter the cell cycle ex vivo, it also hinders their neurogenic potential.
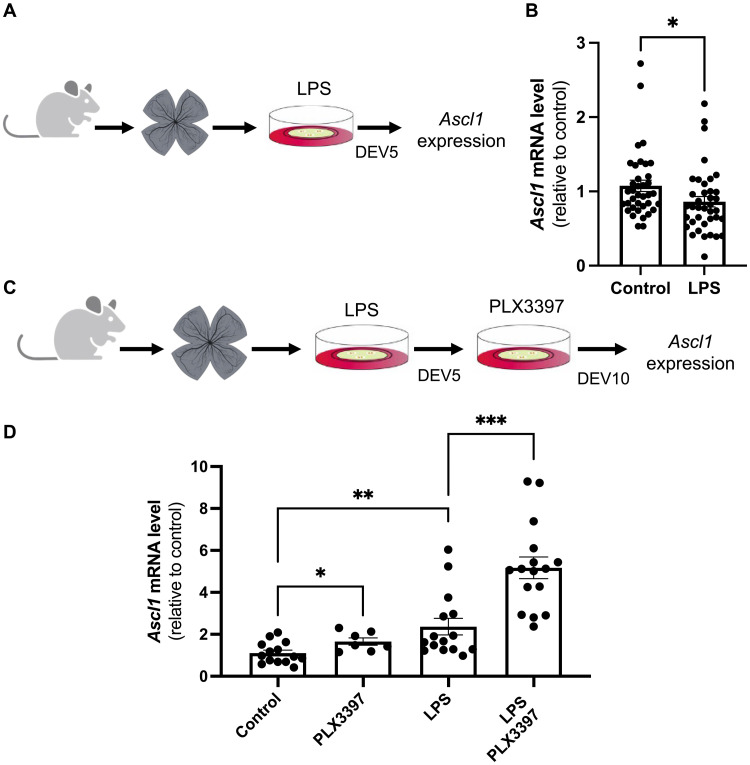
(A) Procedure used in (B). Retinas from wild-type mice were flattened and cultured for 5 days in the presence of LPS. The explants were then processed for qPCR. (B) qPCR analysis of Ascl1 expression at DEV5. (C) Procedure used in (D). Retinas from wild-type mice were flattened and cultured for 5 days in the presence of LPS and then cultured for an additional 5 days in the presence of PLX3397. Other batches of explants were also cultured for 10 days but were either untreated (control) or only treated with LPS for the first 5 days or only with PLX3397 for the last 5 days. Retinal explants were then processed for qPCR. (D) qPCR analysis of Ascl1 gene expression at DEV10. Data are represented as means ± SEM, and each point represents one retinal explant. *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test). (A) and (C) created using BioRender.com.
Given the above result, we hypothesized that sequentially administrating pro- and anti-inflammatory treatments may promote, in a stepwise manner, the proliferation of Müller cells and then their acquisition of neurogenic competence. We thus cultured retinal explants for 5 days (DEV0-5) with LPS and then with PLX3397 for five additional days (DEV5-10) (Fig. 8, C and D). First, we observed that ablating microglia from DEV6 to DEV10 without prior LPS exposure or removing LPS at DEV5 without further treatment was sufficient to increase, although modestly, the expression level of Ascl1 observed at DEV10. Notably, its expression was found up-regulated by more than fourfold after sequential LPS/PLX3397 treatment. Together, these data suggest that attenuating inflammation after an immune challenge may allow proliferative Müller cells to become neurogenic.
TNF-α is detrimental to the neurogenic potential of Müller cells
We next sought to identify the molecular mechanism allowing Müller cells to recover a neurogenic potential upon inflammation attenuation in our sequential immunomodulation setup. Recent data suggest that TNF-α may relay the microglia-dependent inhibition of ASCL1-induced neurogenesis in the injured mouse retina (60). We thus examined its expression dynamic upon LPS/PLX3397 sequential treatment (Fig. 9A). We found that the LPS-dependent increase in Tnf-α was severely reduced following PLX3397 treatment. Tnf-α and Ascl1 levels were thus anticorrelated upon sequential treatment. This makes TNF-α a potential candidate for mediating the LPS-driven Ascl1 repression. Consistent with this hypothesis, we found that TNF-α exposure phenocopied LPS treatment, resulting in a significant inhibition of Ascl1 expression after 5 days of explant culture (Fig. 9B). Furthermore, TNF-α/PLX3397 sequential exposure mimicked the LPS/PLX3397 regimen, with Ascl1 levels being enhanced at DEV10 (Fig. 9C compared to Fig. 8D). Last, the Ascl1 up-regulation driven by sequential exposure to LPS and PLX3397 was fully abolished by adding back TNF-α concurrently with PLX3397 (Fig. 9D). Collectively, these findings are consistent with TNF-α being responsible, at least in part, not only for the proliferative but also for the antineurogenic effects of activated microglia on Müller glial cells.
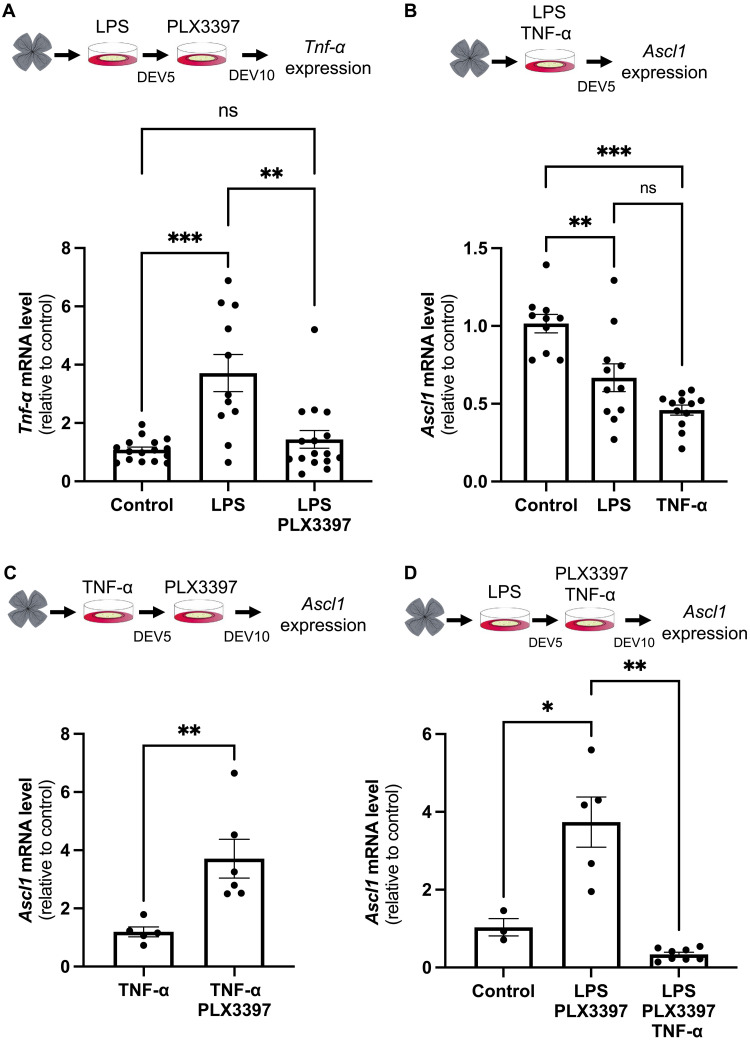
(A) Retinas from wild-type mice were flattened and cultured for 5 days in the presence of LPS and then cultured for an additional 5 days in the presence of PLX3397. Other batches of explants were also cultured for 10 days but were either untreated (control) or only treated with LPS for the first 5 days. Retinal explants were processed for qPCR analysis of Tnf-α expression at DEV10. (B) Retinas from wild-type mice were flattened and cultured for 5 days in the presence of LPS or TNF-α. Retinal explants were processed for qPCR analysis of Ascl1 expression at DEV5. (C) Retinas from wild-type mice were flattened and cultured for 5 days in the presence of TNF-α and then cultured for 5 days with or without PLX3397. Retinal explants were processed for qPCR analysis of Ascl1 expression at DEV10. (D) Retinas from wild-type mice were flattened and cultured for 5 days in the presence of LPS and then cultured for 5 days in the presence of PLX3397 or PLX3397 plus TNF-α. Retinal explants were processed for qPCR analysis of Ascl1 expression at DEV10. Data are represented as means ± SEM, and each point represents one retinal explant. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (Mann-Whitney test). Created using BioRender.com.
DISCUSSION
In this study, we first sought to shed light on the molecular cues underlying the differential response of Müller cells to injury in young Xenopus tadpoles compared to older ones. Through transcriptomic analysis and immunomodulation assays, our findings suggest that the limited reactivation of Müller cells upon retinal degeneration in young tadpoles is not due to an intrinsic barrier but may rather be attributed to the paucity of resident microglia at these developmental stages, resulting in an inadequate/insufficient inflammatory microenvironment. Moreover, our study offers insights into the comparison of regenerative mechanisms in Xenopus and mammals. Notably, we observed in mice that microglia-mediated inflammatory signaling not only fosters the proliferation of Müller cells, as in Xenopus, but is also essential for their known responsiveness to mitogenic factors like YAP or EGF. This finding highlights inflammation as a pro-proliferative trigger for both amphibian and murine Müller cells. However, we showed that attenuating inflammation upon immune challenge is a prerequisite for the subsequent neurogenic phase of regeneration. Last, our work unveils the versatile impact of TNF-α in the mouse retina, serving both pro-proliferative and antineurogenic functions (Fig. 10).
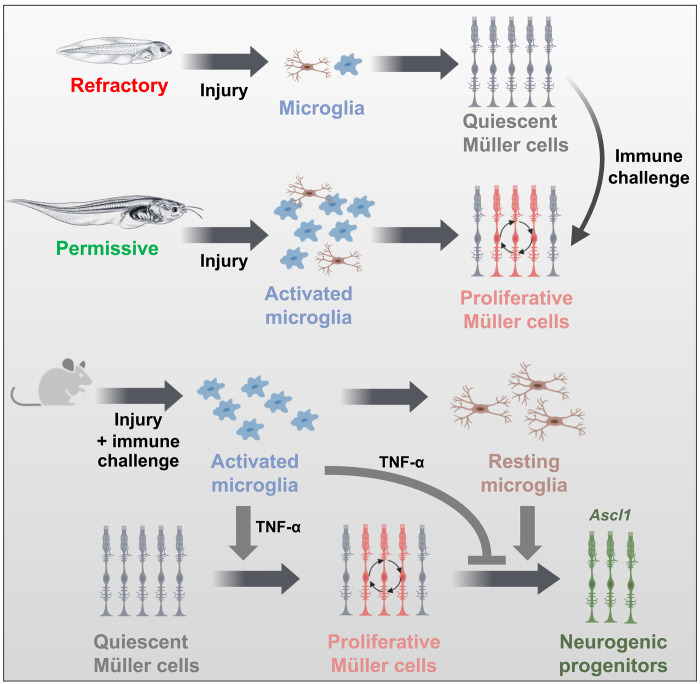
In young Xenopus tadpoles (refractory stages), there are very few microglia in the retina, which could account for the very limited proliferative response of Müller cells in a variety of retinal injury models. In contrast, microglia are more abundant in old tadpoles (permissive stages) and are required for Müller glia to reenter into the cell cycle upon injury. At refractory stages, Müller cells can become proliferative following retinal damage if an immune challenge is provided. In mice, an immune challenge also promotes the proliferation of Müller cells but inhibits their neurogenic potential. An inflammatory state must be sequentially activated and attenuated to achieve both proliferation and neurogenic gene expression. Last, we identified TNF-α as at least one of the cytokines that mediates both proliferative and antineurogenic effects of microglia on Müller cells. Created using BioRender.com.
The recent focus on extrinsic factors in the field of retinal regeneration revealed the critical role played by the immune microenvironment of Müller glia. In nonmammalian species, such as zebrafish and chick, microglia have emerged as key regulators of Müller glia regenerative potential, exerting a positive influence on their proliferative activity and the formation of Müller glia–derived progenitors (14–19). Our research further demonstrates that the activation of microglia/macrophages is indispensable for the proper proliferative response of Müller cells to injury in old Xenopus tadpoles.
While exploring responses of Müller cells in young tadpoles, we observed an intriguing phenomenon: These cells display limited ability to reenter into the cell cycle, if in the rho crispant model (this study) or after mechanical injury or nitroreductase (NTR)–driven rod ablation (3). This challenged our initial expectations that young Müller cells would be more prone to exit quiescence than older ones. It was shown that young mouse Müller cells exhibit regenerative competence following growth factor treatment, which then declines with age (44). The present study led us to propose that this unanticipated refractoriness of Müller cells is not the result of an intrinsic lack of competence to respond to injury. Instead, it may be linked to the scarcity of microglia populating the retina at early tadpole stages and to the subsequent insufficient inflammatory reaction following damage. Recent findings in zebrafish revealed two waves of retinal colonization by microglia with distinct origins (61). The first wave, originating from the rostral blood island, gives rise to a transient microglial population during embryogenesis. The second wave during the larval life is produced by the ventral wall of the dorsal aorta and allows definitive adult microglia to settle within the retina in parallel with the gradual disappearance of the embryonic microglia. This result contrasts with the prevailing notion in mice that microglia stem from a single source, the yolk sac, which is equivalent to the fish blood island (62, 63). If microglia ontogeny in Xenopus occurs as in zebrafish in two waves from distinct origins has not been studied yet. This hypothesis, however, gains strength from the observed low microglia numbers in young tadpoles, a period that might represent a transitional step of retinal colonization by transient and definitive microglial populations. Lineage tracing experiments are now required to validate or invalidate this hypothesis. Besides, analyses in zebrafish suggested distinct intrinsic genetic programs of embryonic and adult microglia populations (64). Therefore, apart from quantitative aspects, microglial cells in young and old Xenopus tadpoles might also differ in their identity and functional capabilities. If this contributes to their inability to stimulate the cycle reentry of Müller cells at early stages remains to be determined. Although immune challenges caused by intraocular injections of CoCl2, zymosan, or heat-killed bacteria are sufficient to increase Müller glia proliferation in young tadpoles, we currently do not know if this is due to the consequent increase in microglial cell number or due to the infiltration of cells with distinct phenotypes and immune properties.
Comparison of acute and chronic retinal injuries in zebrafish revealed different nature of the inflammatory responses. Acute retinal damage promotes a strong pro-inflammatory reaction and an important proliferative response of Müller glia. In contrast, chronic injury triggers only a mild pro-inflammatory response together with a robust anti-inflammatory one and leads to fewer Müller cells reentering the cell cycle (17). This prompts consideration of if the differential activation of Müller cells at refractory stages in the CoCl2 model compared to the rho crispant one might be due to the acute versus chronic nature of the applied injury paradigm. However, we believe that we can rule out this hypothesis as other acute injuries (mechanical injury and NTR genetic conditional rod ablation) trigger limited proliferative responses of Müller cells in young tadpoles (3), as observed in the rho crispant chronic model. Instead, we lean toward the hypothesis that the varying amplitude of the inflammatory reaction is the determining factor.
The immune response has been proposed as detrimental to mammalian regenerative capacity (21, 22, 65). In line with this idea and in contrast to observations in fish and birds, microglia ablation in the ANT mouse model [forced expression of Ascl1 followed by N-methyl-d-aspartate (NMDA) injury along with histone deacetylase inhibition with Trichostatin A] enhances regeneration from Müller glia (60). This has led to the proposal that the neuroinflammatory response to injury plays a pivotal role in the disparity of retinal regenerative potential between nonmammalian vertebrates and mammals. Of note, this model mostly enables direct production of neurons by Müller glia without inducing a proliferative phase (66). Microglia ablation experiments in ANT mice thus demonstrate the adverse effect of inflammation on neurogenesis but leave open the question of its impact on murine Müller glia proliferation because this step is bypassed. In our retinal explant model, we confirmed the antineurogenic effect of microglial cells. However, our data also highlighted their pro-proliferative function, as observed in Xenopus (this study) and in other nonmammalian species (14–19). We therefore propose that the mitogenic effect of the inflammatory reaction is shared by all vertebrates. Its negative impact on the neurogenic phase might be shared as well, as inferred from studies in zebrafish subjected to photoreceptor degeneration. In this model, distinct regeneration outcomes are observed depending on the timing of immunosuppression (14, 67): Before retinal cell loss, it prevents retinal regeneration, while post-injury, it accelerates its kinetics.
If activated microglia exert a positive effect on Müller glia cell cycle reentry, then one might question why Müller cells do not spontaneously and efficiently proliferate in degenerative mouse models exhibiting inflammatory reaction. In our retinal explants, some Müller cells do reenter the cell cycle. This might be triggered by the few microglial cells present in explants because this proliferative subpopulation disappears following treatment with PLX3397. An immune challenge, which stimulates cytokine production, significantly enhances the number of Müller glia that exit quiescence. This suggests that either a minimum magnitude of the inflammatory reaction or a specific polarization state of microglia is required for Müller glia proliferative behavior. Addressing if activating microglia in vivo could similarly affect the proliferation of Müller cells is a crucial question for future research.
Our data indicate that the role of microglia in the mouse extends beyond the facilitation of Müller glia proliferative behavior; these cells appear to be indispensable. This is evidenced by the inefficiency of YAP or EGF to stimulate the proliferation of Müller cells in their absence. If microglia-dependent signaling acts downstream these pathways or is necessary to make Müller cells competent to respond to them remains to be determined. In any case, such a microglia requirement for YAP action in Müller cells unravels an unexpected noncell autonomous component in the control of Müller cell proliferation by this factor. The stimulation of Müller glia–derived progenitor genesis by activation of Wnt signaling in the chick retina was similarly shown to fail upon monocyte depletion (68). In contrast, the same group showed that application of heparin-binding EGF-like growth factor (HB-EGF) completely rescued Müller glia–derived progenitor generation in damaged retinas lacking microglia (68). Why, in our case, was EGF treatment not able to bypass the lack of microglial cells, while HB-EGF in the chick retina did? Several potential explanations can be raised including species-specific effects (mouse versus chick), different methods of microglia ablation (PLX3397 treatment versus injections of clodronate liposomes), or distinct injury paradigms (retinal explant versus NMDA-driven excitotoxicity). The use of EGF instead of HB-EGF could also underlie such apparent discrepancy. It was shown in different models that these ligands can bind to different receptor complexes (69). In line with this, HB-EGF does not stimulate the proliferation of ciliary marginal zone progenitors in the chick retina (70) while EGF does (71). In any case, all these data support the notion that the immune microenvironment plays a pivotal role in regulating pathways that are essential for Müller glia proliferation. Therefore, for any treatment intending to stimulate the complete regeneration process in mammals, it is critical to be aware that inhibiting inflammatory signaling may impede the initial steps of Müller cell reactivation.
Despite the need for inflammatory signaling to initiate the proliferative phase of the regeneration process, several lines of evidence from studies in the brain or retina also suggest that its resolution in a timely manner is a prerequisite for subsequent commitment toward neurogenesis (21, 22, 65). Taking into account these dual effects of inflammation, we implemented the sequential LPS-PLX3397 treatment on mouse retinal explants. This allowed both the induction of a proliferative response from Müller cells and the up-regulation of the neurogenic gene Ascl1. These two phenomena may be directly linked as the S phase entry of Müller glia was shown to be necessary for Ascl1 expression in a rat retinal explant model of injury (72). Our retinal explant model does not permit long-term culture, thus precluding the observation of potential neuronal replacement. Setting up similar sequential procedures in vivo using a model that allows for lineage tracing of Müller cells is thus needed to assess effective regeneration.
Several inflammatory cytokines, including TNF-α and IL-6, were shown to promote Müller glia cell cycle reentry in the injured chick and/or fish retina (16, 52–54, 73). In our mouse explant model, IL-6 did not induce the proliferation of Müller cells, while TNF-α did, as previously reported in primary murine Müller cell cultures (74). This reflects both conserved and potential species-specific responsiveness of these cells to diverse cytokines. Our data further indicate that TNF-α also mimics the antineurogenic effect of microglia, repressing Ascl1 expression as LPS does. Of note, TNF-α was previously shown to inhibit neurogenesis downstream Ascl1 in the ANT mouse model (60). Our data, combined with those from Todd et al. (60), therefore suggest that TNF-α might counteract the neurogenic process both upstream and downstream ASCL1. Overall, we propose that TNF-α stands out as a pivotal cytokine within the murine inflammatory milieu, regulating both the proliferative and the neurogenic phases of Müller glia–dependent retinal regeneration, albeit in an inverse manner. Of note, the cell of origin of TNF-α release following retinal injury is still a matter of debate. This cytokine was proposed to be produced not only by microglia but also by dying neurons in the zebrafish retina and by mammalian Müller glia themselves (53, 75–77). However, single-cell RNA sequencing (RNA-seq) experiments recently revealed that Tnf-α is exclusively expressed by microglia following NMDA-driven injury in the mouse retina (78). Also unresolved is the nature of the signaling pathways that mediate Müller cell responses downstream TNF-α receptors. TNF-α is known to induce the intracellular nuclear factor κB (NF-κB) pathway in murine Müller cells (76, 79), and inhibition of NF-κB following NMDA damage significantly enhances the reprogramming of Ascl1-overexpressing Müller glia into neuron-like cells (80). It is thus tempting to speculate that the antineurogenic effect we observed in retinal explants treated with TNF-α may involve the NF-κB signaling pathway. Besides, which signaling pathways are responsible for TNF-α–dependent stimulation of Müller glia cell cycle reentry remains to be investigated. In primary cultures of Müller cells, TNF-α promotes proliferation through the JAK/STAT (Janus kinase/signal transducer and activator of transcription) and MAPK (mitogen-activated protein kinase) pathways (74). Further research is needed to explore this signaling axis as a possible means of communication between microglia and Müller cells in the context of retinal regeneration and its interplay with the NF-κB signaling pathway.
MATERIALS AND METHODS
Ethics statement
All animal experiments have been carried out in accordance with the European Community Council Directive of 22 September 2010 (2010/63/EEC). Animal care and experimentations were conducted in accordance with institutional guidelines, under the institutional licenses C 91-471-102 and D 91-272-105. The study protocols were approved by the institutional animal care committee CEEA (comité d’éthique en expérimentation animale) no. 59 and received an authorization by the Direction Départementale de la Protection des Populations under the references APAFIS#5938-20160704l3l 04812 v2 and APAFIS#998-2015062510022908v2 for in vivo Xenopus experiments.
Animals
Wild-type C57BL/6JRj mice were kept at 21°C, under a 12-hour light/12-hour dark cycle, with food and water supplied ad libitum. All experiments involving adult mice were performed with male or female mice that were 4 to 8 weeks of age. X. laevis tadpoles were obtained by conventional methods using hormone-induced egg laying and in vitro fertilization. Tadpoles were maintained under standard aquarium conditions at 18° to 20°C and staged according to (81).
Microinjections in Xenopus
rho crispant F0 tadpoles were generated as previously described (4, 82). Briefly, the single guide RNA (250 to 500 pg) against rhodopsin (rho) and the Cas9 protein (5 ng, New England Biolabs) were injected at the one-cell stage. Controls were injected with Cas9 protein only. Embryos were then raised under standard conditions until the desired stage.
Intraocular injections and pharmacological treatments in Xenopus
Intraocular injections were performed as previously described (82). Briefly, tadpoles were anesthetized with 0.005% benzocaine (Sigma-Aldrich) for 5 to 10 min and placed on a wet tissue in a petri dish with dorsal view up. Using a Picospritzer III microinjector (Parker, United States), tadpoles were intraocularly injected with two 30-nl drops of 25 mM CoCl2 (Sigma-Aldrich), zymosan (10 mg/ml; Sigma-Aldrich), or Escherichia coli preheated for 1 hour at 100°C. For immunosuppression, Dex (7.5 mg/ml; Sigma-Aldrich) and PLX3397 (0.5 μM; CliniSciences) were added to the tank water. The treatment started 1 day before CoCl2 injection and continued for 6 days. rho crispant tadpoles were immersed in the Dex/PLX3397 solution for five consecutive days. The tank solution was changed daily. The solvent of the drugs [Modified Barth’s Saline stock solution, dimethyl sulfoxide (DMSO), or ethanol] were systematically used as negative controls.
Xenopus retina dissections and flat-mounted eyes
Tadpoles were euthanized in 0.01% benzocaine (Sigma-Aldrich) and then transferred into a petri dish containing 1X Hanks’ balanced salt solution (HBSS) (Gibco). The eyes were surgically removed with sharp forceps under the stereomicroscope. For flat-mounted eye experiments, the lenses were removed and the free-floating retinas were collected in 1X phosphate-buffered saline (PBS) and then fixed in 4% paraformaldehyde for 30 min. For RNA extraction, both the lens and the retinal pigment epithelium (RPE) were carefully removed. The neural retinas were then collected and kept at −80°C until use.
BrdU/EdU labeling and immunostaining on Xenopus retinal tissues
Tadpoles were immersed in a solution containing 0.5 mM BrdU (Roche) or 0.5 mM EdU (Invitrogen). Following the indicated time period, they were euthanized in 0.01% benzocaine (Sigma-Aldrich), fixed in 4% paraformaldehyde for 2 hours at room temperature, dehydrated, embedded in paraffin, and sectioned (11 μm) with a Microm HM 340E microtome (Thermo Scientific). Antigen retrieval was performed by boiling the sections in 10 mM sodium citrate with 0.05% Tween 20 for 9 min. For BrdU immunostaining, slides were then incubated in 2 N HCl for 45 min at room temperature, as a second unmasking step. After 1 hour of blocking in Dako diluent (Agilent) plus 0.2% Triton X-100, slides were incubated with primary antibodies overnight at 4°C, rinsed three times in 1X PBS supplemented with 0.1% Triton X-100 for 10 min, and then incubated for 2 hours at room temperature with secondary antibodies. The same immunostaining protocol, without the antigen retrieval step, was used on fixed flat-mounted retinas. All used antibodies are listed in table S2. EdU incorporation was detected using the Click-iT EdU Imaging Kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations. For double labeling, EdU staining was done first, followed by immunostaining. Cell nuclei were counterstained with Hoechst (10 μg/mL, Sigma-Aldrich), and samples were mounted using the FluorSave Reagent (Millipore).
EdU labeling and immunostaining on mouse retinal explants
Neural retinas from enucleated eyes were dissected in HBSS (Gibco) by removing the anterior segment, vitreous body, sclera, and RPE. The retina was then cut radially into four equal-sized pieces and flat mounted onto a microporous membrane (13 mm in diameter; Merck Millipore) in a 12-well culture plate, with the ganglion cell layer facing upward. Each well contained 1 ml of a culture medium, consisting of DMEM (Dulbecco’s modified Eagle’s medium) GlutaMAX d-glucose (4.5 g/liter; Gibco) supplemented with 1% fetal bovine serum, 0.2% NaHCO3, 5 mM Hepes, 2% B27, 1% N2, and 1X penicillin-streptomycin. The culture medium containing LPS from E. coli O111:B4 (5 μg/ml; Sigma-Aldrich), recombinant human IL-6 (60 ng/ml; R&D Systems), TNF-α (60 ng/ml; Preprotech), 10 μM PLX3397 (pexidartinib, CliniSciences), 10 μM PLX5622 (CliniSciences), or vehicle (DMSO) was added to the explant culture. For viral transduction, 1011 vg of AAV-GFP or AAV-YAP5SA was applied on the explants (57). Explants were maintained at 37°C in a humidified incubator with 5% CO2. Half of the culture medium was changed every other day. One drop of the medium was added daily over the retinal explants to avoid drying. For proliferation assays, EdU (2.5 ng/ml) was applied during the indicated time period before fixation. Retinal explants were fixed in 1X PBS and 4% paraformaldehyde for 45 min at room temperature. Explants were washed in 1X PBS and then incubated for 30 min at room temperature in a blocking solution, which consisted of Dako diluent (Agilent) and 0.5% Triton X-100 (Sigma-Aldrich). Explants were then incubated in mouse monoclonal primary antibodies for 48 to 72 hours at 4°C under agitation, washed thoroughly in PBS and 0.5% Triton X-100, and incubated with fluorescent secondary antibodies for 24 to 48 hours at 4°C upon agitation. Explants were lastly washed in 1X PBS and 0.5% Triton X-100 and whole mounted on slides or blocked for 1 hour at room temperature in 1X PBS containing 5% normal goat serum (Invitrogen Inc.) and 0.5% Triton X-100 (Sigma-Aldrich, United Kingdom). They were then subjected to EdU labeling using the Click-iT EdU Imaging Kit (Thermo Fisher Scientific). All used antibodies are listed in table S2.
RNA extraction and RT-qPCR
Total RNA was isolated from dissected Xenopus retinas (n = 30 for stages 45 to 48 and n = 5 to 10 for stages 56 to 62) using TRIzol (Invitrogen), according to the manufacturer’s protocol. RNA was then purified with the NucleoSpin RNA kit (Machere-Nagel). In mice, RNA was extracted from one mouse neural retinal explant per condition using the RNeasy Micro Kit (QIAGEN). RNA concentrations were assessed using the NanoDrop 2000c UV-Vis spectrophotometer (Thermo Fisher Scientific). Total RNA was reverse transcribed in the presence of oligo-(dT)12-18 using SuperScript II reagents (Thermo Fisher Scientific). Quantitative polymerase chain reaction (qPCR) reactions were performed in triplicate on 1 ng of mouse cDNA or 10 ng of Xenopus cDNA in the presence of EvaGreen (Bio-Rad) on a CFX96 Real-Time PCR Detection System (Bio-Rad) or a QuantStudio5 (Applied Biosystems). For the mouse, differential expression analysis was performed using the ΔΔCt method using the geometric mean of Rps26, Srp72, and Tbp as endogenous controls (83). The relative expression of each gene in each sample was calculated using the mean of the controls as the reference. For Xenopus paired samples, differential expression analysis was represented as −ΔCt using the geometric mean of odc1 and eef1a1 as endogenous controls. Primers are listed in table S3. Reverse transcription qPCR (RT-qPCR) experiments were performed on 3 to 20 mouse retinal explants and at least six independent Xenopus retinal extracts per condition.
RNA-seq analysis and data availability
RNA extracted from dissected retinas at refractory and permissive stages (47 to 48 and 58 to 62, respectively) were used to prepare mRNA stranded libraries and sequenced for 2x150 cycles on a NextSeq500/550 at the I2BC sequencing facility. After demultiplexing, adapter trimming, and quality control, 27 million to 48 million reads were uniquely mapped with STAR (84) in two-pass on the X. laevis V9.2 genome with an average mapped length of 294 nt, using the annotation and newly found junctions as described in (85) (table S4). Read counts per gene was computed using HTseq-count (“htseq-count -f bam -t exon -i gene_id --additional-attr gene_name -r pos -s reverse --secondary-alignments ignore -m intersection-strict --nonunique none”) (86). All statistical analyses were performed in R (87). Differential expression analysis was performed on reads counts/gene with the package DESEQ2 (88), comparing rho crispant with control retinas at refractory or permissive stages. GSEA analysis on LFC ranked gene was performed with the package clusterProfiler (89) and P value adjusted for multiple testing (Benjamini–Hochberg method). Overrepresentation analysis of Gene Ontology term was performed using TopGO ( 10.18129/B9.bioc.topGO). All R scripts are available on GitHub. Datasets are accessible from ArrayExpress (accession E-MTAB-13881).
Microscopy and quantification
Fluorescence images were acquired using an Apotome-equipped Axio Imager M2 microscope (Zeiss) or an LSM710 confocal microscope (Zeiss). Images were processed using the Zen (Zeiss), ImageJ (90), and Photoshop CS5 (Adobe) software. Quantification of labeled cells in Xenopus retinal sections was performed by manual counting of 5 to 10 sections per retina, and an average number was calculated. The mean number of labeled cells in mouse retinal explants was calculated from eight different fields of 104 μm2 per retina (two in each quarter) and using at least five explants per condition.
Statistics
All experiments were performed at least in duplicate. In graphs, data are presented as means ± SEM. Statistical analyses were performed with the GraphPad Prism 10 software, using the nonparametric Mann-Whitney-Wilcoxon U test or the Wilcoxon matched-paired test, as specified in the figure legends. Of note, for multiple comparisons, we also used the Mann-Whitney-Wilcoxon U test, which is recommended for nonparametric planned comparisons (91). All analyses were two-tailed, and a P value less than 0.05 (P < 0.05) was considered statistically significant.
Acknowledgments
We are grateful to C. Cozma, A. Fichot, C. Hottin, and A. Mouloudi for technical support and F. Sennlaub for scientific discussion. This work has benefited from the facilities and expertise of the I2BC sequencing facility supported by IBiSA, Région Île de France, Plan Cancer, CNRS and Paris-Saclay University, the TEFOR Paris-Saclay’s zootechnics service for the maintenance of Xenopus, and the zootechnics service of NeuroPSI for the maintenance of mice. We also acknowledge the GenOuest bioinformatics core facility (https://www.genouest.org/) for providing the computing infrastructure and the vectorology platform of the Institut de la Vision for the AAV production. Some figures were created with schemas from BioRender.com.
Funding: This work was supported by the Association Retina France (M.P.), AFM (Association Française contre les Myopathies) (M.P.), FMR (Fondation maladies rares) (M.P.), BBS (Association Bardet-Biedl syndrome) (M.P.), Fondation de France (M.P.), and UNADEV (Union Nationale des Aveugles et Déficients Visuels), in partnership with ITMO NNP (Institut Thématique Multi-Organisme Neurosciences, sciences cognitives, neurologie, psychiatrie)/AVIESAN (Alliance nationale pour les sciences de la vie et de la santé) (M.P.). S.L., J.B., and J.E.R. are supported by Retina France.
Author contributions: Conceptualization and supervision: C.B., J.B., M.P., and P.P. Investigation: A.C., A.D., C.B., D.G.-G., D.K., D.V.W., J.B., J.R., J.L., K.P., L.S., L.V.-G., S.L., S.P., P.P., and X.S.-S. Data analysis: all authors. RNA-seq analysis: Y.A. Writing: C.B., J.B., M.L., M.P., and P.P. Writing—review and editing: K.P., J.L., D.G.-G, L.V.-G., J.E.R., and Y.A. Funding acquisition: M.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq datasets are accessible from ArrayExpress (accession E-MTAB-13881). The data that support the findings of this study are available in table S5.
REFERENCES AND NOTES
Articles from Science Advances are provided here courtesy of American Association for the Advancement of Science
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/168875789
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
DNA Damage Response in Proliferating Müller Glia in the Mammalian Retina.
Invest Ophthalmol Vis Sci, 57(3):1169-1182, 01 Mar 2016
Cited by: 15 articles | PMID: 26975029
Retinal microglia signaling affects Müller cell behavior in the zebrafish following laser injury induction.
Glia, 67(6):1150-1166, 22 Feb 2019
Cited by: 51 articles | PMID: 30794326
Insights on the Regeneration Potential of Müller Glia in the Mammalian Retina.
Cells, 10(8):1957, 31 Jul 2021
Cited by: 20 articles | PMID: 34440726 | PMCID: PMC8394255
Review Free full text in Europe PMC
Microglial-induced Müller cell gliosis is attenuated by progesterone in a mouse model of retinitis pigmentosa.
Glia, 66(2):295-310, 16 Oct 2017
Cited by: 37 articles | PMID: 29034506

 1
,*¶
1
,*¶

