Abstract
Free full text

Evaluation of blood MSI burden dynamics to trace immune checkpoint inhibitor therapy efficacy through the course of treatment
Abstract
Analysis of serial liquid biopsy (LB) samples has been found to be a promising approach for the monitoring of tumor dynamics in the course of therapy for patients with colorectal cancer (CRC). Currently, somatic mutations are used for tracing the dynamics of the tumor via LB. However, the analysis of the dynamic changes in the molecular signatures such as microsatellite instability (MSI) is not currently used. We hypothesized that changes in blood MSI burden (bMSI) could be registered using serial LB sampling in the course of immune checkpoint inhibitors (ICI), and that its changes could potentially correlate with treatment outcomes. We report the preliminary findings of the observational trial launched to study (NCT06414304) the dynamics of bMSI in 9 MSI-positive CRC patients receiving ICI. NGS-based MSI testing was performed on both pre-treatment FFPE and serial LB samples. For patients who had detectable bMSI burden in any of the LB samples (n =
= 8, 89%), median bMSI was 1.4% (range, 0.01-40%). Among patients with detectable MSI in available FFPE samples, median MSI burden was 29.3% (range, 10–40%). bMSI detected in baseline LB and FFPE samples were positively correlated (Pearson’s R 0.47). Maximal variant allele frequencies of driver mutations observed in LB were also positively correlated with bMSI burden (Pearson’s R 0.7). Patients who had clinical benefit had undetectable bMSI burden at follow-up. Our results provide the rationale for further validation of bMSI as a predictive biomarker of ICI in MSI-positive patients.
8, 89%), median bMSI was 1.4% (range, 0.01-40%). Among patients with detectable MSI in available FFPE samples, median MSI burden was 29.3% (range, 10–40%). bMSI detected in baseline LB and FFPE samples were positively correlated (Pearson’s R 0.47). Maximal variant allele frequencies of driver mutations observed in LB were also positively correlated with bMSI burden (Pearson’s R 0.7). Patients who had clinical benefit had undetectable bMSI burden at follow-up. Our results provide the rationale for further validation of bMSI as a predictive biomarker of ICI in MSI-positive patients.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73952-1.
Background
Colorectal cancer (CRC) remains one of the leading causes of cancer-related mortality globally1. Although most patients are diagnosed with early-stage disease, the 5-year survival rate is 71%2. Notwithstanding the advancements in screening, diagnosis, and treatment modalities, there is still an ongoing need for innovative approaches to improve patient outcomes.
Immunotherapy (IT) with immune checkpoint inhibitors (ICI) has emerged as promising treatment for CRC3. However, ICI have only demonstrated efficacy in microsatellite instability (MSI) or mismatch repair deficiency (dMMR) positive CRC4. In stage IV CRC, the frequency of MSI is relatively low, at 4%, compared to 10–20% in stages I-III5,6, allowing IT to become the preferred strategy in the early stages7.
MSI can be associated with Lynch syndrome8–10, however, in CRC, most cases of MSI are sporadic, arising from frequently co-occurring epigenetic inactivation of MLH1 and BRAF V600E mutations11–13, found in up to 45% of CRC with sporadic MSI14,15.
The standard methods for assessing MSI and dMMR are polymerase chain reaction (PCR) and 4-antibody immunohistochemistry (IHC). In clinical practice, limitations of MSI/dMMR testing include false negative or false positive results in cases when the sample quality is suboptimal, as well as variable concordance between the two methods. Furthermore, IHC can yield inconclusive results, such as the loss of only one protein9,16. Next-generation sequencing (NGS) is an emerging approach that allows to expand the range of analyzed markers9. NGS allows to dynamically monitor tumor variability through the analysis of mutations in circulating tumor DNA (ctDNA)17,18. NGS-based analysis of liquid biopsy (LB) has several advantages, as it allows to evaluate the dynamics of tumor variability, as well as monitor the response to therapy19. There is a discrepancy in assessing patient responses to therapy using ctDNA. Current approaches are based on an evaluation of total variant allele frequency (VAF) or ctDNA copies per mL of plasma or VAF of the driver mutation.
The literature encompasses findings on MSI assessment in ctDNA through high-throughput sequencing employing diverse methodologies20–22. The majority of computational NGS-based approaches for MSI analysis rely on the classification of each single short tandem repeat (STR) on stable/unstable with further accumulation of MSI score as the percent of unstable loci23. Numerous studies demonstrated outstanding sensitivity of diverse software utilizing this approach for detecting MSI in tumor and/or LB samples. Nevertheless, per-STR binary classification puts an intrinsic restraint on the use of tracing MSI score dynamics throughout longitudinal monitoring of LB, as the score reflects the breadth of instability across STRs rather than the burden thereof in DNA fragments derived from exhibiting intra-tumor heterogeneity and contamination with normal tissue24.
In order to investigate dynamics of bMSI of CRC patients receiving ICI’s, we initiated a multicenter observational clinical trial. Here, we describe outcomes of the first nine patients enrolled in the study.
Methods
Ethics statement
The study was approved by the Ethics Committee at the Blokhin National Medical Research Center of Oncology Research Center (№13-02-2023) and was carried out in accordance with the Declaration of Helsinki. Written informed consents were received from the patients before starting any procedures related to the study.
Study design and participants
This is a prospective multicenter observational study (ClinicalTrials ID: NCT06414304) evaluating the dynamics of MSI burden in serial LB samples from patients receiving ICI due to the presence of MSI/dMMR.
Eligible patients were aged ≥
≥ 18 years with MSI/dMMR-positive confirmed colorectal adenocarcinoma with measurable disease as per Response Evaluation Criteria in Solid Tumor version 1.1 (RECIST v1.1) by local investigator/radiology assessment, Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1, and adequate organ function were eligible for the study. Patients had to be candidates for the treatment with ICI regardless of the setting. Patients not willing or unable to participate in blood plasma collection were also excluded.
18 years with MSI/dMMR-positive confirmed colorectal adenocarcinoma with measurable disease as per Response Evaluation Criteria in Solid Tumor version 1.1 (RECIST v1.1) by local investigator/radiology assessment, Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1, and adequate organ function were eligible for the study. Patients had to be candidates for the treatment with ICI regardless of the setting. Patients not willing or unable to participate in blood plasma collection were also excluded.
Participants described here underwent treatment and study related procedures at the N.N. Blokhin National Medical Research Center or Moscow Multidisciplinary Clinical Center “Kommunarka” (Moscow, Russia) from 2022 to 2024. Patients received prolgolimab (anti-PD-1 monoclonal antibody25) in neoadjuvant settings under investigational protocol or nivolumab for the systemic treatment of metastatic disease.
Medical records including radiological and pathological response data were collected for all patients.
Collection of clinical samples and diagnostic procedures
Participants had to provide archival formalin-fixed paraffin-embedded (FFPE) blocks (‘tumor baseline’) and a blood plasma (‘blood baseline’ or ‘BL’) sample derived prior to the start of ICI. Additionally, blood plasma samples were collected two (W2) and four weeks (W4) following the initiation of ICI, as well as at every control visit (F1 and F2).
As part of the study, patients underwent confirmatory MSI/dMMR testing and NGS analysis of primary tumor samples and all serial LB samples.
Venous blood samples were collected into 10 mL EDTA tubes (GradBioMed, Russia). The tubes were stored at room temperature and transferred to the laboratory within 5 days.
MSI testing
Confirmatory MSI/dMMR testing with gold standard methods (5-loci PCR or 4-antibody IHC was performed, depending on the preceding method used prior to the study recruitment), on primary FFPE samples. In cases when initial MSI testing was performed via NGS, both PCR and IHC were used. In cases when IHC resulted in the loss of only two of the four MMR proteins, the result was considered equivocal as per ESMO guidelines9.
NGS-based MSI testing was performed on both FFPE and serial LB samples.
DNA extraction and sequencing
Tumor DNA was isolated from archival FFPE blocks using the ExtractDNA FFPE kit (Evrogen JSC, Russia) according to the manufacturer’s protocol.
Blood plasma samples were centrifuged for 15 min at 1900 g and 4 °C. The supernatants were collected and centrifuged for 10 min at 16,000 g and 4 °C, and were then stored at -80 °C. ctDNA from blood plasma was isolated using the QIAamp MinElute ccfDNA Midi Kit (Qiagen, Hilden, Germany). DNA was quantified using the Qubit 2.0 fluorometer and the Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol.
Amplification of target regions was performed using the Solo-test Atlas Pro (OncoAtlas LLC, Russia) panel covering clinically significant regions of 38 commonly altered genes (SNV, indel, CNV) in cancer, not including Lynch syndrome genes, as well as 56 STRs for MSI detection. Pooled libraries were sequenced employing Genolab sequencer (GeneMind Biosciences, China).
Sequence data analysis was conducted as described previously26,27. In short, reads were aligned to the human genome assembly GRCh37.p13. Samtools was employed to conduct an initial assessment of the technical properties of the identified variants, including variant site coverage depth, observed alternative allele counts, observed alternative allele frequency28. Additional technical annotations of detected variants were performed using FreeBayes29, Mutect230, SGA31, and SiNVICT32. MSIsensor2 was used to call count of unstable STRs33. MSI burden was calculated as cumulative prevalence of k-mers corresponding to altered STR across all STRs within a single sample34. Data analysis (including variant calling and MSI analysis) was conducted within Solo AVES platform tailored to Solo-test Atlas Pro panel (validation studies will be published separately) and provided by test-kit supplier. Variant calling, VAF calculation, MSI detection and bMSI fraction calculation were additionally validated prior to the current serial case report using the mixing experiments of the paired samples (Supplementary table S1, Supplementary Fig. S1). Oncoprints were made with Oncoprinter35–37.
Mutation classification
Mutations were classified as oncogenic, uncertain significance (VUS) or neutral according to community guidelines38. OncoKB, JAX and a proprietary database were used for the identification of driver mutations39,40. Neutral or VUS mutations were considered passenger.
Statistical analysis
Descriptive measures (i.e., median) were derived for continuous variables and categorical variables were expressed as % frequency distribution. Pearson correlation coefficient and Fisher’s exact test were used for statistical analysis. A p value of < 0.05 was considered as significant with two-sided statistical tests. Analysis was carried out using the SciPy library for Python.
0.05 was considered as significant with two-sided statistical tests. Analysis was carried out using the SciPy library for Python.
Results
Clinical characteristics and outcomes
Nine patients were recruited to the study. The patients’ median age at study enrollment was 56 years (range, 28–77). Seven patients had colon cancer and 2 had rectal cancer. Most CRC patients had right-sided localization of the primary tumor (before splenic flexure), one had left-sided localization, and one was diagnosed with synchronous tumors of the ascending and sigmoid colon (Table 1). Patients who had localized disease at the time of study enrollment (n =
= 7) were assigned to the neoadjuvant treatment with prolgolimab under investigational protocol. Two patients had advanced/metastatic disease and were assigned to systemic treatment with nivolumab monotherapy.
7) were assigned to the neoadjuvant treatment with prolgolimab under investigational protocol. Two patients had advanced/metastatic disease and were assigned to systemic treatment with nivolumab monotherapy.
Table 1
Сlinical characteristics of the first nine patients included in the clinical trial.
| Patient ID | Sex | Primary tumor location | TNM | Tumor stage | Drug | Therapy setting | Method used for initial MSI/dMMR analysis | BRAF V600E status | Radiological response | Pathological response | Reason for withdrawal |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Male | Left | T3N2M0 | III | Prolgolimab | Neoadjuvant | PCR | wt | PR | mPR | Completion of the planned course of treatment |
| Case 2 | Male | Right | T4bN1M0 | III | Nivolumab | Palliative | PCR | wt | SD | NA | Refusal of treatment |
| Case 3 | Female | Right | T3N1M0 | III | Prolgolimab | Neoadjuvant | PCR | V600E | Censored | NA | Death |
| Case 4 | Female | Right/Left | T3N2Mx | III | Prolgolimab | Neoadjuvant | PCR | wt | CR | NA | Completion of the planned course of treatment |
| Case 5 | Male | Rectum | T4N2аM0 | III | Prolgolimab | Neoadjuvant | PCR | V600E | PR | NA | Completion of the planned course of treatment |
| Case 6 | Male | Right | T3N2M0 | III | Prolgolimab | Neoadjuvant | PCR | wt | PR | pCR | Completion of the planned course of treatment |
| Case 7 | Female | Right | T3N0M0 | II | Prolgolimab | Neoadjuvant | PCR | V600E* | CR | pCR | Completion of the planned course of treatment |
| Case 8 | Male | Rectum | T4aN1bM0 | III | Prolgolimab | Neoadjuvant | PCR | wt | PR | pCR | Completion of the planned course of treatment |
| Case 9 | Male | Right | T3N1M1 | IV | Nivolumab | Palliative | PCR | V600E** | PD | NA | Death |
BRAF V600E status is listed according to the patients’ medical records. *For case 7, BRAF mutation was previously found by PCR, but was undetectable by NGS analysis. **For case 9, BRAF mutation was only identified via NGS described here. Abbreviations: N/A - not available, MSS - microsatellite stable, MSI - microsatellite instability, SD - stable disease, (m)PR - (major) partial response, (p)CR - (pathological) complete response, PD - progressive disease, wt - wild type, PCR - polymerase chain reaction, IHC - immunohistochemistry.
As per treatment guidelines, molecular testing of the commonly altered oncogenes in CRC had been performed prior to the study recruitment for all of the patients. According to the medical records and/or subsequent NGS FFPE and/or LB analysis, the BRAF p.V600E variant was identified in 4 patients with a median age of 64 years (range, 41–77), while other 5 patients with wild-type BRAF had a median age of 48 (range, 28–66). All patients had MSI detected by PCR. Confirmatory IHC testing was performed for 4 (44.4%) patients, as for others, no baseline FFPE was available (n =
= 3), or the available FFPE sample had suboptimal tumor content (n
3), or the available FFPE sample had suboptimal tumor content (n =
= 2). Results of PCR and IHC were concordant in 50% of the tested cases. Although no germline analysis was conducted, according to the medical records, no patients had a reported family or personal history of Lynch syndrome-related cancer.
2). Results of PCR and IHC were concordant in 50% of the tested cases. Although no germline analysis was conducted, according to the medical records, no patients had a reported family or personal history of Lynch syndrome-related cancer.
Of 7 patients receiving neoadjuvant treatment, 2 (29%) had complete response (CR), 4 (57%) had partial response (PR) and 1 (14%) was censored. Of 2 patients receiving systemic therapy one had stable disease (SD) and one had disease progression (PD). Of 4 patients who underwent surgery, 3 had complete pathological response (pCR) and 1 had major pathological response (mPR).
Mutational profile and blood MSI burden analysis
Baseline FFPE samples were available for 5 (56%) at the time of analysis. Of those, 4 samples were classified as MSI-positive by NGS (80% agreement with IHC/PCR; 100% agreement with PCR) (Table S2). Among the 9 baseline LB samples, 7 (77%) were MSI-positive. Among the 5 available tumor-plasma paired samples, 4 had concordant MSI status determined by NGS (80% agreement). MSI status established in ctDNA was also concordant with the results obtained by standard methods (8 of 9, 89% agreement).
Among the 5 tumor samples mutations were detected in 4 samples. A total of 16 mutations were detected in 5 available FFPE samples, with a median number of mutations of 4 (range, 0–5). Of 9 BL samples, 7 had at least one mutation, with a total of 7 mutations identified (median, 1; range, 0–2).
During blood ctDNA monitoring (described in the section below) additional 3 mutations were detected. In summary, 21 mutations were found, 12 of them were classified as driver and 9 as passenger. The detected mutations were typical for the CRC mutational landscape, including BRAF mutations41 (Fig. 1).
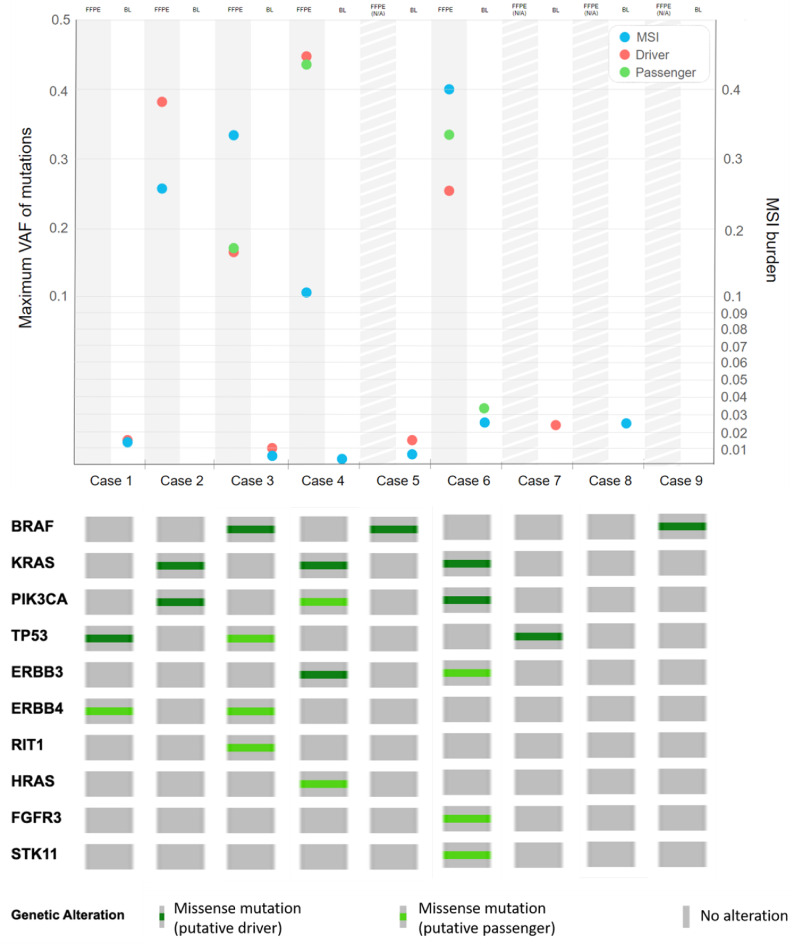
Results of NGS analysis. Maximum VAF of mutations and MSI burden in baseline FFPE (when available) and baseline blood plasma (BL) samples (top) and mutations detected at any time point (i.e., in any of the patients’ samples) for all of the patients included in the analysis (bottom). Abbreviations: VAF - variant allele frequency, BL - baseline blood plasma sample, FFPE N/A - FFPE not available.
Correlation of bMSI burden with clinical responses
For most patients (n =
= 5; 56%), LB samples were collected at 5 time points, while for the remaining patients, there were a median of 2.5 time points (range from 2 to 4). For 6 out of 7 patients receiving neoadjuvant therapy, sample collection was stopped due to the completion of therapy. Of 2 patients receiving treatment in the palliative setting, 1 had PD and died before F1, and 1 achieved SD and refused treatment after F2. One patient receiving neoadjuvant therapy died before reaching W4 time point and was censored (Table S2).
5; 56%), LB samples were collected at 5 time points, while for the remaining patients, there were a median of 2.5 time points (range from 2 to 4). For 6 out of 7 patients receiving neoadjuvant therapy, sample collection was stopped due to the completion of therapy. Of 2 patients receiving treatment in the palliative setting, 1 had PD and died before F1, and 1 achieved SD and refused treatment after F2. One patient receiving neoadjuvant therapy died before reaching W4 time point and was censored (Table S2).
The dynamics of bMSI and VAF of the observed mutations were generally consistent with the patients’ clinical outcomes (Fig. 2). For patients who had detectable bMSI burden in any of the LB samples (n =
= 8, 89%), median bMSI burden was 1.4% (range, 0.01-40%). Among patients with detectable MSI in FFPE samples (n
8, 89%), median bMSI burden was 1.4% (range, 0.01-40%). Among patients with detectable MSI in FFPE samples (n =
= 4, 80% of available FFPE), median MSI burden was 29.3% (range, 10–40%). MSI burden detected in baseline LB (bMSI) and FFPE samples were positively correlated (Pearson’s R 0.47). bMSI burden and maximal VAFs of driver mutations observed in LB were also positively correlated (Pearson’s R 0.7).
4, 80% of available FFPE), median MSI burden was 29.3% (range, 10–40%). MSI burden detected in baseline LB (bMSI) and FFPE samples were positively correlated (Pearson’s R 0.47). bMSI burden and maximal VAFs of driver mutations observed in LB were also positively correlated (Pearson’s R 0.7).
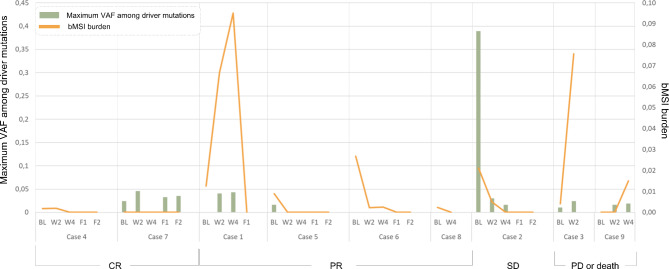
Dynamics of bMSI and maximal VAF of driver mutations observed in each of the patients’ serial blood plasma samples. Patient cases are grouped by the radiological responses to ICI.
For 2 patients (Cases 3, 9) who died or had PD, a clear increase of both bMSI and cumulative VAFs at available time points (W2 for Case 3, W4 for Case 9) was observed. Case 1 initially had an increase in bMSI and VAFs at W2 (+ 5.4% and +
5.4% and + 4% for bMSI and maximum VAF of driver, respectively, as compared to baseline LB samples) and W4 (+
4% for bMSI and maximum VAF of driver, respectively, as compared to baseline LB samples) and W4 (+ 2.9% and +
2.9% and + 0.23% for bMSI maxVAF of driver, respectively, as compared to W2 samples), followed by a dramatic decrease to zero values at F1 point (154th day of treatment), which was consistent with the clinical observations of a major PR. Although for Case 7 no signs of bMSI along with no decrease in the VAF of driver mutations were observed, the patient achieved a CR (radiological and pathological) to the therapy. A decrease of both bMSI and mutational VAF to zero values observed in three cases (Cases 5, 6, 8) was consistent with pCR. For Case 8, only baseline and W4 LB samples were available at the time of analysis. Overall, patients who had CR, PR or SD had undetectable bMSI burden at follow-up. Radiological and pathological responses were consistent for Cases 1, 7 (Tables S2-S3, Fig.S2).
0.23% for bMSI maxVAF of driver, respectively, as compared to W2 samples), followed by a dramatic decrease to zero values at F1 point (154th day of treatment), which was consistent with the clinical observations of a major PR. Although for Case 7 no signs of bMSI along with no decrease in the VAF of driver mutations were observed, the patient achieved a CR (radiological and pathological) to the therapy. A decrease of both bMSI and mutational VAF to zero values observed in three cases (Cases 5, 6, 8) was consistent with pCR. For Case 8, only baseline and W4 LB samples were available at the time of analysis. Overall, patients who had CR, PR or SD had undetectable bMSI burden at follow-up. Radiological and pathological responses were consistent for Cases 1, 7 (Tables S2-S3, Fig.S2).
In order to compare the dynamics of ctDNA parameters with radiological responses, contingency table analysis was performed. Pairs of computed tomography (CT) results, carried out in an interval of 1–3 months, were selected. CT results changes within pairs were stratified into 5 categories: “>20% increase”, “<20% increase”, “no changes”, “<20% decrease”, “>20% decrease”. For every second СT result in a pair the closest ctDNA analysis in an interval of -3/+4 weeks was chosen. The result of ctDNA analysis was also interpreted as a change from previous analysis and also stratified into 5 groups: “increase for more than 50%”, “increase for less than 50%”, “no changes”, “decrease for less than 50%”, “decrease for more than 50%”. “No changes” was determined as change less than 20%, or, for bMSI only, absolute change less than 0.1. Fisher’s exact test did not reveal a statistically significant relationship between CT and ctDNA results (Fig. 3).
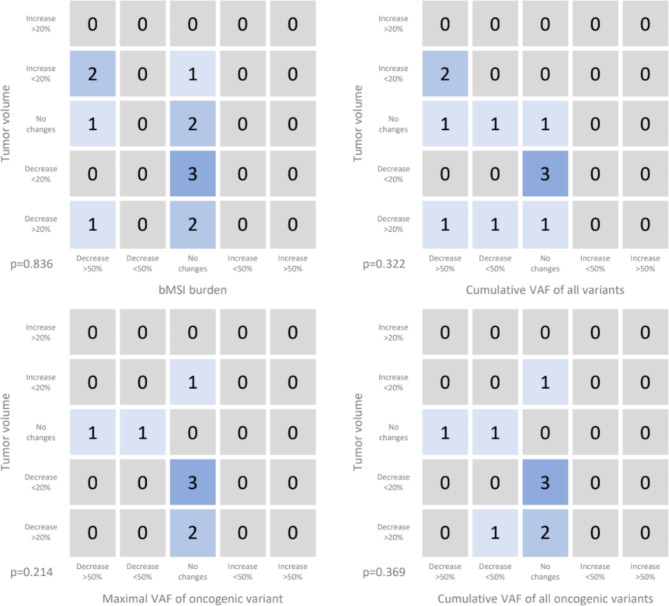
Connection of tumor volume radiological changes with changes in MSI-score and VAFs in the blood plasma. One point was added to the corresponding cell of the table if the dynamics of CT and ctDNA parameters coincided. The following ctDNA parameters were used: bMSI, cumulative VAF of all variants, maximal VAF of oncogenic variant, cumulative VAF of all oncogenic variants.
Discussion
Here, we describe preliminary data on the relationship between the dynamics of bMSI and mutational status dynamics following ctDNA NGS analysis and the response to anti-PD-1 IT. Patients participating in the study received IT in either the neoadjuvant setting (n =
= 7, stages II/III), or as systemic therapy for metastatic disease (n
7, stages II/III), or as systemic therapy for metastatic disease (n =
= 2, stage IV). It is considered that the release of ctDNA is contingent upon tumor burden, vascularity, and location42. Detecting ctDNA is hindered during the early stages of disease43,44. The location of metastatic sites is also thought to influence the ctDNA level45. Nevertheless, preliminary results of our study suggest the clinical utility of monitoring bMSI and mutational dynamics in serial LB samples in both early and late stages of CRC.
2, stage IV). It is considered that the release of ctDNA is contingent upon tumor burden, vascularity, and location42. Detecting ctDNA is hindered during the early stages of disease43,44. The location of metastatic sites is also thought to influence the ctDNA level45. Nevertheless, preliminary results of our study suggest the clinical utility of monitoring bMSI and mutational dynamics in serial LB samples in both early and late stages of CRC.
In routine clinical practice, treatment decisions are made based on the results of radiological assessments of the target lesions. However, standard imaging can miss early progression or response, and does not fully capture the changes within the tumor. Studies show that ctDNA analysis might be a suitable addition, as the appearance of ctDNA can oftentimes precede the radiological progression, and changes in ctDNA reflect the dynamic changes in the tumor in the course of therapy46–48. The promise of ctDNA analysis as an additional tool for monitoring therapy responses and guiding therapeutic decisions has been widely studied. For instance, in previous studies of neoadjuvant therapy, ctDNA dynamics demonstrated a better association with treatment efficacy and pathological response compared with CT in both CRC and other tumor types19,49. In the metastatic CRC, the rechallenge of anti-EGFR therapy can be guided by the analysis of ctDNA dynamics50. In breast cancer, switching to fulvestrant after the rise of ESR1 mutation in ctDNA in the course of aromatase inhibitor therapy has been shown to result in significant clinical benefit51. To the best of our knowledge, clinical trials are yet to clarify the role of ICI discontinuation and rechallenge in the treatment of patients with MSI-positive tumors. Nonetheless, ctDNA monitoring appears to be a promising tool aiding the identification of hyper- and pseudoprogression, a persistent clinical problem52–54.
From a methodological point of view, current approaches for the analysis of ctDNA for monitoring tumor dynamics is based on the assessment of VAF of driver mutations19. Given that only a subset of MSI-positive tumors harbor BRAF mutations and that the molecular profiles of MSI vs. MSS tumors appear to differ55, VAF-based evaluation of tumor changes in ctDNA might be limited. Using a different approach, such as the analysis of genomic signatures in ctDNA, may be helpful in overcoming the limitations associated with the lack of driver mutations in MSI-positive tumors. This approach has only been validated in relation to bTMB, and not bMSI. In a meta-analysis by Wei et al., bTMB was not found to be predictive of ICI benefit56. Given that MSI exhibits spatial and temporal heterogeneity in metastatic disease57, we reasoned that bMSI monitoring might be an alternative approach for molecular-guided clinical decisions during the course of ICI for patients with MSI-positive tumors. Additionally, methodologically, this approach might be easier, as bMSI analysis may not require molecular barcoding.
Analysis of ctDNA allowed for accurate detection of bMSI, which was highly concordant with VAF of the observed mutations, however, bMSI analysis exhibited higher accuracy in relation to treatment outcomes. In terms of therapy response monitoring, analysis of bMSI dynamics in serial plasma samples and VAFs changes were generally consistent with response to the therapy. For patients with negative treatment outcomes, bMSI rapidly increased in the course of treatment, while for patients with clinical benefit bMSI was undetectable on the first follow-up visit after starting ICI. The only patient with undetectable level of MSI in ctDNA had stage II disease, which is consistent with observations that at earlier stages sensitivity of ctDNA tests is lowered (Case 7). Although generally correlating with the treatment outcomes, a standalone analysis of maximal VAF of driver mutations, a methodology commonly used for the analysis of tumor dynamics using blood plasma, was not consistent with treatment outcomes for all of the patients, partially due to the fact that not all of the patients had detectable driver mutations.
Nevertheless, although unlike VAF analysis, bMSI dynamics were consistent with the results of radiological assessment in all cases (Fig. 2), the study was unable to statistically confirm the connection between dynamics of the ctDNA parameters and the dynamics of objective responses registered with CT. This may not only be due to the small sample size, but the mismatches in the testing time via СT and ctDNA. Additionally, bMSI correlated with VAF of the observed mutations in serial LB samples, indicating that determination of bMSI is highly dependent on the total ctDNA fraction. At the same time, in 3 (out of 5) cases with known pathological response, the clearance of bMSI to zero coincided with pCR (Cases 5, 6, 8), while radiological responses coincided with pathological in only two cases (Case 1, 7), indicating that the dynamics of bMSI in ctDNA may have a predictive value only for pathological responses19,49, but at the same time may be used in the absence of detectable mutations. Analysis of the relationship between MSI dynamics and the development of resistance to ICI in the metastatic cases was complicated by either the loss of follow-up (Case 2) or rapid progression of disease (Case 9).
Despite the limited patient cohort, our results illuminate the promising application of MSI monitoring in the blood plasma for tracking tumor responses and disease progression in the course of ICI therapy. Analysis of bMSI in serial samples may overperform the traditional analysis of the VAF of driver mutations, and serve as a complementary approach to standard imaging studies for monitoring tumor response.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
| LB | Liquid biopsy |
| CRC | Colorectal cancer |
| MSI | Microsatellite instability |
| bMSI | Blood MSI burden |
| ICI | Immune checkpoint inhibitors |
| IT | Immunotherapy |
| dMMR | Mismatch repair deficiency |
| PCR | Polymerase chain reaction |
| IHC | Immunohistochemistry |
| NGS | Next-generation sequencing |
| ctDNA | Circulating tumor DNA |
| VAF | Variant allele frequency |
| STR | Short tandem repeat |
| RECIST v1.1 | Response Evaluation Criteria in Solid Tumor version 1.1 |
| ECOG | Eastern Cooperative Oncology Group |
| FFPE | Formalin-fixed paraffin-embedded |
| VUS | Variant of uncertain significance |
| CR | Complete response |
| PR | Partial response |
| SD | Stable disease |
| PD | Disease progression |
| pCR | Complete pathological response |
| mPR | Major pathological response |
| N/A | Not available |
| MSS | Microsatellite stable |
| wt | Wild type |
| BL | Baseline blood plasma sample |
| CT | Computed tomography |
Author contributions
Olesya Kuznetsova, Daria Kravchuk, Tatiana Grigoreva, Victoria Yudina, Laima Belyaeva, and Vladislav Nikulin carried out the studies, participated in collecting data. Egor Veselovsky, Alexandra Lebedeva, Ekaterina Belova, Anastasiia Taraskina, Alexandra Kavun, and Maxim Ivanov performed data analysis and drafted the manuscript. Alexey Tryakin, Mikhail Fedyanin, Vladislav Mileyko, and Maxim Ivanov provided administrative support. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the Russian Science Foundation (grant No. 22-75-10154, https://rscf.ru/project/22-75-10154/).
Data availability
Data will be made available from the corresponding author on reasonable request.
Declarations
Egor Veselovsky, Alexandra Lebedeva, Olesya Kuznetsova, Ekaterina Belova, Anastasia Taraskina, Tatyana Grigoreva, Alexandra Kavun, Laima Belyaeva, Vladislav Mileyko, Maxim Ivanov are currently employed by OncoAtlas LLC. Other co-authors have no conflicts of interest to disclose.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/169166649
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Clinical Trials
- (2 citations) ClinicalTrials.gov - NCT06414304
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Performance of Next-Generation Sequencing for the Detection of Microsatellite Instability in Colorectal Cancer With Deficient DNA Mismatch Repair.
Gastroenterology, 161(3):814-826.e7, 13 May 2021
Cited by: 24 articles | PMID: 33992635
Prediction of Benefit from Checkpoint Inhibitors in Mismatch Repair Deficient Metastatic Colorectal Cancer: Role of Tumor Infiltrating Lymphocytes.
Oncologist, 25(6):481-487, 22 Jan 2020
Cited by: 61 articles | PMID: 31967692 | PMCID: PMC7288636
Plasma-based microsatellite instability detection strategy to guide immune checkpoint blockade treatment.
J Immunother Cancer, 8(2):e001297, 01 Nov 2020
Cited by: 19 articles | PMID: 33172882 | PMCID: PMC7656957
Funding
Funders who supported this work.
Russian Science Foundation (1)
Grant ID: No 22-75-10154

 1,3
1,3

