Abstract
Background
Phenotypic variability within families with epilepsy is often observed, even when relatives share the same monogenic cause. We aimed to investigate whether common polygenic risk for epilepsy could explain the penetrance and phenotypic expression of rare pathogenic variants in familial epilepsies.Methods
We studied 58 clinically heterogeneous families with genetic epilepsy with febrile seizures plus (GEFS+). Relatives were coded as either unaffected or affected with epilepsy, and graded according to phenotype severity: no seizures, febrile seizures (FS) only, febrile seizures plus (FS+), generalised/focal epilepsy, or developmental and epileptic encephalopathy (DEE). Epilepsy polygenic risk scores (PRSs) were tested for association with epilepsy phenotype. Within families, the mean PRS difference was compared between pairs concordant versus discordant for phenotype severity. Statistical analyses were performed using mixed-effect regression models.Findings
304 individuals segregating a known, or presumed, rare variant of large effect, were studied. Within families, higher epilepsy polygenic risk was associated with an epilepsy diagnosis (OR = 1.39, 95% CI 1.08, 1.80, padj = 0.040). Relatives with a more severe phenotype had a mean pairwise PRS difference of +0.19 higher than relatives with a milder phenotype (padj = 0.010). The difference increased with greater phenotype discordance between relatives. As the cohort included two rare variants with >30 relatives each, variant-specific genotype-phenotype associations could also be analysed. Whilst the epilepsy PRS effect was strong for relatives segregating the GABRG2 p.Arg82Gln pathogenic variant (padj = 0.0010), the effect was not significant for SCN1B p.Cys121Trp.Interpretation
We provide support for genetic background modifying the penetrance and phenotypic expression of rare variants associated with 'monogenic' epilepsies. In GEFS+ families, relatives with higher epilepsy PRSs were more likely to show penetrance (epilepsy diagnosis) and a more severe phenotype. Variant-specific analyses suggest that some rare variants may be more susceptible to PRS modification, carrying important genetic counselling and disease prognostication implications for patients.Funding
National Health and Medical Research Council of Australia, Medical Research Future Fund of Australia.Free full text

Investigating the effect of polygenic background on epilepsy phenotype in ‘monogenic’ families
Summary
Background
Phenotypic variability within families with epilepsy is often observed, even when relatives share the same monogenic cause. We aimed to investigate whether common polygenic risk for epilepsy could explain the penetrance and phenotypic expression of rare pathogenic variants in familial epilepsies.
Methods
We studied 58 clinically heterogeneous families with genetic epilepsy with febrile seizures plus (GEFS+). Relatives were coded as either unaffected or affected with epilepsy, and graded according to phenotype severity: no seizures, febrile seizures (FS) only, febrile seizures plus (FS+), generalised/focal epilepsy, or developmental and epileptic encephalopathy (DEE). Epilepsy polygenic risk scores (PRSs) were tested for association with epilepsy phenotype. Within families, the mean PRS difference was compared between pairs concordant versus discordant for phenotype severity. Statistical analyses were performed using mixed-effect regression models.
Findings
304 individuals segregating a known, or presumed, rare variant of large effect, were studied. Within families, higher epilepsy polygenic risk was associated with an epilepsy diagnosis (OR = 1.39, 95% CI 1.08, 1.80, padj = 0.040). Relatives with a more severe phenotype had a mean pairwise PRS difference of +0.19 higher than relatives with a milder phenotype (padj = 0.010). The difference increased with greater phenotype discordance between relatives. As the cohort included two rare variants with >30 relatives each, variant-specific genotype–phenotype associations could also be analysed. Whilst the epilepsy PRS effect was strong for relatives segregating the GABRG2 p.Arg82Gln pathogenic variant (padj = 0.0010), the effect was not significant for SCN1B p.Cys121Trp.
Interpretation
We provide support for genetic background modifying the penetrance and phenotypic expression of rare variants associated with ‘monogenic’ epilepsies. In GEFS+ families, relatives with higher epilepsy PRSs were more likely to show penetrance (epilepsy diagnosis) and a more severe phenotype. Variant-specific analyses suggest that some rare variants may be more susceptible to PRS modification, carrying important genetic counselling and disease prognostication implications for patients.
Funding
National Health and Medical Research Council of Australia, Medical Research Future Fund of Australia.
Introduction
In monogenic disease, incomplete penetrance and variable phenotype expressivity are typically observed. This is true even when individuals have inherited the same rare pathogenic variant of major effect. Epilepsy is no exception.
Whilst the overall genetic architecture of the epilepsies is complex, the number of over 1000 established monogenic causes, sets epilepsy apart from many other complex diseases.1,2 Most epilepsy genes are associated with a broad phenotypic spectrum, even in families segregating a pathogenic variant.3 This creates confusion and concern for patients and clinicians, particularly with regards to disease prognostication.
Genetic modifiers have long been proposed to at least partially explain phenotypic heterogeneity of monogenic diseases. Therefore, modifiers are important to detect, not just for disease prognosis, but because they may reveal novel therapeutic avenues. Despite promising findings in murine models, limited progress towards identifying human modifier genes has been made.4 Recently, however, multiple studies of specific human diseases, such as coronary artery disease and cancer, highlighted that common polygenic background could modify the penetrance of monogenic variants.5,6
In the epilepsies, interrogation of common variant burden has recently become feasible, through the application of polygenic risk scores (PRSs) derived from large genome-wide association studies (GWASs).7 The common focal and generalised epilepsies have largely been regarded as following complex inheritance, invoking a polygenic basis with possible environmental contributions. It was unsurprising, therefore, to find that epilepsy PRSs were enriched in independently sampled patients with common focal and generalised epilepsies.8 Intriguingly, however, cohorts with rare epilepsies of known or presumed monogenic cause, such as the developmental and epileptic encephalopathies (DEEs)9 and large families with epilepsy,10 were also enriched for epilepsy PRSs.
With common genetic variation playing a role in monogenic epilepsies, we sought to determine whether common variant burden could explain (at least partially) the elusive genetic modifiers influencing phenotype in the epilepsies. Specifically, we asked whether increased epilepsy polygenic risk is associated with epilepsy diagnosis (penetrance) and increasing phenotype severity (variable expressivity) in families with monogenic epilepsies.
Methods
Cohort
We selected families with genetic epilepsy with febrile seizures plus (GEFS+) to seek evidence for common epilepsy risk variants modifying phenotypes because of the marked intra-familial phenotypic variability and the number of GEFS+ families with monogenic causes.3 We studied families from the University of Melbourne's (UoM's) Epilepsy Genetics Program and the Epi4K Consortium11 that met the following criteria for GEFS+3: 1) families where ≥2 relatives had a GEFS+ phenotype, of which at least one individual had febrile seizures plus (FS+), or 2) if the family contained no individuals with FS+, then three or more individuals with well-recognised GEFS+ phenotypes. Well-recognised GEFS+ phenotypes include febrile seizures (FS), FS+, FS/FS+ with generalised or focal seizures, epilepsy with myoclonic-atonic seizures (EMAtS), and Dravet syndrome.3,12,13
We also included five individuals with GEFS+ phenotypes from Australia, the United Kingdom, and United States who shared the SCN1B p.Cys121Trp pathogenic variant from a distant common ancestor with nine GEFS+ families.14 For our statistical analyses 41 relatives, from these 14 distantly-related families, were treated as one family.
Population control data were obtained from the QSkin Sun and Health Study (QSkin), a prospective cohort study of men and women aged 40–69 years, randomly sampled from the Australian state of Queensland.15
Ethics
All participants, or their parents or legal guardians, provided signed informed consent according to local IRB requirements. This study was approved by Austin Health Human Research Ethics Committee (H2007/02961) and Walter and Eliza Hall Institute Ethics Committee (G20/01). The QSkin Sun and Health Study was approved by QIMR Berghofer Human Research Ethics Committee (P1309, P2034), including amendments to share data with this study.
Phenotyping
Phenotypes were classified according to the International League Against Epilepsy classification16 incorporating seizure semiology, EEG, and brain MRI results, where available. Family members were grouped into those with and without an epilepsy diagnosis (binary scale) and according to the following phenotype severity scale: 1 = no seizures, 2 = FS only, 3 = FS+, 4 = generalised/focal/unclassified/mixed epilepsy, or 5 = DEE.
FS were defined as seizures associated with fever of at least 38°C, occurring between 6 months and 6 years of age.17 Simple and complex FS were not distinguished. A classification of FS+ was made when FS occurred outside the 6 months to 6 years age range or when an afebrile tonic-clonic seizure occurred in addition to seizures with fever. Individuals with an EMAtS or Dravet syndrome diagnosis were classified as having a DEE.
Overall, individuals were considered affected with epilepsy if they had a phenotype scale of 3, 4 or 5. Individuals with no seizures or FS only were considered unaffected for epilepsy (Table 1). Noncarrier, unaffected founders and married-in parents were excluded from the cohort (Supplementary Figure S1).
Table 1
Individual phenotypes within the GEFS+ cohort.
| Epilepsy | N | Phenotype | Severity scale | N (%) |
|---|---|---|---|---|
| No | 149 | No seizures | 1 | 66 (21.7%) |
| FS only | 2 | 83 (27.3%) | ||
| Yes | 155 | FS+ | 3 | 47 (15.4%) |
| Generalised epilepsy (n = 45, 50%) Focal epilepsy (n = 34, 36%) Unclassified/Mixed epilepsy (n = 13, 14%) | 4 | 92 (30.3%) | ||
| DEE | 5 | 16 (5.3%) | ||
| Total | 304 | 304 | ||
Abbreviations: N, number; FS, febrile seizures; FS+, febrile seizures plus; DEE, developmental and epileptic encephalopathy.
We included families where a heterozygous rare variant (RV) of large effect was either known or presumed to segregate with GEFS+ phenotypes, consistent with autosomal dominant inheritance.
Individuals were explicitly coded as RV carriers (yes/no) according to records from the UoM's Epilepsy Genetics Program and Epi4K Consortium. Variants were classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines. We included RVs that were classified as “pathogenic” and “likely pathogenic”.18 We also included variants that met criteria for “uncertain significance” but segregated with GEFS+ phenotypes in families in a biologically plausible gene. This included variants that have been previously published as “risk variants”19,20 and those in established epilepsy genes that have not been previously associated with GEFS+ specifically. No families were known to segregate pathogenic, or any of the established recurrent risk, copy number variants (e.g., the 15q13.3 microdeletion) for epilepsy.
Genetic data processing
GEFS+ families from the UoM's Epilepsy Genetics Program and population controls were genotyped using the Illumina Global Screening Array-24 (San Diego, CA, USA). GEFS+ families from the Epi4K Consortium were genotyped on the Illumina HumanCore or Human Multi-Ethnic arrays. We harmonised across the array types as previously described10 and performed standard quality control (QC) measures using PLINK v1.9.21
We excluded SNPs that exhibited high rates of missingness (>2%) or low minor allele frequency (MAF<5%). We also excluded samples with a high proportion of missing genotypes (>2%) or those with sex mismatch between genetically inferred and the reported sex assigned at birth. We generated pairwise identity-by-descent estimates to confirm pedigree structures. Principal component (PC) analysis for ancestry was performed on our GEFS+ cohort, merged with 1000 Genomes data, using PC-Air to account for related samples.22 Samples that did not cluster within ± four standard deviations (SDs) of the mean PC1 and PC2 for the European super population, were excluded.
SNP imputation to the HRC r1.1 2016 (GRCh37/hg19) reference panel was performed using Minimac4 as implemented on the Michigan Imputation Server 23 with pre-imputation phasing using Eagle v2.4.23 Post-imputation, we selected high-quality imputed and genotyped SNPs (imputation quality scores R2 > 0.9) and repeated the above standard QC measures.
PRSs were calculated using PRSice-224 for all samples using SNP effect sizes from the most recent epilepsy GWAS.7 We utilised the European-derived summary statistics that included cases with generalised (n = 6,952), focal (n = 14,939) and unclassified (n = 5,668) epilepsy.
We considered the ‘all’ epilepsy GWAS the most relevant for our PRS model due to it incorporating a mixture of epilepsy phenotypes, as is typically observed in GEFS+ families.3 Sensitivity analyses were performed using PRSs for focal epilepsy, generalised epilepsy7 and FS25 (Supplementary Table S1). All PRS models were calculated using a significance threshold of p < 0.5 for SNP inclusion. We chose this lenient threshold as it has previously been shown to be optimal for epilepsy.8,10 PRS values were standardised to a normal distribution with a mean of zero and SD of one; this was done based on the population control group that had a raw PRS SD of 1.78.
Of note, 14/58 GEFS+ families contributed a single individual to the ‘all’ epilepsy GWAS (0.05% cases).7 Recognising that this may be a source of bias,26 we repeated our ‘all’ epilepsy analysis excluding the 14 impacted families (Supplementary Table S2).
Statistics
Polygenic risk in GEFS+ families
Statistical analyses were performed to compare the mean PRS values for GEFS+ family members without an epilepsy diagnosis (no seizures or FS) to those with epilepsy (FS+, generalised, focal, unclassified, mixed epilepsy or DEE); i.e., the binary phenotype scale. To account for familial data, we used logistic mixed-effects regression models as implemented in the lme4 R package.27 The model treated epilepsy as a binary outcome variable with PRSs as the exposure variable, adjusted for sex assigned at birth, RV status, FS status, cohort (UoM or Epi4K), the first five ancestry PCs as fixed-effects and family identifier (FID) as a random-effects covariate with an assumed normal distribution.
Putative confounders for inclusion as fixed-effects were identified based on (modified) disjunctive cause criterion where we included variables that may be causal contributors to the exposure (PRS) or outcome (epilepsy) or both (Supplementary Figure S2). The first five ancestry PCs were chosen as they account for >98% of the variance. The reported p-value was Bonferroni-adjusted for the four PRS models tested (Supplementary Table S1).
We then stratified the entire cohort by PRS decile. Decile breakpoints were determined from the PRS distribution of population controls. Standardised odds ratio (OR) estimates were derived from logistic mixed-effects regression models as described above to test for enrichment in the top decile of PRS distribution. Next, we stratified the GEFS+ cohort according to presence or absence of a RV and repeated the logistic mixed-effects regression analysis for both groups.
Pairwise phenotype severity analyses
We were particularly interested in exploring phenotypic heterogeneity within families. For this analysis, we utilised the severity phenotype scale (i.e., ranging from 1 [no seizures] to 5 [DEE]). To determine whether higher epilepsy PRSs were associated with greater phenotype severity, we calculated the PRS difference between all relative pairs within each family (Supplementary Figure S1).
The PRS difference between concordant pairs was calculated by randomly taking the PRS value of one relative from the other; for these pairs there was no a priori order. A random order that best reflected a mean PRS difference of zero was determined by performing 10,000 simulations with pair orders randomly switched each time resulting in a normal distribution (Supplementary Figure S3).
The PRS difference was then calculated for all discordant pairs. Here, the relative order was determined by the phenotype scale due to the a priori hypothesis that epilepsy PRSs would be positively associated with clinical severity. As such, the PRS for the relative with the milder phenotype (lower scale) was always taken from the PRS of the relative with the more severe phenotype (higher scale). These discordant pairs were further coded by degree of phenotypic discordance; the degree of discordance could range from 1 to 4. For example, for a pair comprising one relative with FS only (phenotype scale 2) and the other FS+ (phenotype scale 3) the level of discordance was 1-grade. A pair comprising one relative with no seizures (phenotype scale 1) and the other a DEE (phenotype scale 5), the degree of discordance was 4-grades.
Statistical comparisons were made using linear mixed models with kinship included as a fixed-effect covariate and family identifier (FID) as a random-effects covariate with an assumed normal distribution. As the distribution of the resulting test statistic was unknown, we randomly re-ordered the discordance group label for relative pairs 105 times to generate an empirical null distribution against which to compare the pairwise PRS test statistic. The proportion of test statistics that were greater than or equal to the original linear mixed model estimates provided an empirical p-value for the null hypothesis. Reported p-values were Bonferroni-adjusted for the five test comparisons.
Variant-specific analyses
The pairwise analysis approach was further utilised to explore the association between epilepsy PRSs and the two pathogenic RVs with the largest sample size: GABRG2 p.Arg82Gln and SCN1B p.Cys121Trp. Here the mean PRS difference between concordant and discordant relative pairs were compared for the ‘all’, generalised and focal epilepsy PRS models. Statistical comparisons were made using linear regression models with pairwise kinship values included as a fixed-effect covariate. Reported p-values were Bonferroni-adjusted for the six test comparisons.
Putative genetic interactions
The variant-specific analyses revealed positive findings for the GABRG2 p.Arg82Gln RV. To identify individual SNPs from the epilepsy PRS that could be driving this association, we first annotated each SNP for an associated gene symbol using the variant effect predictor (VEP) application.28 We then identified the SNPs associated with “GABA receptor activity” (GO:0016917) genes according to the Gene Ontology database.29,30 SNP genotypes for each GABA receptor associated SNP, for all individuals carrying the GABRG2 p.Arg82Gln RV, were extracted using PLINK v1.9.21 Hierarchical clustering analysis of 29 SNPs was performed using the ComplexHeatmap package31 in R, using default parameters.
All statistical analyses were performed using R version 3.6.3. Relevant code can be found here: https://github.com/bahlolab/GEFS-PRS-manuscript.
Role of funders
The funders played no role in study design, data collection, data analyses, interpretation, or writing of this manuscript.
Results
Cohort
We studied 304 individuals from 58 families that met the clinical criteria for GEFS+, were consistent with autosomal dominant inheritance, and had genetic data available. The cohort comprised an almost equal number of unaffected relatives (n = 149) and affected (n = 155) family members with epilepsy (Table 1). The median family size was 3 (IQR 2–6).
Twenty-eight of 58 GEFS+ families (48%) had a known heterozygous RV (Supplementary Table S3). Twenty-four families had a causal pathogenic variant; another four had RVs identified as a potentially relevant to their familial epilepsy. The clinical pattern and family sizes were similar regardless of RV status (Supplementary Figures S4 and S5).
Polygenic risk in GEFS+ families
Within families, higher ‘all’ epilepsy PRSs were associated with epilepsy status. For every 1-SD increase in epilepsy PRS, relatives were approximately 40% more likely to be diagnosed with epilepsy than not (OR = 1.39, 95% CI 1.08, 1.80, padj = 0.040, Wald test), where the mean epilepsy PRS for unaffected relatives was −0.0094 (SD 0.96) compared to 0.31 (SD 1.05) for affected relatives (Fig. 1). We note, however, that despite the positive shift in PRS distribution for family members with epilepsy compared to those without, the distributions do largely overlap limiting current clinical application (ROC AUC = 74.9%, 95% CI 69.4%, 80.4%; Supplementary Figure S6).
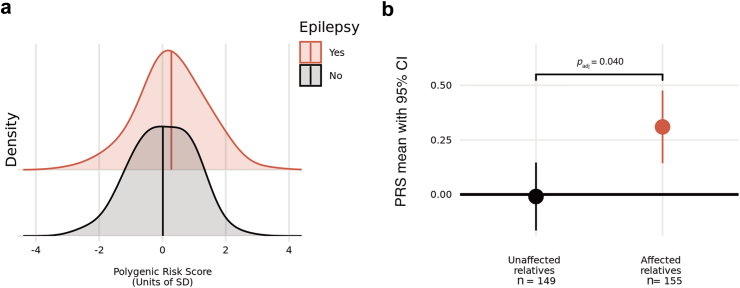
a: Normalised distributions of epilepsy PRS in GEFS+ cohort stratified by epilepsy status, b: Mean epilepsy PRS values with 95% confidence intervals for cohort stratified by epilepsy status relative to the control population sample (bold horizontal line at y = 0). Note, the unaffected relative group without epilepsy includes individuals with no seizures and those with febrile seizures only. The epilepsy PRS model was comprised of 39,074 SNPs. The logistic mixed-effects regression model used PRS as exposure variable, sex assigned at birth, cohort, RV, FS and the first five ancestry PCs as fixed-effects covariates, and family identifier as random-effects covariate. The reported p-value was adjusted for the four PRS models tested (see methods; Supplementary Table S1).
Relatives in the highest decile of ‘all’ epilepsy polygenic risk were 3.5 times more likely to have an epilepsy diagnosis compared to all other relatives (OR = 3.52, 95% CI 1.50, 8.22, p = 0.0037, Wald test; Supplementary Table S4a). When using the lowest decile of the ‘all’ epilepsy PRS as the reference compared to all other deciles of epilepsy polygenic risk, it was only the highest decile that was significantly enriched for relatives with epilepsy compared to those without (OR = 4.44, 95% CI 1.05, 18.7, p = 0.041, Wald test; Supplementary Table S4b).
When families were stratified by RV status, the positive epilepsy PRS association with epilepsy status was not significant; likely due to the smaller sample sizes (Supplementary Figure S7).
Pairwise phenotype severity analyses
Our cohort comprised a total of 2399 relative pairs. Of these, 784 (33%) relatives had concordant phenotypes (Supplementary Table S5) and a mean PRS difference of zero (Supplementary Figure S3).
For discordant pairs, relatives with the more severe phenotype had a higher epilepsy PRS compared to their less severely affected counterpart; the mean PRS difference was +0.19 (padj = 0.010, permutation-derived, Fig. 2a). The difference increased in an additive trend with degree of phenotype discordance (Fig. 2b). Whilst the mean PRS difference was zero for discordant relatives separated by only 1-grade of severity, the difference was +0.27 for relatives separated by 2-grades, and then +0.41 for relative pairs with a 3-grade difference in phenotype severity.
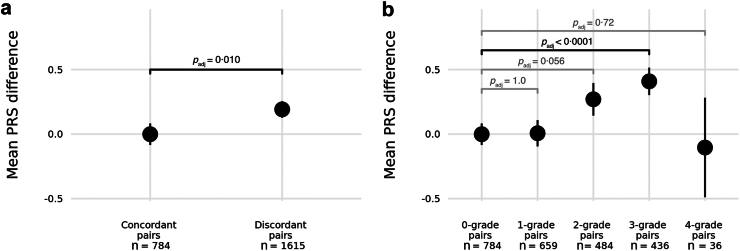
Mean epilepsy PRS difference with 95% confidence intervals between pairs of relatives within GEFS+ families. a: Comparison between relative pairs with concordant versus discordant phenotypes. b: Comparison between relative pairs grouped by degree of phenotype discordance where 0-grade difference captures concordant phenotypes. Statistical comparisons were made using linear mixed models with kinship included as a fixed-effect covariate and family identifier (FID) as a random-effects covariate. Permutation derived p-values were adjusted for five test comparisons.
The positive trend did not hold for the group of relative pairs with greatest phenotypic difference (4-grade). This group comprised a much smaller sample size of only 36 relative pairs, from just 12 families, where one family member had no seizures whilst the other had a DEE (Supplementary Table S5). More than half of these phenotypically disparate pairs (20/36, 56%) are carriers of the SCN1B p.Cys121Trp founder variant. Notably, one of the SCN1B p.Cys121Trp relatives with DEE is bi-allelic with a known second RV in SCN1B (c.449-3C>A), thus representing a different genetic model of recessive inheritance.
Variant-specific analyses
We repeated the pairwise analyses in experiments comparing relatives carrying one of two RVs: SCN1B p.Cys121Trp and GABRG2 p.Arg82Gln. These two pathogenic variants accounted for the largest number of RV positive samples (n = 41 SCN1B p.Cys121Trp; n = 32 GABRG2 p.Arg82Gln) (Supplementary Table S3), thus providing sample sizes sufficient for independent analysis.
Relatives sharing the GABRG2 p.Arg82Gln variant (496 relative pairs; 128 concordant, 368 discordant) demonstrated a strong positive association between epilepsy PRS and phenotype severity (Fig. 3). On average, relatives with the more severe phenotype had a higher epilepsy PRS compared to those with a less severe phenotype (+0.54 mean PRS difference; padj = 0.0010, permutation-derived). Pairwise epilepsy PRS differences for the SCN1B p.Cys121Trp founder variant (820 pairs; 176 concordant, 644 discordant) showed a similar but non-significant trend (Table 2).
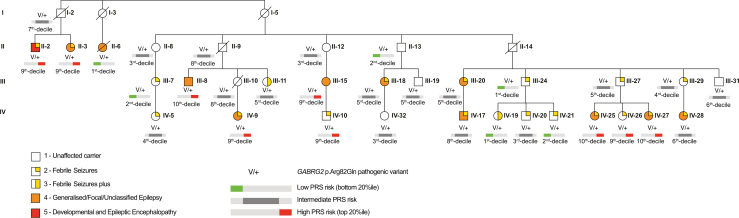
Pedigree of family with the GABRG2 p.Arg82Gln rare variant. Modified from version published in Marini et al., 2003 to only include relatives with both the RV and DNA available for PRS generation. Individual pedigree identifiers have been kept the same for all relatives that are included in both papers. Eight of the eleven relatives coded as severity scale 4 (orange symbol) have childhood absence epilepsy (a generalised epilepsy syndrome). The exceptions are individuals II-6 and IV-9 with temporal lobe epilepsy (a focal epilepsy syndrome) and III-15 with unclassified epilepsy. Individual II-2 coded as severity scale 5 (red symbol) has a diagnosis of epilepsy with myoclonic-atonic seizures (EMAtS).
Table 2
Variant-specific mean pairwise PRS difference result comparisons for three epilepsy PRS models.
| Rare-variant | PRS model | Mean PRS difference ±SD | Unadjusted p-values | Adjusted p-values | |
|---|---|---|---|---|---|
| Concordant pairs | Discordant pairs | ||||
| GABRG2 p.Arg82Gln | ‘All’ epilepsy PRS | 0.00 ± 1.24 | 0.54 ± 1.38 | 0.00017 | 0.0010 |
| Generalised epilepsy PRS | 0.00 ± 1.20 | 0.52 ± 1.33 | 0.00017 | 0.0010 | |
| Focal epilepsy PRS | 0.00 ± 1.16 | 0.25 ± 1.34 | 0.047 | 0.28 | |
| SCN1B p.Cys121Trp | ‘All’ epilepsy PRS | 0.00 ± 1.22 | 0.14 ± 1.34 | 0.10 | 0.60 |
| Generalised epilepsy PRS | 0.00 ± 1.43 | 0.00 ± 1.46 | 0.46 | 1.0 | |
| Focal epilepsy PRS | 0.00 ± 1.65 | 0.27 ± 1.66 | 0.029 | 0.17 | |
The GABRG2 p.Arg82Gln are based on 128 relative pairs concordant for phenotype and 368 discordant. The SCN1B p.Cys121Trp are based on 176 relative pairs concordant for phenotype and 644 discordant. Statistical comparisons were made using linear models with kinship included as a fixed-effect covariate. Permutation derived p-values were adjusted for six test comparisons.
Stratifying the variant-specific discordant pairs by degree of phenotype disparity, a similar trend to the overall cohort result was seen for SCN1B p.Cys121Trp where the positive association failed to hold for the most disparate relatives (i.e., 4-grade pairs). This was not surprising as SCN1B p.Cys121Trp relatives contributed the majority of pairs to this group. In contrast, however, for GABRG2 p.Arg82Gln pairs, the positive trend held across all four degrees of phenotypic disparity, albeit with even smaller numbers (Supplementary Figure S8).
Next, noting that the epilepsy phenotypes associated with the two RVs differed, we also performed variant-specific analyses using the generalised and focal epilepsy PRS models. GABRG2 p.Arg82Gln relatives were more likely to have a generalised epilepsy syndrome diagnosis (specifically childhood absence epilepsy)32 compared to relatives with the SCN1B p.Cys121Trp RV who were more likely to have a focal epilepsy syndrome diagnosis (specifically temporal lobe epilepsy)14 (Supplementary Table S6).
For the GABRG2 family, the generalised epilepsy PRS model performed almost as well as the ‘all’ epilepsy model, however, the focal PRS model did not. For the SCN1B relatives, we saw the opposite with the focal epilepsy PRS model performing better than the ‘all’ epilepsy model whilst the generalised model showed no effect (Table 2).
Putative genetic interactions
We were interested in exploring which of the common variants contributing to the ‘all’ epilepsy PRS might be driving the GABRG2 p.Arg82Gln RV association. We specifically hypothesised an interaction effect between GABRG2 p.Arg82Gln with one or more biologically related genes. Hierarchical cluster analysis of 29 common SNPs associated with “GABA receptor activity” (GO:0016917) revealed rs12898904 in GABRB3 on chromosome 15q12 as the top candidate associated with epilepsy phenotype (Supplementary Figure S9).
Discussion
Epilepsy is highly genetically and phenotypically heterogeneous.2 Epilepsy genes often have a broad phenotypic spectrum, ranging from self-limited epilepsies to DEEs, which may be due to different functional effects (e.g., loss-of-function versus gain-of-function).33 To account for allelic heterogeneity in the search for genetic modifiers, we took a familial approach, allowing us to control for the monogenic cause of major effect, at both the gene and variant level, to determine whether polygenic background plays a role in phenotypic variation.
We explored whether common variant burden explained phenotypic penetrance and severity in families with GEFS+ with a known or presumed pathogenic variant of major effect. Family members with higher epilepsy PRSs were more likely to have epilepsy and greater phenotypic severity. For every 1-SD increase in epilepsy PRS, relatives were approximately 40% more likely to have epilepsy; and in the top decile of the epilepsy PRS distribution, we observed a 4-fold enrichment of relatives with an epilepsy diagnosis. On average, relatives with a more severe phenotype had higher epilepsy PRSs compared to less severely affected relatives and the PRS difference increased with severity disparity.
We specifically targeted GEFS+, a syndrome characterised by phenotypic heterogeneity.3 GEFS+ families typically show autosomal dominant inheritance patterns, with several genes identified. Our cohort included two large families with a known cause; SCN1B p.Cys121Trp and GABRG2 p.Arg82Gln. Due to the large number of relatives in each, we also performed variant-specific analyses and found that higher ‘all’ epilepsy polygenic risk was associated with greater phenotype severity for carriers of GABRG2 p.Arg82Gln.
Interestingly, the pattern of epilepsy phenotypes within each family differed. Individuals with the SCN1B RV were more likely to have temporal lobe epilepsy (focal epilepsy), whilst the GABRG2 RV carriers were more likely to have childhood absence epilepsy (generalised epilepsy). This led us to explore whether common risk variants specific to generalised or focal epilepsy had differing phenotypic impacts. We found an enrichment for common focal epilepsy risk variants in SCN1B p.Cys121Trp carriers with more severe phenotypes. One exception was an individual who had a second RV in SCN1B contributing to a rare recessive SCN1B DEE phenotype.34 Taken together, common variants, specific to focal epilepsy risk, and rare variants are likely to contribute to the phenotypic heterogeneity in GEFS+ associated with SCN1B p.Cys121Trp.
The phenotypic heterogeneity in GEFS+ caused by GABRG2 p.Arg82Gln is strongly associated with common risk variants for both ‘all’ epilepsy and generalised epilepsy. This variant, previously annotated as p.Arg43Gln, segregates in a GEFS+ family with predominantly FS and childhood absence epilepsy.35 In 2003, an attempt to identify genetic modifiers by linkage analysis to explain this phenotypic heterogeneity revealed putative loci on chromosomes 10q21, 13p11-q12, 14q22-q23 and 15q11-q13,32 however, no specific variants were identified. A knock-in GABRG2 p.Arg82Gln mouse model (known as Gabrg2 R43Q) recapitulated the FS and childhood absence epilepsy phenotypes observed in the GEFS+ family.36 Subsequently the Gabrg2 R43Q mouse was back-crossed to strains with different seizure susceptibility. This revealed that only the knock-in mice on a DBA/2J “seizure susceptible” background developed spike-and-wave discharges; knock-in mice on a C57BL/6J “seizure resistant” background had no spontaneous spike-and-wave activity.37 This result mirrors our observation that the penetrance of the RV depends on epilepsy risk conferred by genetic background as captured by the ‘all’ epilepsy PRS.
The ‘all’ epilepsy PRS model utlised in this study incorporated >39,000 common SNPs. Determining which of these SNPs, or associated genes, are contributing to the observed genetic modification is an important, though challenging, next step. For GABRG2 p.Arg82Gln, we hypothesised an interplay between one or more PRS variants involved in the same biological pathway. Thus, our exploratory analysis focussed on SNPs in the epilepsy PRS associated with “GABA receptor activity” genes. This revealed a single SNP in GABRB3 as a top interaction candidate. The plausibility of a rare GABRB3 modifier variant (in linkage disequilibrium with rs12898904) for generalised epilepsy is supported by reports of rare GABRB3 variants being associated with absence epilepsies and more severe epilepsy syndromes.38,39 Furthermore, GABRB3 was included in one of the four “childhood absence epilepsy linkage” regions (15q11-q13) reported in this family in 2003.32 Systematic analysis of gene–gene interaction or epistasis in complex disease has been limited, largely due to the substantial multiple-testing burden intrinsic to genome-wide epistasis screening.40 Leveraging our results to target specific SNPs will help navigate the multiple-testing burden of any hypothesis-free approach.
The correlation of higher epilepsy PRSs and more severe phenotype did not hold true in all cases. For example, two individuals with the GABRG2 p.Arg82Gln variant coupled with a high PRS only had FS (Fig. 3; IV-10 and IV-26). These ‘genomically resilient’ individuals may harbor protective variants and represent another avenue for future research. In one reported GEFS+ family, a second variant in SCN1A modified pathogenicity of the pathogenic SCN1A variant shared by all family members. The second rare variant, exclusively shared by unaffected carriers, counteracted the loss-of-function effect of the pathogenic variant.41 Mouse models have also revealed other potential modifier genes for GEFS+. Specifically, knock-in mice, heterozygous for an Scn1A GEFS+ missense variant (p.Arg1648His), demonstrated dramatic phenotypic variability when combined with variants in Scn2a, Kcnq2, and Scn8a.42
Our study has limitations. Overall, our sample size was small and we cannot rule out result bias from unmeasured confounders. As with most PRS analyses, we remained restricted to individuals of European ancestry. This is because the epilepsy GWAS from which the PRSs were derived comprised >90% European ancestry cases.7 Removal of non-European ancestry families further reduced our cohort's size. To counter this, we included families with both known and presumed RVs of major effect to ensure our sample size was as large as possible. As a result, the families without a known RV, may not be “monogenic”. In this case, the enrichment for epilepsy PRSs potentially reflects the contribution of common epilepsy risk variants to complex inheritance as opposed to acting as a genetic modifier on a gene of major effect. However, by investigating two specific RVs, our results are shown to support epilepsy PRSs playing a role in modifying RV penetrance and possibly epilepsy phenotype. Our differing results for SCN1B and GABRG2 suggests that certain genes and variants may be more susceptible to common variant genetic modification, however, these were also the two largest families and may not reflect effects in smaller families with different RVs. As such, it is important to note that epilepsy PRSs are not yet at the stage of clinical application, even for families with GEFS+; they remain underpowered for the purposes of disease prediction and for determining who will develop which type of epilepsy.
The mechanisms underlying phenotypic variability of most monogenic disorders remain poorly understood. Whilst epilepsy modifying genes have been hypothesised, they have been elusive. We provide evidence that, collectively, common epilepsy risk variants play a role, although specific variants have not been identified. This work shifts the current paradigm of epilepsy genetics – no rare pathogenic variant is likely to be acting alone. Interpretation of rare pathogenic variants in the context of polygenic background promises to provide an additional level of stratification for epilepsy risk in patients and their relatives. Furthermore, identifying specific modifier genes or pathways in the future has the potential to inform prognostic and genetic counselling, development of novel therapeutics and potentially disease prevention.
Contributors
K.L.O, I.E.S, C.A.E., B.E.G., S.F.B., M.B. contributed to the conception and design of the study. K.L.O., C.A.E., B.E.G. contributed to the curation, quality control and analysis of genetic data. K.L.O, I.E.S, B.E.G., C.A.E., S.F.B., Epi4K Consortium contributed to participant recruitment and phenotyping. K.L.O, C.A.E., M.B. performed and interpreted statistical analyses. K.L.O, C.A.E., M.B. have directly accessed and verified the underlying data reported in the manuscript. K.L.O. wrote the first manuscript draft. All co-authors contributed to manuscript revisions and read and approved the final version.
Data sharing statement
Data used in this study is not available due to privacy and ethical restrictions.
Declaration of interests
M.B. has received payment for thesis examination from University of Sydney, Australian National University, University of Melbourne; has received support for registration to the Genetics Society of AustralAsia and GeneMappers conferences; is/was a member of the Australian Academy of Health and Medical Sciences Reports Committee, the GenV Steering Committee, the Australian Academy of Health and Medical Sciences Australian Learned Academies Data Internetworking Network (ALADIN) Project Steering Committee, American Epilepsy Society Basic Sciences Committee, the Viertel Foundation Medical Advisory Board and a board member for the Australian Genome Research Facility. S.F.B has received educational grants from UCB Pharma, Eisai, SEER, Chiesi, LivaNova; consulting fees from Praxis Precision Medicines, Sequiris; speaker honoraria from Eisai, DeltaMed; serves as the Chief Medical Officer for the Epilepsy Foundation (Victoria); and has a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies. I.E.S has served on scientific advisory boards for Bellberry Ltd, BioMarin, Chiesi, Eisai, Encoded Therapeutics, Garvan Institute of Medical Research, Knopp Biosciences, Longboard Pharmaceuticals, UCB, Takeda Pharmaceuticals; has received speaker honoraria from Akumentis Pharma, Biocodex, BioMarin, Chiesi, Eisai, GlaxoSmithKline, Liva Nova, Nutricia, Stoke Therapeutics, UCB, Zuellig Pharma; has received funding for travel from Biomarin, Eisai, GlaxoSmithKline, Stoke Therapeutics, UCB; has served as an investigator for Anavex Life Sciences, Cerebral Therapeutics, Cerecin Inc, Cereval Therapeutics, Eisai, Encoded Therapeutics, EpiMinder Inc, ES-Therapeutics, GW Pharmaceuticals, Marinus Pharmaceuticals, Neuren Pharmaceuticals, Neurocrine BioSciences, Ovid Therapeutics, Takeda Pharmaceuticals, UCB, Ultragenyx, Xenon Pharmaceuticals, Zogenix, Zynerba Pharmaceuticals; and has consulted for Biohaven Pharmaceuticals, Care Beyond Diagnosis, Cerecin Inc, Eisai, Epilepsy Consortium, Longboard Pharmaceuticals, UCB, Zynerba Pharmaceuticals; and is a Non-Executive Director of Bellberry Ltd and a Director of the Australian Academy of Health and Medical Sciences and the Australian Council of Learned Academies Ltd. She may accrue future revenue on pending patent WO2009/086591: Diagnostic And Therapeutic Methods For EFMR (Epilepsy And Mental Retardation Limited To Females); has a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; has a patent molecular diagnostic/theranostic target for benign familial infantile epilepsy (BFIE) [PRRT2] with royalties paid. C.A.E has received consulting fees from Epiminder Pty Ltd. The Epi4K Consortium was supported by an NINDS National Institute of Health Grant [U01NS077367].
Acknowledgements
We thank the patients and their families for participating in our research program. We also thank Professor Sanjay Sisodiya for contributing an individual patient and Dr Kasper Lolk for statistical advice. Furthermore, we thank the participants in the QSKIN Study for contributing genetic data to this study as population controls.
S.F.B and I.E.S were supported by a National Health and Medical Research Council (NHMRC) of Australia Program Grant [APP1091593], Synergy grant [APP2010562) and Investigator Grants [SFB 1196637; IES 1172897]. M.B. and I.E.S. were supported by a MRFF Genomics Health Futures Missions grant [MRF2007707] and M.B. was supported by an NHMRC Investigator Grant [APP1195236]. K.L.O was supported by an Australian Government Research Training Program Scholarship [APP533086] provided by the Australian Commonwealth Government and the University of Melbourne. CAE was supported by the National Institute of Neurological Disorders And Stroke of the National Institutes of Health Award Number K23NS121520. The QSKIN Study was supported by NHMRC Grants [APP1185416, APP1073898, APP1063061]. This work was also supported by the Victorian Government’s Operational Infrastructure Support Program and the NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS).
Footnotes
Appendix ASupplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105404.
Appendix A. Supplementary data
References
Articles from eBioMedicine are provided here courtesy of Elsevier
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/170005187
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Common risk variants for epilepsy are enriched in families previously targeted for rare monogenic variant discovery.
EBioMedicine, 81:104079, 27 May 2022
Cited by: 11 articles | PMID: 35636315 | PMCID: PMC9156876
Generalized epilepsy with febrile seizures plus (GEFS+): clinical spectrum in seven Italian families unrelated to SCN1A, SCN1B, and GABRG2 gene mutations.
Epilepsia, 45(2):149-158, 01 Feb 2004
Cited by: 32 articles | PMID: 14738422
The role of common genetic variation in presumed monogenic epilepsies.
EBioMedicine, 81:104098, 06 Jun 2022
Cited by: 12 articles | PMID: 35679801 | PMCID: PMC9188960
SCN1B Genetic Variants: A Review of the Spectrum of Clinical Phenotypes and a Report of Early Myoclonic Encephalopathy.
Children (Basel), 9(10):1507, 01 Oct 2022
Cited by: 3 articles | PMID: 36291443 | PMCID: PMC9600564
Review Free full text in Europe PMC




