Abstract
Free full text

Short Repeat Ribonucleic Acid Reduces Cytotoxicity by Preventing the Aggregation of TDP-43 and Its 25 KDa Carboxy-Terminal Fragment
Abstract
TAR DNA/RNA-binding protein 43 kDa (TDP-43) proteinopathy is a hallmark of neurodegenerative disorders, such as amyotrophic lateral sclerosis, in which cytoplasmic aggregates containing TDP-43 and its C-terminal fragments, such as TDP-25, are observed in degenerative neuronal cells. However, few reports have focused on small molecules that can reduce their aggregation and cytotoxicity. Here, we show that short RNA repeats of GGGGCC and AAAAUU are aggregation suppressors of TDP-43 and TDP-25. TDP-25 interacts with these RNAs, as well as TDP-43, despite the lack of major RNA-recognition motifs using fluorescence cross-correlation spectroscopy. Expression of these RNAs significantly decreases the number of cells harboring cytoplasmic aggregates of TDP-43 and TDP-25 and ameliorates cell death by TDP-25 and mislocalized TDP-43 without altering the cellular transcriptome of molecular chaperones. Consequently, short RNA repeats of GGGGCC and AAAAUU can maintain proteostasis by preventing the aggregation of TDP-43 and TDP-25.
Introduction
TAR DNA/RNA-binding protein 43 kDa (TDP-43) is the key focus of research to understand neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).1,2 A hallmark of these disorders is cytoplasmic aggregates of TDP-43 and its carboxy-terminal fragments (CTFs). TDP-43 carries two RNA/DNA-recognition motifs (RRM1 & 2), each of which contains two highly conserved short sequence motifs required for nucleic acid binding,3 and a C-terminal glycine-rich intrinsically disordered region (IDR). Many of the amino acid substitutive mutations in TDP-43 associated with ALS and FTD are conserved in the IDR.4 TDP-43 is mainly localized in the nucleus but is also a nuclear-cytoplasmic shuttling protein, which maintains cellular homeostasis regulated by the quality of RNA such as mRNA and small RNA.5 A 25 kDa TDP-43 CTF (TDP-25; amino acids 220–414),6 which lacks RRM1 and a portion of RRM2 including major amino acids that are required for canonical RNA recognition, is highly prone to aggregation and leads to cell death. TDP-25 aggregates sequestrate full-length TDP-43; therefore, the loss-of-function of TDP-43 may be associated with cytotoxicity in addition to the gain-of-function of these aggregates.7 Aggregates of TDP-43 and TDP-25 that can be viewed by a microscope and contain amyloid-like fibrils8,9 in the cytoplasm may not necessarily be essential for neuronal degeneration in ALS.10,11 Mislocalization of TDP-43 contributes to the cytotoxicity in ALS through the regulation of various cellular functions such as gene expression, mRNA maturation, repression of alternate splicing, cryptic exon splicing, and alternate polyadenylation.12,13 The long-term mislocalization of TDP-43 in the cytoplasm produces its cytoplasmic aggregates, leading to the disruption of physiological function (e.g., sequestration of cellular proteins such as other ALS-associated and autophagy-regulating proteins); thus, the homeostatic level regulated by proteins (proteostasis) could be declined.14,15 Therefore, motor neuron degeneration may be involved in the mislocalization of TDP-43 with the formation of its parallel aggregate.11
RNA-binding proteins (RNP), such as TDP-43, and RNA interact and induce condensation of RNPs through phase separation.16 In contrast, RNA degradation induces aggregation of not only TDP-43 but also TDP-2517,18 even though TDP-25 lacks major RNA-binding sequences. The binding of TDP-43 to RNA antagonizes aggregation and phase transitions.19,20 Furthermore, the idea that RNAs such as rRNA, tRNA, and so on, might have chaperone-like activity has been proposed.21−24 Therefore, RNA regulates the aggregation and condensation of RNPs with their phase transition and plays a role in the regulation of the intermolecular assembly using the physicochemical properties of RNA. However, which RNA regulates such aggregation of ALS/FTD-causative TDP-43 and then plays a role in cytoprotection remains unclear. Here, we show that GGGGCC and AAAAUU hexanucleotide repeat RNAs (r(G4C2) and r(A4U2), respectively) can reduce proteotoxicity by preventing the aggregation of TDP-43 and TDP-25 in a cellular model. The C-terminal domain of TDP-43 was found to recognize the structure of G-quadruplex (Gq) in RNA,25 and Gq DNA protects the aggregation of denatured proteins.26 Therefore, we first focused on whether TDP-25 binds to the four repeats r(G4C2) (hereafter G4), which can be folded as a Gq and is included in a major ALS-causative transcript, C9orf72. Then, we demonstrate whether non-ALS-associated and short r(G4C2) plays a cytoprotective role in TDP-25 aggregation and mislocalized TDP-43 with its aggregation. Furthermore, we showed that the four repeat r(A4U2) (hereafter, A4), which is generally used as a control sequence for r(G4C2) repeats,27,28 also binds to TDP-43 and TDP-25, and suppresses their aggregation, in addition to reducing the cytotoxicity caused by their mislocalization and aggregation. The following results suggest that these repeat RNAs may have chaperone-like activities that protect protein aggregation and maintain intracellular proteostasis.
Results
GGGGCC and AAAAUU Hexanucleotide Repeats Bind to TDP-43 and TDP-25 in Cell Lysates
To analyze the interaction between TDP-43/TDP-25 and RNA, we employed a single molecule detection method, fluorescence cross-correlation spectroscopy (FCCS), which can quantitatively determine the strength of interaction between diffuse fluorescent species in solution from the fluorescence fluctuation that occurs by passing fluorescent molecules through the confocal detection volume (Figure Figure11a).29,30 After RNA labeled with a chemical near-infrared fluorescent dye, Alexa Fluor 647 (AF), in lysates of murine neuroblastoma Neuro2a cells expressing green fluorescent protein (GFP)-tagged TDP-25 (T25), GFP-tagged TDP-43 (T43), or GFP monomers as the tag was mixed, FCCS was performed. As we have previously shown,18 the relative cross-correlation amplitude (RCA) of the AF-labeled 24-mer of uracil guanine (UG) repeat RNA (AF-UG), a consensus TDP-43-recognizing sequence, with T43 was high, whereas those with GFP and T25 were not observed (Figure Figure11b), indicating that the interaction between T25 and AF-UG may be either absent or very weak. The RCA when AF-labeled G4 (AF-G4) in a coexisting state of the hairpin (HP) and Gq structures (Gq + HP)28,31 was mixed with T43 or T25 was significantly positive; this suggests that AF-G4 can interact with not only T43 but also T25. The RCA of T43 and T25 with only HP-formed AF-G4 was also positive (Figure Figure11b), suggesting that the structure of Gq may not be required for interaction with T43 and T25. Next, to investigate whether T25 also interacts with other RNAs that are not different from G4, we tested AF-labeled A4(AF-A4), which is often used as a control sequence for r(G4C2).27,28 Although we had expected that T25 had not bound to AF-A4, the RCA of T43 and T25 with AF-A4 was positive but lower than that with AF-G4 in a mixed state of the hairpin (HP) and Gq structures (Figure Figure11b), suggesting that TDP-43 and TDP-25 can interact with G4 and A4, regardless of their steric structures. Furthermore, as UG repeats did not bind to TDP-25, the interaction between TDP-25 and G4 and A4 may be specific.

Interaction of the G4C2/A4U2-repeat RNA with TDP-43 and TDP-25 in cell lysates using fluorescence cross-correlation spectroscopy. (a) Illustration of the detection of molecular interactions from the relative cross-correlation amplitude (RCA) using fluorescence cross-correlation spectroscopy. (b) RCA values when AF-labeled RNAs (UG, G4, and A4) that were folded in 100 mM KCl (Gq + HP) or 100 mM LiCl (HP) were mixed in lysates of Neuro2a cells expressing GFP monomers (GFP), GFP-TDP-43 (T43), and GFP-TDP-25 (T25) with 100 mM KCl (n = 3; dots and bars indicate independent trials and mean ± SE). (c) RCA values between AF-labeled RNAs (UG, G4, and A4) and T43/T25 in cell lysates at 100, 150, 200, and 250 mM KCl concentrations. G4 (Gq + HP) was folded in the presence of 100 mM KCl, and then mixed with the cell lysate. The dashed line indicates zero. Concentration-dependent RCA values of ATTO488- and ATTO647N-labeled double-strand DNA as a positive control (PC-DNA) and the mixture of ATTO488- and ATTO647N-labeled primers as a negative control (NC-DNA) in the presence of 100–250 mM KCl are represented in Figure S1a. (d) RCA values between AF-labeled RNAs (UG, G4, and A4) and T43/T25 in cell lysates at 0, 5, 10, and 15 mM MgCl2 concentrations. The dashed line indicates zero. Concentration-dependent RCA values of PC-DNA and NC-DNA in the presence of 0–15 mM MgCl2 are represented in Figure S1b. (e) RCA values between AF-labeled RNAs (UG, G4, and A4) and T43/T25 in cell lysates at 0, 5, and 10% 1,6-hexanediol concentrations. The dashed line indicates zero. Concentration-dependent RCA values of PC-DNA and NC-DNA in the presence of 0, 5, and 10% 1,6-hexanediol are represented in Figure S2a. (f) RCA values when AF-labeled RNAs (G4, G2, and A2) were used. G4 (Gq + HP) was folded in the presence of 100 mM KCl, and then mixed with the cell lysate including 100 mM KCl. G2 (HP) was folded in the presence of 100 mM LiCl, then mixed with the cell lysate including 100 mM KCl (n = 3; mean ± SE). (b, d) Significance: *p < 0.05, **p < 0.01, ***p < 0.001, and ns (p ≥ 0.05).
Next, to demonstrate biochemical properties in the interaction between TDP-43/TDP-25 and G4/A4, the interaction strength in the presence of high concentrations of KCl (150, 200, and 250 mM) was analyzed using FCCS. The RCA of neither dual color-fluorescent DNA32 as a positive control nor the mixture of the dual fluorescent dyes as a negative control changed at the high concentrations of KCl compared to that at 100 mM (Figure S1a), indicating that the FCCS detection system did not change at high KCl concentration. The RCA between T43 and AF-UG dramatically declined with the increase in the KCl concentration (cyan line in Figure Figure11c), suggesting the contribution of electrostatic interaction. The RCA between T25 and AF-UG did not emerge at high KCl concentrations (orange line in Figure Figure11c). The RCAs between T43 and AF-G4/AF-A4 prominently declined with an increase in KCl concentration (blue and green lines in Figure Figure11c); however, those between T25 and AF-G4/AF-A4 declined little by little with an increase in KCl concentration (pink and red lines in Figure Figure11c). Furthermore, at 150 and 200 mM KCl concentrations, the RCA between T25 and AF-G4/AF-A4 was still positive and higher than that between T43 and AF-G4/AF-A4 (Figure Figure11c), suggesting that both hydrophobic and electrostatic interactions may contribute significantly to the interactions between TDP-25 and G4/A4; however, the electrostatic interaction may relatively highly contribute to that between TDP-43 and G4/A4 compared with TDP-25. These trends were similarly observed with the addition of MgCl2 (5, 10, and 15 mM), which generates the divalent cation Mg2+ (Figure Figure11d and Figure S1b), and with the addition of heparin (Figure S1c). The divalent cation Mg2+ functioned effectively at concentrations 1/10 or lower compared to the monovalent cation K+, indicating inhibition of charge-based electrostatic interactions. By contrast, adding the aliphatic alcohol 1,6-hexanediol (HD), which interferes with weak hydrophobic interactions, exhibited an inverse correlation of RCA values compared with increasing concentrations of KCl and MgCl2 (Figure Figure11e and Figure S2a). Circular dichroism analysis confirmed that no dramatic changes in the G4/A4 conformation were observed upon increasing the concentration of HD (Figure S2b,c). Therefore, the biochemical RNA interaction mechanism, especially the balance of electrostatic and hydrophobic interaction, may differ between TDP-43 and TDP-25.
Next, to determine whether the shorter repeats were adopted for this interaction, two repeats of r(G4C2) (G2) and r(A4U2) (A2) were used. Although the RCAs between T43/T25 and AF-labeled G2 (AF-G2) were positive, the RCA between T43/T25 and AF-labeled A2 (AF-A2) was not observed (Figure Figure11e). Therefore, four repeats of r(A4U2) may be required to interact with TDP-43/TDP-25. Furthermore, the Gq structure of r(G4C2) repeats is not necessarily required for this interaction.
Direct Interaction between GGGGCC/AAAAUU Hexanucleotide Repeats and TDP-43/TDP-25
The above analysis was performed in cell lysate. Although this is an advantage of FCCS because the biomolecular interaction can be measured in a solution mixture containing fluorescent molecules, it is not yet clear whether T25 interacts with RNA directly or cellular RNA-binding proteins in complexes of T25 interact with RNA. To confirm the direct interaction with these RNAs, we prepared purified GFP, T43, and T25 through tandem affinity purification using polyhistidine and strep tags. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining showed that highly purified GFP was obtained; however, several cellular proteins were included in the purified T43 and T25 samples (Figure Figure22a). Since T43 is an RNP and binds to RNA with Gq-foldable sequences,25,33 we identified the copurified proteins with T25 using peptide mass fingerprinting with mass spectroscopy (PMF-MS). Five major proteins copurified with T25 were identified—heat shock protein 70 kDa (HSP70), mitochondrial pyruvate carboxylase (mPC), tubulin, and actin—by PMF-MS, and a GFP-containing fragment was confirmed by Western blotting. Bands corresponding to mPC, tubulin, and actin were also observed with T43 (Figure Figure22a). As HSP70 and mPC have been reported to have RNA-binding properties,34,35 we confirmed that G4/A4 did not bind to these copurified cellular proteins using FCCS. AF-G4/AF-A4 was mixed in the lysate of cells expressing HSP70, β-tubulin, α-actin, or mPC tagged with GFP or yellow fluorescent protein (YFP), and following FCCS analysis under the same conditions as T43/T25 did not show their significant interaction (Figure S3). Since HSP70 is an abundant molecular chaperone and is frequently coprecipitated, the interaction with HSP70 may contribute to maintaining the solubility of TDP-25. In other words, it may be difficult to recover TDP-25 without HSP70 in the soluble fraction. Because the endogenous TDP-43 band was not observed in the purified T25 sample (Figure Figure22a), and the interaction between TDP-25 and TDP-43 was not detected using FCCS in cell lysate,18 it is unlikely that the purified T25 sample contains endogenous TDP-43. Since AF-UG did not bind to T25 in the cell lysate experiments (Figure Figure11b), it is unlikely that G4/A4 are recognized by TDP-43 bound to TDP-25. Consequently, even if other RNPs were included in the purified T25 sample, they may not contribute to the interaction analysis with G4/A4. The interaction of purified GFP, T43, and T25 with AF-G4/AF-A4 was then analyzed using FCCS. No positive RCA of the GFP, T43, and T25, when mixed with just AF dye as a negative control, was observed; however, positive RCA of T43 and T25, when mixed with AF-G4 and AF-A4, was observed, and it was higher with AF-G4 than with AF-A4 (Figure Figure22b and Figure S4). Dissociation constants (Kd) of these interactions determined from the FCCS analysis were from the submicromolar to micromolar level (Figure Figure22c). These Kd ranges are considered reasonable for RNA-protein interactions in solution. Consequently, TDP-43 and TDP-25 interact directly with G4 and A4 and not through other RNPs.
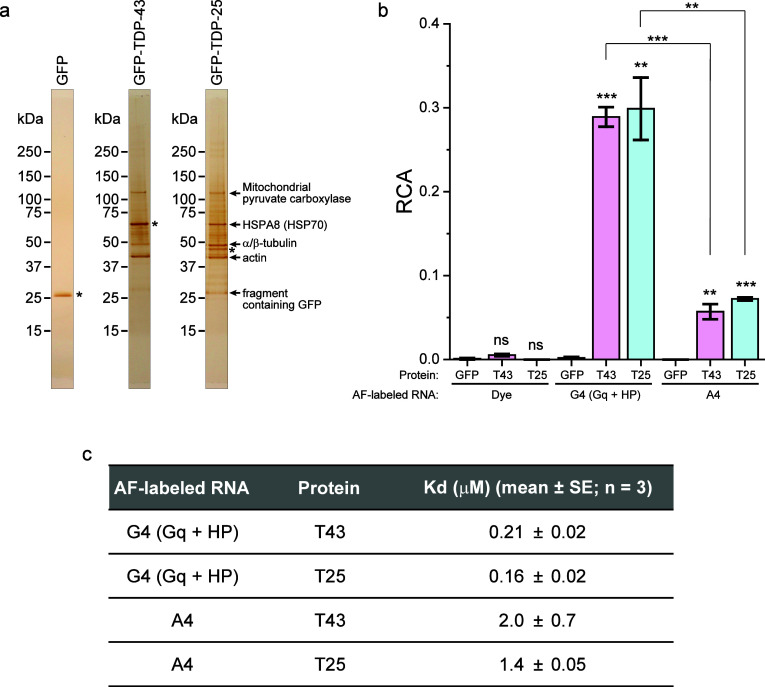
Direct interaction of the G4C2/A4U2-repeat RNA with purified TDP-43 and TDP-25. (a) SDS-PAGE of purified GFP, GFP-TDP-43, and GFP-TDP-25, followed by silver staining. Asterisks show purified target proteins. (b) RCA values when AF-labeled RNAs (G4, and A4) folded in 100 mM KCl were mixed with purified GFP monomers (GFP), GFP-TDP-43 (T43), and GFP-TDP-25 (T25) with 100 mM KCl (n = 3; mean ± SE). Significance: **p < 0.01, ***p < 0.001, and ns (p ≥ 0.05). The typical normalized cross-correlation functions are represented in Figure S4. (c) Dissociation constant (Kd) values between T43/T25 (proteins) and AF-labeled G4/A4 (RNA) calculated from Figure Figure22b (n = 3; mean ± SE).
Both Truncated RRM2 and IDR Are Responsible for the Interaction between GGGGCC/AAAAUU Hexanucleotide Repeats and C-Terminal Fragments of TDP-43
How does TDP-25 recognize RNA even though it lacks many of the RRMs? To identify the contribution of truncated RRM2 and IDR in TDP-43 CTFs to the interaction with G4/A4, the interactivity of two additional truncated variants of TDP-25 (F220 and C274;36Figure Figure33a) with G4/A4 was analyzed using FCCS in Neuro2a cell lysates. Neither F220 nor C274 interacted with G4 in a mixture of Gq and HP, HP-fold G4, and A4 (Figure Figure33b), suggesting that both truncated RRM2 and IDR are responsible for the interaction with G4/A4 not through highly conserved nucleic acid binding sequences such as the RRM.

Truncated RRM2 is required for the interaction with G4C2/A4U2-repeat RNA. (a) Primary structures of human TDP-43 (1–414 amino acids), TDP-25 (220–414 amino acids), F220 (220–262 amino acids), and C274 (274–262 amino acids). Numbers show the amino acid position. (b) RCA values when AF-labeled RNAs (G4, and A4) that were folded in 100 mM KCl (Gq + HP) or 100 mM LiCl (HP) were mixed with GFP monomers (GFP), GFP-TDP-43 (T43), GFP-TDP-25 (T25), GFP-F220 (220), and GFP-C274 (274) with 100 mM KCl (n = 3; mean ± SE). Significance: *p < 0.05, **p < 0.01, ***p < 0.001, and ns (p ≥ 0.05).
GGGGCC/AAAAUU Hexanucleotide Repeats Prevent Aggregation of TDP-25 and Ameliorate Its Cytotoxicity
Next, to demonstrate the activity of G4 and A4 that prevent the aggregation of TDP-25, G4/A4 tagged with bacteriophage-derived M13 sequences (M13-G4 and M13-A4, respectively) were expressed in Neuro2a cells by transfection of plasmid DNA that transcribes such RNAs under the human H1 promoter. The expression of M13-G4, M13-A4, and M13-tagged 24-mer UG repeat RNA (M13-UG), and only the M13 tag as a control (M13N) in Neuro2a cells was confirmed by reverse transcription PCR (RT-PCR) (Figure S5a). The population of cells expressing M13-G4 and M13-A4 that harbor T25 aggregates in the cytoplasm was decreased, while that of cells expressing M13N and M13-UG was not (Figure Figure44a). Although the expression levels of T25 in the soluble fraction of the cell lysates were not changed in cells expressing M13-G4, M13-A4, M13-UG, or M13N compared to those in empty vector transfection, the levels of T25 in the insoluble fraction significantly decreased in cells expressing M13-G4 and M13-A4 (Figure Figure44b and Figure S6a). The total abundance of T25 in whole cell lysates was not changed in the expression of M13-G4 and M13-A4 (Figure S6b). These results suggest that G4/A4 can be an aggregation suppressor for TDP-25, similar to a cytoplasmic molecular chaperone HSP70 for TDP-25 aggregation.37 Furthermore, RNA that binds to TDP-43, as in the case of UG repeat RNA, is not a sufficient condition for the prevention of T25 aggregation. Although these G4/A4 are supposed to, the next question is whether the G4/A4 introduced into cells disturbs the intracellular transcriptome, resulting in the indirect suppression of T25 aggregation through the upregulation of stress-induced proteins, such as heat shock proteins. To this end, transcriptomic analysis was performed using RNA-seq when M13-G4, M13-A4, and M13-UG were expressed with T25 in Neuro2a cells. No transcripts with a >2- and <0.5-fold change in the significant expression ratio (p < 0.05) were observed in both mRNA and noncoding RNA (ncRNA) in the comparison of M13-G4/A4 with M13-UG (Figure Figure44c). Therefore, it is unlikely that up-regulated chaperone-like genes by expression of M13-G4/A4 inhibit T25 aggregation indirectly; in other words, we suggest that these RNAs directly prevent T25 aggregation.
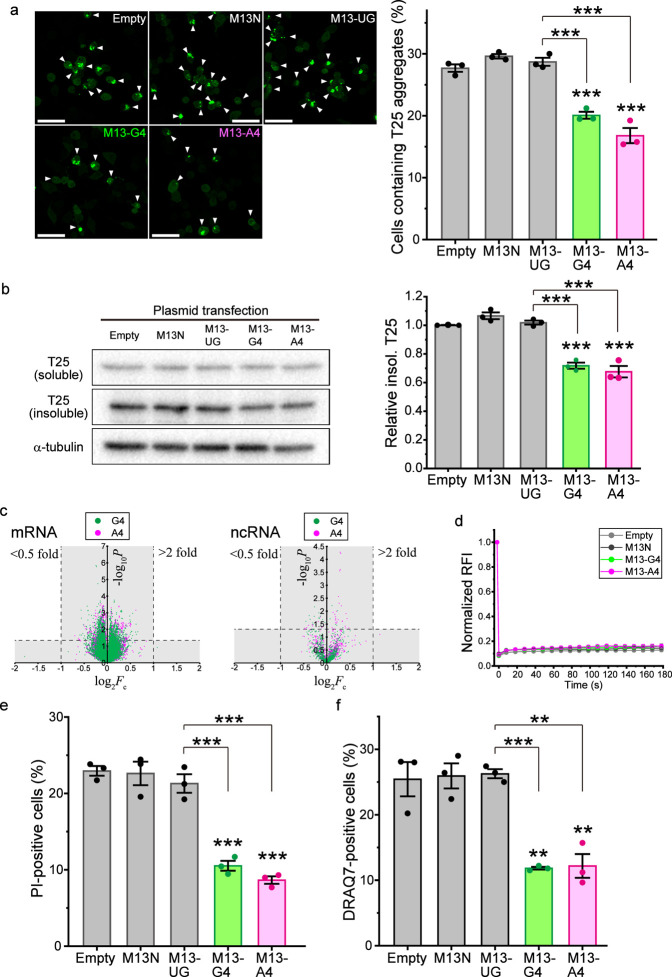
Reduced aggregates of GFP-TDP-25 in Neuro2a cells expressing G4C2/A4U2-repeat RNA. (a) Confocal fluorescence images of GFP-TDP-25 (T25) in Neuro2a cells (left). The percentage of cells containing cytoplasmic aggregates of T25 when an empty vector (empty) or M13-tagged RNA was expressed (M13N, M13-UG, M13-G4, and M13-A4) (right) (the number of trials = 3; mean ± SE). (b) Western blotting of Neuro2a cells expressing T25 when M13-tagged RNAs were expressed. Typical blots of T25 in soluble and insoluble fraction, and α-tubulin in the soluble fraction (left). The quantified intensity ratio of T25 in the insoluble fraction compared with that in the empty vector transfection (the number of trials = 3; mean ± SE) (right). The gel loading the insoluble fraction of the cell lysate stained with Coomassie brilliant blue as an internal control was represented in Figure S6a. The blots of total T25 abundance in whole cell lysate were represented in Figure S6b. (c) Volcano plot of mRNA (left) and ncRNA (right) expression change when M13-G4 or -A4 was expressed in Neuro2a cells compared with M13-UG (the number of trials = 4). Areas of nonsignificant change are shaded in gray (0.5–2-fold change and p ≥ 0.05). (d) Normalized relative fluorescence intensity (RFI) indicating mobile fraction of T25 in the cytoplasmic aggregates determined using FRAP (measured cell number = 9; mean ± SE). (e) Population of T25-expressing cells stained with propidium iodide (PI) as a dead cell marker (the number of trials = 3; mean ± SE). (f) Population of T25-expressing cells stained with DRAQ7 as a dead cell marker using FACS (the number of trials = 3; mean ± SE). (a, b, e, f) Significance: *p < 0.05, **p < 0.01, and ***p < 0.001.
Are the biophysical molecular dynamics of TDP-25 within the aggregates then changed in the expression of G4/A4? To investigate this issue, we performed fluorescence recovery after photobleaching (FRAP) analysis in live Neuro2a cells was performed. TDP-25 was quite immobile and formed amyloids in the aggregates;18,36 however, the expression of M13-G4/A4 did not alter the immobility of T25 (Figure Figure44d), suggesting that G4/A4 may act on TDP-25 in the diffusive states in the cytoplasm to prevent aggregation before the formation of deposited aggregates. As a possible reason why G4/A4 cannot act on the deposited aggregates, HSP70 overexpression does not dramatically increase the mobile population of TDP-25 in the aggregates.18,37 HSP70 is colocalized and dynamically interacts with TDP-25 aggregates;37 however, RNA is not stained inside the cytoplasmic TDP-25 aggregates.18 This unremarkable difference in TDP-25 mobility in the aggregates, even when overexpressed HSP70 interacts with the aggregates, is consistent with no significant changes in TDP-25 mobility in the aggregates when M13-G4 and M13-A4 were expressed, as shown here.
Next, to demonstrate that G4/A4 without any tag directly prevents TDP-25 aggregation, the aggregate formation of T25 in Neuro2a cells introduced with RNA was analyzed. Transfection of synthetic 24-mer UG repeat RNA (UG) and siRNA against the Renilla luciferase mRNA (siRL)38 as controls preintroduction of T25-expression vector did not change the population of cells containing T25 aggregates in the cytoplasm compared to mock transfection; however, the direct transfection of synthetic G4/A4 preintroduction of T25-expression vector significantly decreased the proportion of cells containing T25 aggregates (Figure S7a,b). The levels of T25 in the insoluble fraction decreased in G4- and A4-transfected cells (Figure S7b,c). These results confirm the hypothesis that G4/A4 can be used as an aggregation suppressor for T25 by introducing it into cells. Furthermore, the transfection of synthetic G4/A4 postintroduction of T25-expression vector significantly decreased the proportion of cells containing T25 aggregates and the amount of T25 in the insoluble fraction (Figures S7f–k); however, the efficiency of reduction of T25 aggregates during G4/A4 to UG transfection was higher with pre-T25 transfection than with post-T25 transfection (Figures S7). Therefore, G4/A4 may inhibit the aggregation and assembly process before TDP-25 forms aggregates in the cytoplasm that can be viewed by microscopes. These RNAs would maintain the oligomeric states of TDP-25 and/or prevent its intermolecular assembly.
To demonstrate whether M13-G4 and M13-A4 expression ameliorates TDP-25 cytotoxicity, we quantified the population of propidium iodide (PI)-stained cells using fluorescence microscopy and DRAQ7-stained cells using fluorescence-activated cell sorting (FACS), indicating dead cells. The population of PI- and DRAQ7-positive and T25-expressing cells was dramatically reduced in M13-G4/A4-expressing cells compared to M13N or M13-UG expression and empty vector transfection as controls (Figure Figure44e,f), suggesting that G4/A4 plays a role in the reduction of cytotoxicity in TDP-25-expressing cells.
Because G2 but not A2 interacted with TDP-25 (Figure Figure11d), we next investigated whether G2/A2 can reduce the TDP-25 aggregate. M13-tagged G2/A2 (M13-G2/A2) was expressed as much as M13-G4/A4 in Neuro2a cells (Figure S8a). M13-G2/A2 expression in Neuro2a cells did not change the population of cells that harbor T25 aggregates and the levels of T25 in the insoluble fraction (Figure S8b). G2 is believed to be too short to form the Gq structure efficiently, while it is not entirely incapable of forming Gq as intermolecular assemblies. The interaction strength of AF-G2 with T25 was reduced by approximately 60% compared to AF-G4 (Figure Figure11e). Therefore, for effective inhibition of the TDP-25 aggregation by r(G4C2) in the cell, a certain degree of interaction strength with TDP-25, which corresponds to that of four repeats of r(G4C2), may be required. Furthermore, r(A4U2) may also require four repeats to efficiently suppress the TDP-25 aggregation.
GGGGCC/AAAAUU Hexanucleotide Repeats Prevent Aggregation of TDP-43 and Ameliorate Its Cytotoxicity
Although TDP-25 is highly prone to aggregation compared to TDP-43 and its CTFs,7,10,36,39 the mislocalization of full-length TDP-43 with its aggregation is thought to be more associated with pathogenicity.11,40,41 However, wild-type TDP-43 is not mislocalized and does not form any cytoplasmic aggregates in Neuro2a cells.18 Thus, we chose a variant of TDP-43 that lacks the ability of nuclear localization and exporting signal sequences and carries amino acid substitutions of C173S/C175S (TDP-43CS).42 Furthermore, because GFP-tagged TDP-43CS (T43CS) did not form cytoplasmic aggregates in Neuro2a cells, we used 293 cells in which T43CS formed cytoplasmic aggregates efficiently. The expression of M13-G4, M13-A4, and M13N in 293 cells was confirmed using RT-PCR (Figure S5b). The population of cells that harbor T43CS aggregates in the cytoplasm was significantly reduced in cells expressing M13-G4 and M13-A4 compared to that of cells expressing M13-UG, M13N, and empty-vector-transfected cells as controls (Figure Figure55a). Next, to biochemically detect the reduction in aggregates by expression of M13-G4/A4, these cells were lysed in a buffer containing 0.1% SDS and then fractionated using centrifugation. Almost all T43CS was recovered in the soluble fraction, and no significant change in the T43CS amount among the soluble and insoluble fractions and whole cell lysates was observed in M13-G4/A4-expressed cells (Figure Figure55b and Figure S9a,b). The aggregates of T43CS may be more loosely assembled than those of T25. However, the filter retardation assay showed that the amount of T43CS trapped on the membrane was decreased by M13-G4/A4 expression (Figure Figure55b), suggesting that T43CS may not exist as complete monomers and may probably exist as oligomers and/or soluble aggregates that were larger than the pore size of the membrane (>0.2 μm); furthermore, they can be solubilized by SDS, and G4/A4 decreases oligomers and/or soluble aggregates of T43CS likely by affecting its oligomerization process.

Reduced aggregates of GFP-tagged TDP-43CS in 293 cells expressing G4C2/A4U2-repeat RNA. (a) Confocal fluorescence images of GFP-tagged TDP-43CS (T43CS) in 293 cells. The white arrows indicate the cells containing the aggregates (left). The percentage of cells containing cytoplasmic aggregates of T43CS when empty vector (empty) or M13-tagged RNA was expressed (M13N, M13-UG, M13-G4, and M13-A4) (right) (the number of trials = 3; mean ± SE). (b) Western blotting (top) and filter retardation (FR) (bottom) of 293 cells expressing T43CS when M13-tagged RNAs were expressed. (c) Quantified intensity ratio of T43CS in the insoluble fraction compared with that in the empty vector transfection (the number of trials = 3; mean ± SE). The gel loading the insoluble fraction of the cell lysate stained with Coomassie brilliant blue as an internal control was represented in Figure S8a. The blots of total T43CS abundance in whole cell lysate were represented in Figure S8b. (d) Quantified intensity ratio of the retarded T43CS on the membrane compared with the empty vector transfection (the number of trials = 3; mean ± SE). (e) Volcano plot of mRNA (left) and ncRNA (right) expression change when M13-G4 or M13-A4 was expressed in 293 cells compared with M13-UG (the number of trials = 4). Areas of nonsignificant change are shaded in gray (0.5–2-fold change and p ≥ 0.05). Transcripts that exhibited significant changes upon expression of M13-G4 or M13-A4 are described in Table S4. (f) Normalized relative fluorescence intensity (RFI) indicating mobile fraction of T43CS in the cytoplasmic aggregates determined using FRAP (measured cell number = 7; mean ± SE). (g) Population of T43CS-expressing cells stained with propidium iodide (PI) as a dead cell marker (the number of trials = 3; mean ± SE). (h) Population of T43CS-expressing cells stained with DRAQ7 as a dead cell marker using FACS (the number of trials = 3; mean ± SE). (a, c, d, g, h) Significance: *p < 0.05, **p < 0.01, and ***p < 0.001.
Likewise, for 293 cells, transcriptomic analysis was performed using RNA-seq when G4/A4 were expressed. No transcripts with a >2- and <0.5-fold change in the significant expression ratio (p < 0.05) were observed in ncRNA. Although several transcript levels in mRNA were altered in 293 cells by expression of M13-G4/A4 in contrast to Neuro2a cells, none of the transcripts had distinct chaperone functions (Figure Figure55e). Therefore, it is unlikely that up-regulated chaperone-like genes by G4/A4 expression inhibit T43CS aggregation indirectly; in other words, we suggest that these RNAs directly prevent TDP-43CS aggregation as TDP-25.
FRAP analysis revealed that T43CS was considerably immobile in the aggregates as T25, and the expression of M13-G4/A4 did not alter the immobility of T43CS (Figure Figure55f), suggesting that G4/A4 may also act on TDP-43CS in diffusive states in the cytoplasm and likely not in the aggregates deposited. This is consistent with the findings of the Western blot and filter retardation assays. Furthermore, because the cytoplasmic mislocalization of TDP-43 is cytotoxic,11 to demonstrate whether G4/A4 expression ameliorates cytotoxicity, we quantified the population of propidium iodide (PI)-stained cells using fluorescence microscopy and DRAQ7-stained cells using FACS. The population of PI- and DRAQ7-positive and T43CS-expressing cells was dramatically reduced in M13-G4/A4-expressing cells compared to M13N expression and empty vector transfection as controls (Figure Figure55g and h), suggesting that G4/A4 plays a role in the reduction of cytotoxicity in TDP-43CS-expressing cells. Therefore, G4/A4 could allow cells with cytoplasmic mislocalization of TDP-43 to be protected with a decrease of its oligomers/soluble aggregates.
Discussion
We demonstrate that G4/A4 has an antiaggregation activity against TDP-43/TDP-25 and decreases cytotoxicity by these aggregates. Not only electrostatic but also hydrophobic interactions may contribute to the binding between TDP-25 and G4/A4 unlike TDP-43 (Figure Figure11). Both truncated RRM2 and IDR in TDP-25 may be required for the recognition of G4 and A4 (Figure Figure33). These results suggest that G4/A4 may not recognize a distinct RNA recognition motif such as RRMs. G4 can fold in a mixed state between Gq and HP structure in the presence of potassium ion;28 however, A4 is predicted to exhibit no optimal secondary structure thermodynamically by an RNAfold prediction server,43 suggesting that specific structures or folding states of RNA would not be necessarily involved in the aggregation-suppressing activity. Furthermore, since UG-repeat RNA did not prevent the aggregation of TDP-43CS and TDP-25 (Figures Figures44 and and55 and Figure S7), the sequence specificity of RNA acting as an aggregation suppressor may be surely required. The exogenous introduction of G4/A4 did not affect the intracellular transcriptome in Neuro2a (Figure Figure44c) but only slightly in 293 cells (Figure Figure55e), suggesting that transcriptome changes due to G4/A4 expression may slightly depend on cell lines but basically do not result in its dramatic changes. Therefore, G4/A4 ameliorates and probably prevents a proteostasis collapse in the cell by directly preventing TDP-43 and TDP-25 aggregation without altering the overall expression of TDP-43 and TDP-25.
Repeat sequences of r(G4C2)/r(A4U2) have been observed in transcripts using bioinformatic local alignment search tools such as BLAST. These transcripts that carry repeats of r(G4C2)/r(A4U2) would orchestrate the prevention of the aggregation of TDP-43 and its CTFs in the cell. The most frequent repeats of r(G4C2) and even of nonpathogenic C9orf72 transcripts are two. Although 4–23 repeats of r(G4C2) in the C9orf72 transcript are associated with the formation of a cellular condensate—granule of RNP—more than 23 repeats lead to ALS/FTD-associated phase transitions of RNP, such as TDP-43 because the multiple and tandem Gq may work as a scaffold of RNP assembly.44,45 Because the stable and abundant Gq structure can likely lead to such phase transitions, the binding ability of TDP-43 and TDP-25 not only to Gq but also to the HP structure may be an important physicochemical aspect for the regulation of their aggregation/condensation. As described above, although here we use a short minimum repeat of r(G4C2) to be able to form the Gq structure intramolecularly, increasing Gq structure in the cells is possibly a double-edged sword if it is used as a therapeutic drug as it can encourage the abnormal assembly of RNPs such as TDP-43. In contrast, the r(A4U2) repeat may not have a specific tertiary structure such as Gq. Therefore, the r(A4U2) repeat would be safer than the r(G4C2) repeat. Furthermore, because the pathogenic expansion of r(G4C2) hexanucleotide repeats (>~20) in C9orf72 transcripts produces toxic aggregates of dipeptide repeats by repeat-associated non-ATG translation,46,47 they would rather be cytotoxic; however, the nonpathogenic short hexanucleotide repeats, G4/A4, may not be involved in dipeptide repeats by repeat-associated non-ATG translation. This difference in repeat length would be possible as the threshold for switching between the antiaggregation activity and toxicity of r(G4C2) hexanucleotide repeats.
In conclusion, this is the first demonstration of aggregation-suppressing RNA against aggregation-prone TDP-43 and TDP-25 in the cell. We were able to demonstrate that four repeats of r(G4C2) and r(A4U2) have antiaggregation and cytoprotective activity against pathogenic TDP-43 and TDP-25. These two RNAs may maintain the proteostasis associated with TDP-43 and its CTFs. This report will pave the way for new therapeutic and preventive strategies for ALS/FTD (i.e., such as oligonucleotide therapeutics).
Significance
Protein aggregation of TDP-43 and its C-terminal fragments is associated with disease progression in neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTD). These protein aggregates are so cytotoxic that suppression of aggregate formation enables the reduction of cell death and degeneration that underlies diseases such as motor neurons. Here, using a binding assay in vitro and cellular expression of short RNAs, four repeats of GGGGCC and AAAAUU RNA sequence were validated as aggregation suppressors for TDP-43 and its 25 kDa C-terminal fragment, which for the first time links antiaggregation RNAs for TDP-43 and its fragment. Given the use of these short repeat RNAs in antiaggregation therapy, these studies will have a direct impact on the inhibition of disease progression of ALS and FTD.
Methods
Chemicals
HEPES, Tris base, potassium chloride, sodium chloride, lithium chloride, and magnesium chloride were purchased from Fujifilm Wako Chemicals (Osaka, Japan). Heparin sodium salt from porcine intestinal mucosa and 1,6-hexanediol was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan).
Cell Lines
Murine neuroblastoma Neuro-2a cells (#CCL-131; ATCC, Manassas, VA, USA) and Flp-In T-REx 293 cells (Thermo Fisher, Waltham, MA, USA) were maintained in DMEM (D5796; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FBS (12676029, Thermo Fisher), 100 U/mL penicillin G (Fujifilm Wako Chemicals), and 0.1 μg/mL streptomycin (Fujifilm Wako Chemicals) as described previously.18
Construction of Plasmids
The plasmids for the expression of monomeric enhanced GFP (meGFP)-tagged TDP-25 (pGFP-TDP-25), 220–262 amino acids CTF (F220; truncated RRM2), and 274–414 amino acids CTF (C274) were used as per the protocol established in previous reports.18,36 The synthetic DNA fragment for a variant of TDP-43 that lacks nuclear localization and exporting signal sequences and carries amino acid substitutions of C173S/C175S (TDP-43CS-mNLS/NES),42 was ordered via DNA fragment synthesis services (Eurofins Genomics, Tokyo, Japan). The fragment was inserted into the HindIII and BamHI sites of pmeGFP-N1 (pTDP43CS-GFP). For purification of meGFP-tagged wild type TDP-43, synthetic cDNA fragment for polyhistidine tag and Strep-tag (hereafter, underlined primer sequences denote restriction enzyme sites for cloning) (5′-TGTACAGTCACCATCATCACCATCACTGGAGCCACCCGCAGTTCGAAAAATAAGCGGCCGC-3′) was inserted into the BsrGI and NotI sites of wild type TDP-43 fused with meGFP at the C-terminal, as used in our previous study (pTDP43-GFP-HS).18 For purification of GFP-TDP25 and GFP monomers, synthetic cDNA fragment for polyhistidine tag and Strep-tag (5′-GCTAGCCACCATGCACCATCATCACCATCACTGGAGCCACCCGCAGTTCGAAAAACTACCGGT-3′) was inserted into the NheI and AgeI sites of pGFP-TDP-25 and pmeGFP-N1 (pHS-GFP-TDP-25 and pHS-GFP, respectively). The synthetic DNA fragment for M13-tagged RNA (M13N, M13-UG, M13-G4, M13-A4, M13-G2, and M13-A2) (Table S1) was ordered using DNA oligonucleotide synthesis services (Thermo Fisher, Waltham, MA, USA). The fragments were inserted into the BglII and HindIII sites of pSUPERIOR.neo (pSUPn-M13N, M13-UG, pSUPn-M13-G4, pSUPn-M13-A4, pSUPn-M13-G2, and pSUPn-M13-A2). An expression plasmid for GFP-tagged HSP70 (pHSP70-GFP) was modified from that for red fluorescent protein-tagged HSP70 in a previous report.37 Expression plasmids for yellow fluorescent protein (YFP)-tagged β-actin (pEYFP-Actin) and GFP-tagged α-tubulin (pEGFP-Tub) were purchased from Takara Bio (Shiga, Japan). An expression plasmid for GFP-tagged mitochondrial pyruvate carboxylase (pCMV3-PC-GFPSpark; HG15615-ACG) was purchased from Sino Biological Japan (Kanagawa, Japan).
RNA Synthesis
All synthetic RNAs were ordered using RNA oligonucleotide synthesis services (AJINOMOTO BioPharma, Osaka, Japan). All sequences used in this study are presented in Table S2. For fluorescence assay, the 5′ end of the RNAs were labeled with Alexa Fluor 647, a fluorescent dye, followed by purified by high performance liquid chromatography using oligonucleotide synthesis and purification services (AJINOMOTO BioPharma). For introduction into the cells, synthetic RNAs with no modification (AJINOMOTO BioPharma) were used. The sequence of siRNA against renilla luciferase mRNA (siRL) was obtained from a reference.38
Circular Dichroism (CD)
CD spectra of synthetic RNAs in an HD-containing buffer were recorded in a 1 mm path length cuvette at 20 °C using a spectropolarimeter (J-820, Jasco, Tokyo, Japan) equipped with a Peltier temperature controller in a 220–340 nm measurement range, 0.5 nm pitch wavelength, 1 nm bandwidth, 100 nm/min scanning speed. Spectral contribution from the buffer was subtracted from all CD spectra.
Cell Culture and Transfection
Neuro2a (2.0 × 105) or Flp-In T-REx 293 cells (4.0 × 105) were spread in a 35 mm glass-bottom dish for microscopic experiments (3910–035; IWAKI-AGC Technoglass, Shizuoka, Japan) or a 35 mm plastic dish for cell lysis (150318; Thermo Fisher) 1 day before transfection. Before spreading Flp-In T-REx 293 cells, the dishes were coated with 0.01 mg/mL poly-l-lysine. The mixture of pGFP-TDP-25 (1 μg) or pTDP43CS-GFP (1 μg) and pSUPERIOPR.neo (1 μg), pSUPn-M13N (0.25 μg mixed with 0.75 μg of the empty vector to normalized the RNA expression level), pSUPn-M13-G4 (1 μg), pSUPn-M13-A4 (1 μg), pSUPn-M13-G2 (1 μg), or pSUPn-M13-A2 (1 μg) were transfected using 5.0 μL of Lipofectamine 2000 (Thermo Fisher). After overnight incubation for transfection, the medium was replaced. Neuro2a and Flp-In T-REx 293 cells were cultured for 24 and 48 h after transfection, respectively, before microscopic analysis or cell lysis. For protein purification, Neuro2a cells (3.0 × 106) were spread in a 150 mm plastic dish (TR4003; NIPPON Genetics, Tokyo, Japan) 1 day before transfection. A mixture of 14 μg of salmon sperm DNA (BioDynamics Laboratory Inc., Tokyo, Japan) and 16 μg of expression plasmid DNA (pTDP-43-GFP-HS, pHS-GFP-TDP-25, or pHS-GFP) was diluted in a 150 mM NaCl solution, followed by the addition of 77.5 μg polyethylenimine (PEI) (Fujifilm Wako Chemicals) in a 10 mM Tris-HCl buffer (pH 8.0). The mixture was added to the dishes after 15 min of incubation. After 24 h of incubation, the cells were lysed for protein purification. For purification, 30, 30, and 2 dishes were prepared for TDP-43, TDP-25, and GFP monomers, respectively. For sequential transfection of RNA and plasmids, Neuro2a cells (2.0 × 105) were spread in a 35 mm glass-bottom dish (IWAKI-AGC Technoglass) 1 day before the first transfection. For RNA transfection pre-T25 vector introduction, after the cells were transfected with RNA (25 pmol) using 5.0 μL of Lipofectamine RNAi MAX (Thermo Fisher), they were incubated for 7 h. After the medium exchange, the cells were transfected with pGFP-TDP-25 (1 μg) using 2.5 μL of Lipofectamine 2000 (Thermo Fisher). The cells were cultured for 24 h before microscopic analysis. For RNA transfection post-T25 vector introduction, after the cells were transfected with pGFP-TDP-25 (1 μg) using 2.5 μl of Lipofectamine 2000 (Thermo Fisher), the cells were incubated for 25 h. After the medium exchange, the cells were transfected with RNA (25 pmol) using 5.0 μL of Lipofectamine RNAi MAX (Thermo Fisher). The cells were cultured for 24 h before the microscopic analysis.
Protein Purification
GFP-tagged protein-expressing Neuro2a cells were collected in tubes after trypsinization. Lysis buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 1% Noidet P-40 (Fujifilm Wako Chemicals), and 1× protease inhibitor cocktail (P8340, Sigma-Aldrich) was added to a tube placed on ice, and the supernatant was recovered after centrifugation at 20,400 × g for 10 min at 4 °C. Ni-NTA agarose beads (25 μL slurry for GFP monomers and 75 μL slurry for TDP-43 and TDP25; Fujifilm Wako Chemicals) were added to the supernatant, followed by rotation and incubation for 60 min. The beads were washed three times in a buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 20 mM imidazole, and 0.2% Noidet P-40. After the proteins were eluted in a buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 250 mM imidazole, and 0.2% Noidet P-40, the protein solutions were applied to a spin column containing a Strep-Tactin XT Superflow suspension (Zymo Research, Irvine, CA, USA). After the column was washed in a buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, and 0.2% Noidet P-40, the proteins were eluted in a buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 0.2% Noidet P-40, and 50 mM biotin (Fujifilm Wako Chemicals). The proteins were concentrated in a buffer containing 20 mM HEPES-KOH (pH 7.5) and 0.2% Noidet P-40 using a dialysis spin column (Amicon Ultra-10; Merck, Darmstadt, Germany). The concentrations of purified fluorescent proteins were measured by using fluorescence correlation spectroscopy (FCS). The purified proteins were immediately used for the FCCS measurements. For mass spectroscopic analysis, the purified proteins were separated on a 5–20% gradient gel (ATTO, Tokyo, Japan) using SDS-PAGE, followed by silver staining using silver stain reagents (423413; Cosmo Bio, Tokyo, Japan). The bands of interest were cut and analyzed using MALDI-TOF-MS, followed by peptide mass fingerprinting, which was performed using an outsourced service (Genomine, Pohang, South Korea).
Fluorescence Cross-Correlation Spectroscopy
Before FCCS measurement, 10 μM Alexa Fluor 647-labeled RNAs were heat-denatured and cooled (7 °C/min) in a buffer containing 20 mM Tris-HCl (pH 8.0) and 100 mM KCl for the mixture of Gq and HP or LiCl for the only HP as described previously.31 The tertiary structure of these RNA was confirmed using circular dichroism and nondenatured electrophoresis.31 Neuro2a cells were lysed in a buffer containing 20 mM HEPES-KOH (pH 7.5), 1% Noidet P-40, and a 1 x protease inhibitor cocktail (Sigma-Aldrich). The supernatants of the lysates were recovered after centrifugation at 20,400 × g for 10 min at 4 °C. The supernatant was mixed with 100 nM RNA in KCl, MgCl2, heparin, or HD-containing buffer and incubated for 5 min before the measurement. Double-strand 500 bp DNA as a positive control (PC-DNA) for FCCS was amplified by PCR with ATTO488- and ATTO647N-labeled primers and lambda DNA as a PCR template and was purified using MicroSpin S-400 HR columns (GE Healthcare, Chicago, IL, USA) as previously reported.32 The mixture of ATTO488- and ATTO647N-labeled primers (ATTO488-λDNA-R and ATTO647N-λDNA-F, respectively) was used as a negative control (NC-DNA) for FCCS. The DNA sequences as PC-DNA, ATTO488-λDNA-R, and ATTO647N-λDNA-F are represented in Table S3. FCCS was performed using LSM 510 META + ConfoCor3 (Carl Zeiss) with a C-Apochromat 40 × /1.2NA W. UV–VIS-IR water immersion objective lens (Carl Zeiss) as previously reported.18 The photons were recorded over 100 s. Curve fitting analysis for the acquired auto- and cross-correlation function was performed using ZEN software (Carl Zeiss) with a model for two-component 3D diffusion involving a one-component exponential blinking fraction in the fitting for autocorrelation function, as shown in the following eq 1, and with a model for two-component 3D diffusion without a blinking fraction (T = 0 in eq 1) in the fitting for cross-correlation function.
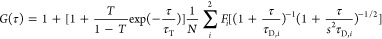
where G(τ) represents an auto- or cross-correlation function of the time interval τ; τD, i represents the diffusion time (i = 1 or 2); Fi represents the fraction of the component (i = 1 or 2); N represents the average number of particles in the confocal detection volume; T and τT represent the exponential blinking fraction and its meantime, respectively (these are zero for cross-correlation function); s represent the structural parameter determined by analyzing the standard fluorescent dyes (ATTO 488 and Cy5) on the same day. The relative cross-correlation amplitude (RCA) was calculated using eq 2.

where NC and NG represent the number of interacting and total green molecules, respectively. Dissociation constants (Kd) were calculated as reported previously.48
RT-PCR
Total RNA of cells expressing M13-tagged RNA was isolated using an RNeasy Mini Kit (QIAGEN, Venlo, Netherlands) according to the manufacturer’s instructions. The RNA was eluted in diethyl pyrocarbonate-treated ultrapure water. To detect M13-tagged RNAs, total RNA (1.9 μg) was used to synthesize first-strand cDNA using the Mir-X miRNA First-Strand Synthesis Kit (Takara Bio Inc.) according to the manufacturer’s instructions. PCR was performed using a 0.25 U PrimeSTAR HS DNA polymerase (Takara Bio Inc.) with 0.2 mM dNTP mixture, 2 pmol of M13-forward primers (5′-gtaaaacgacggccagt-3′), 2 pmol of M13-reverse primers (5′-gagcggataacaatttcacacagg-3′), and template cDNA (corresponding to 15 ng of total RNA). Amplification was performed by using a PC320 thermal cycler (Astec-Bio, Fukuoka, Japan). The products were separated on a 5–20% polyacrylamide gel (ATTO) in Tris-glycine buffer. The gel was stained with SYBR Gold (Thermo Fisher), and gel images were acquired by using a Limited-STAGE II gel imager with blue LED light (AMZ System Science, Osaka, Japan).
Confocal Microscopy and Cell Counting
GFP-expressing cells were observed using an LSM 510 META microscope (Carl Zeiss, Jena, Germany) with a Plan-Apochromat 10×/0.45NA objective (Carl Zeiss). GFP was excited at a wavelength of 488 nm. The excitation beam was divided using an HFT405/488. Fluorescence was corrected via a 505–570 nm band-pass filter (BP505–570) and then acquired using a photomultiplier tube. The pinhole diameter was set to 72 mm. Transmission bright-field images were acquired simultaneously with the GFP fluorescence images. Aggregate-positive cells were counted by using the Fiji-ImageJ software. The population of cells that harbored cytoplasmic aggregates was normalized to the number of GFP-positive cells. Multiple fluorescent images were acquired at randomly selected locations, and then GFP-positive and aggregate-positive cells were counted using the all acquired images. Aggregate-positive cells were counted when the brightness of the structures was higher than that in the cytoplasm. The total number of GFP-positive cells was counted, ranging from 200 to 400 per trial. Statistical analysis of three independent transfections was performed (Student’s t test), and the results are plotted in the graph (trial number = 3; mean ± SE). Propidium iodide (PI) staining for determining dead cells was performed as previously reported.18 The population of PI-positive cells was normalized to the total number of GFP-positive cells. The total number of cells that were counted ranged from 392 to 735 for TDP-25 and from 761 to 1327 for TDP-43 per trial from the randomly acquired multiple fields of fluorescent images. Statistical analysis of three independent trials was performed (Student’s t test), and the results are plotted in the graph (trial number = 3; mean ± SE).
Fluorescence Recovery after Photobleaching
Photobleaching experiments were performed with an LSM 510 META system using a C-Apochromat 40×/1.2NA W Korr. UV–VIS-IR M27 objective lens (Carl Zeiss), as previously reported.18,36,37 GFP was excited and photobleached at 488 nm. The GFP fluorescence was collected through a band-pass filter (BP505–570). The pinhole size was set to 200 μm, the zoom factor was set to 5×, and the interval time for image acquisition was set to 10 s. The X- and Y-scanning sizes were 512 pixels, and GFP was photobleached at 488 nm with 100% transmission for less than 1.92 s. Relative fluorescence intensities (RFIs) were measured using the Fiji-ImageJ software and calculated as reported previously.18,36,37 Normalized RFIs were calculated using the mean RFI of T25 and T43CS in the aggregates with an empty vector transfection just after photobleaching.
Cell Lysis, Solubility Assay, and Filter Retardation Assay
Cells expressing T25 and T43CS were lysed and fractionated as reported previously.18 Briefly, cells were lysed in a lysis buffer containing 25 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 1% Noidet P-40, 0.1% SDS, 0.25 U/mL benzonase (Sigma-Aldrich), and 1× protease inhibitor cocktail (Sigma-Aldrich). After centrifugation (20,400 × g, 10 min, 4 °C), the fractionated supernatants were recovered, and the pellets were solubilized in PBS containing 1 M urea. To recover the whole cell lysates without the fractionation, cells were lysed in PBS containing 1 M urea, 1% SDS, 0.25 U/mL of benzonase, and 1× protease inhibitor cocktail (Sigma-Aldrich). SDS-PAGE sample buffer-mixed lysates were denatured at 98 °C for 5 min. The samples were applied to a 12.5% polyacrylamide gel (ATTO) and subjected to electrophoresis in an SDS-containing buffer. Proteins were transferred onto polyvinylidene difluoride membranes (Cytiva, Marlborough, MA, USA). The membrane was blocked with a PBS-T buffer containing 5% skim milk for 1 h. Horseradish peroxidase-conjugated anti-GFP (#598–7, MBL, Tokyo, Japan) or anti-α-tubulin antibodies (#HRP-66031, Proteintech, Rosemont, IL, USA) were reacted in CanGet Signal Immunoreaction Enhancer Solution 1 (TOYOBO, Osaka, Japan). To detect endogenous HSP70, an anti-HSP70 antibody (#10995–1-AP, Proteintech) was reacted in the blocking buffer and then reacted with a horseradish peroxidase-conjugated antirabbit IgG antibody (#111–035–144, Jackson ImmunoResearch, West Grove, PA, USA). Chemiluminescent signals were acquired using a ChemiDoc MP imager (Bio-Rad, Hercules, CA, USA). As an internal control for the insoluble fraction, the insoluble fractions were applied to a 12.5% polyacrylamide gel (ATTO) and subjected to electrophoresis in an SDS-containing buffer, followed by staining with a Coomassie brilliant blue R-250 (Fujifilm Wako Chemicals). Images of the stained gels were obtained by using a Limited-STAGE II gel imager with transparent white light (AMZ System Science). For the filter retardation assay, the cells were lysed in a PBS buffer containing 1% SDS and 0.25 U/mL of benzonase (Sigma-Aldrich). The lysates were applied to a cellulose acetate membrane with a pore size of 0.2 μm (Advantec Toyo, Tokyo, Japan) by using a Bio-Dot SF blotter (Bio-Rad). The membrane was blocked in PBS-T buffer containing 5% skim milk for 1 h. The GFP on the membrane was detected by Western blotting, as described above. All of the intensities were measured using Fiji-ImageJ. After subtracting the values in the background region where there are no bands, the ratio value of the intensity of T43CS/T25 in the insoluble fraction or trapped on the filter to the control was calculated.
FACS Analysis
RNA-transfected Neuro2a and 293 cells were prepared the same as for cell lysis and confocal microscopy. Cells were trypsinized and suspended in a medium containing FBS. After centrifugation (1,000 × g, 3 min, 25 °C), the cells were suspended in Opti-MEM I (Thermo Fisher). The cell concentration was adjusted to 6.0 × 105 cells/ml after cell counting. Subsequently, 3 μM DRAQ7 (BioStatus, Leicestershire, UK) was added for dead cell staining. Immediately before FACS analysis, the cells were passed through a cell strainer (no. 352235, Corning Incorporated, Corning, NY, USA), and fluorescence of DRAQ7-positive cells was detected using a 668–722 nm band-pass filter with 638 nm excitation on a JSAN desktop cell sorter (Bay Bioscience, Brookline, MA, USA). The percentage of dead cells was determined based on higher fluorescence intensity.
RNA-seq and Data Processing
Total RNA of the cells expressing M13-tagged RNA was recovered in the same manner as that of RT-PCR. The total RNA concentration was measured using a Quantus Fluorometer with a QuantiFluor RNA system (Promega). The quality of total RNA was examined using a fragment analyzer system with an Agilent HS RNA kit (Agilent Technologies). Libraries were prepared using an MGIEasy RNA Directional Library Prep Set (MGI Tech Co., Ltd.) according to the manufacturer’s instructions. The concentration of the libraries was measured using a Qubit 3.0 Fluorometer with a dsDNA HS assay kit (Thermo Fisher). The quality of the libraries was analyzed using a fragment analyzer with a dsDNA 915 reagent kit (Agilent Technologies). Circular DNA was created using the libraries and an MGIEasy Circularization kit (MGI Tech Co., Ltd.), according to the manufacturer’s protocol. DNA Nano Ball (DNB) was prepared using a DNBSEQ-G400RS high-throughput sequencing kit (MGI Tech Co., Ltd.) according to the manufacturer’s protocol. DNBs were sequenced using a DEBSEQ-G400 sequencer (MGI Tech Co., Ltd.). After removing the adaptor sequence using Cutdapt ver. 1.9.1 software, Pair reads that were less than 40 bases and a quality score of less than 20 were removed using a sickle ver. 1.33 software. Mapping was performed by referring to a murine genome reference (GRCm39) using the hisat2 ver. 2.2.1 software. The number of reads was counted using the featureCounts ver. 2.0.0 software. Reads per kilobase per million (RPKM) and transcripts per million (TPM) values were calculated. After removing transcripts with low read counts (<19), TPM values derived from total RNA obtained four times, independently, were averaged. The mean fold change and p-values were calculated for cells expressing M13-G4/A4 in comparison with cells expressing M13-UG. Volcano plots were prepared by using Microsoft Excel.
Quantification and Statistical Analysis
All of the data were presented as mean ± standard error from at least three independent trials. Student’s t tests were performed using Microsoft Excel.
Acknowledgments
A.F. was supported by the Support for Pioneering Research Initiated by the Next Generation (SPRING) program by the Japan Science and Technology Agency (JST) at Hokkaido University (JPMJSP2119). M.K. was supported by a Japan Society for Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (B) (22H02578) and Grant-in-Aid for Challenging Research (Exploratory) (22K19886). A.K. was supported by grants from Japan Agency for Medical Research and Development (AMED) (JP23gm6410028 and JP22ym0126814); by grants from JSPS Grant-in-Aid for Scientific Research (24H02286; 22H04826; 18K06201; and 16KK0156); by a grant from Hokkaido University Office for Developing Future Research Leaders (L-Station); by a grant from Hoansha Foundation; by a grant from Hagiwara Foundation of Japan; and by a grant from Nakatani Foundation. We would like to thank Ms. Asuka Murata and Mr. Koya Yoshizawa for technical support, Editage (www.editage.com) for English language editing, and the Open Facility, Global Facility Center, Creative Research Institution, Hokkaido University for using a ChemiDoc MP imager and a JSAN desktop cell sorter.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.4c00566.
(Figure S1) Interaction strength in the presence of electrostatic interaction inhibitors; (Figure S2) controls of FCCS measurement and RNA conformation when adding 1,6-hexanediol; (Figure S3) binding of AF-G4/A4 to the proteins copurified with GFP-TDP-43/TDP-25; (Figure S4) typical normalized cross-correlation functions (CCFs) when purified proteins were mixed with AF-labeled RNA; (Figure S5) confirmation of the expression levels of M13-tagged RNA using RT-PCR; (Figure S6) total GFP-TDP-25 abundance in cell lysate when expressing M13-G4/A4; (Figure S7) reduced aggregates of GFP-TDP-25 in Neuro2a cells when synthetic RNA was directly introduced; (Figure S8) aggregate formation of GFP-TDP-25 in Neuro2a cells expressing M13-G2/A2; (Figure S9) total GFP-TDP-43CS abundance in cell lysate when expressing M13-G4/A4; (Table S1) cDNA sequence for plasmid DNA; (Table S2) synthetic RNA sequence; (Table S3) DNA sequences for FCCS controls; (Table S4) transcripts that exhibited changes upon expression of M13-G4/A4 in 293 cells (PDF)
Reagents (PDF)
Author Contributions
A.F. performed the experiments, analyzed the data, and edited the manuscript. M.K. partially contributed to the FCCS measurement and its analysis. A.K. conceptualized and supervised this project, performed the experiments, analyzed the data, visualized the data, provided resources, wrote the original manuscript draft, and edited the manuscript. CRediT: Ai Fujimoto conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, resources, validation, visualization, writing - original draft, writing - review & editing; Masataka Kinjo funding acquisition, methodology, resources; Akira Kitamura conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing - original draft, writing - review & editing.
References
- Arai T.; Hasegawa M.; Akiyama H.; Ikeda K.; Nonaka T.; Mori H.; Mann D.; Tsuchiya K.; Yoshida M.; Hashizume Y.; Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and biophysical research communications 2006, 351 (3), 602–11. 10.1016/j.bbrc.2006.10.093. [Abstract] [CrossRef] [Google Scholar]
- Neumann M.; Sampathu D. M.; Kwong L. K.; Truax A. C.; Micsenyi M. C.; Chou T. T.; Bruce J.; Schuck T.; Grossman M.; Clark C. M.; McCluskey L. F.; Miller B. L.; Masliah E.; Mackenzie I. R.; Feldman H.; Feiden W.; Kretzschmar H. A.; Trojanowski J. Q.; Lee V. M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314 (5796), 130–3. 10.1126/science.1134108. [Abstract] [CrossRef] [Google Scholar]
- Francois-Moutal L.; Perez-Miller S.; Scott D. D.; Miranda V. G.; Mollasalehi N.; Khanna M. Structural Insights Into TDP-43 and Effects of Post-translational Modifications. Front. Mol. Neurosci. 2019, 12, 301.10.3389/fnmol.2019.00301. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lagier-Tourenne C.; Polymenidou M.; Cleveland D. W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human molecular genetics 2010, 19 (R1), R46–64. 10.1093/hmg/ddq137. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Buratti E.; Baralle F. E. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010, 7 (4), 420–9. 10.4161/rna.7.4.12205. [Abstract] [CrossRef] [Google Scholar]
- Zhang Y. J.; Xu Y. F.; Cook C.; Gendron T. F.; Roettges P.; Link C. D.; Lin W. L.; Tong J.; Castanedes-Casey M.; Ash P.; Gass J.; Rangachari V.; Buratti E.; Baralle F.; Golde T. E.; Dickson D. W.; Petrucelli L. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 2009, 106 (18), 7607–12. 10.1073/pnas.0900688106. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chhangani D.; Martin-Pena A.; Rincon-Limas D. E. Molecular, functional, and pathological aspects of TDP-43 fragmentation.. iScience 2021, 24 (5), 10245910.1016/j.isci.2021.102459. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li Q.; Babinchak W. M.; Surewicz W. K. Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat. Commun. 2021, 12 (1), 1620.10.1038/s41467-021-21912-y. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Arseni D.; Hasegawa M.; Murzin A. G.; Kametani F.; Arai M.; Yoshida M.; Ryskeldi-Falcon B. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 2022, 601 (7891), 139–143. 10.1038/s41586-021-04199-3. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Berning B. A.; Walker A. K. The Pathobiology of TDP-43 C-Terminal Fragments in ALS and FTLD. Front. Neurosci. 2019, 13, 335.10.3389/fnins.2019.00335. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Suk T. R.; Rousseaux M. W. C. The role of TDP-43 mislocalization in amyotrophic lateral sclerosis. Mol. Neurodegener. 2020, 15 (1), 45.10.1186/s13024-020-00397-1. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Buratti E.; Baralle F. E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 2008, 13, 867–78. 10.2741/2727. [Abstract] [CrossRef] [Google Scholar]
- Polymenidou M.; Lagier-Tourenne C.; Hutt K. R.; Huelga S. C.; Moran J.; Liang T. Y.; Ling S. C.; Sun E.; Wancewicz E.; Mazur C.; Kordasiewicz H.; Sedaghat Y.; Donohue J. P.; Shiue L.; Bennett C. F.; Yeo G. W.; Cleveland D. W. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature neuroscience 2011, 14 (4), 459–68. 10.1038/nn.2779. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Labbadia J.; Morimoto R. I. The biology of proteostasis in aging and disease. Annual review of biochemistry 2015, 84, 435–64. 10.1146/annurev-biochem-060614-033955. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hipp M. S.; Kasturi P.; Hartl F. U. The proteostasis network and its decline in ageing. Nature reviews. Molecular cell biology 2019, 20 (7), 421–435. 10.1038/s41580-019-0101-y. [Abstract] [CrossRef] [Google Scholar]
- Tauber D.; Tauber G.; Parker R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends in biochemical sciences 2020, 45 (9), 764–778. 10.1016/j.tibs.2020.05.002. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Aarum J.; Cabrera C. P.; Jones T. A.; Rajendran S.; Adiutori R.; Giovannoni G.; Barnes M. R.; Malaspina A.; Sheer D. Enzymatic degradation of RNA causes widespread protein aggregation in cell and tissue lysates. EMBO Rep. 2020, 21 (10), e4958510.15252/embr.201949585. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Nakayama Y.; Shibasaki A.; Taki A.; Yuno S.; Takeda K.; Yahara M.; Tanabe N.; Kinjo M. Interaction of RNA with a C-terminal fragment of the amyotrophic lateral sclerosis-associated TDP43 reduces cytotoxicity. Sci. Rep. 2016, 6, 19230.10.1038/srep19230. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mann J. R.; Gleixner A. M.; Mauna J. C.; Gomes E.; DeChellis-Marks M. R.; Needham P. G.; Copley K. E.; Hurtle B.; Portz B.; Pyles N. J.; Guo L.; Calder C. B.; Wills Z. P.; Pandey U. B.; Kofler J. K.; Brodsky J. L.; Thathiah A.; Shorter J.; Donnelly C. J. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 2019, 102 (2), 321–338. 10.1016/j.neuron.2019.01.048. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rengifo-Gonzalez J. C.; El Hage K.; Clement M. J.; Steiner E.; Joshi V.; Craveur P.; Durand D.; Pastre D.; Bouhss A. The cooperative binding of TDP-43 to GU-rich RNA repeats antagonizes TDP-43 aggregation. Elife 2021, 10, e6760510.7554/eLife.67605. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Choi S. I.; Han K. S.; Kim C. W.; Ryu K. S.; Kim B. H.; Kim K. H.; Kim S. I.; Kang T. H.; Shin H. C.; Lim K. H.; Kim H. K.; Hyun J. M.; Seong B. L. Protein solubility and folding enhancement by interaction with RNA. PloS one 2008, 3 (7), e267710.1371/journal.pone.0002677. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Horowitz S.; Bardwell J. C. RNAs as chaperones. RNA Biol. 2016, 13 (12), 1228–1231. 10.1080/15476286.2016.1247147. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kaiser C. M.; Goldman D. H.; Chodera J. D.; Tinoco I. Jr.; Bustamante C. The ribosome modulates nascent protein folding. Science 2011, 334 (6063), 1723–7. 10.1126/science.1209740. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Son A.; Huizar Cabral V.; Huang Z.; Litberg T. J.; Horowitz S. G-quadruplexes rescuing protein folding. Proc. Natl. Acad. Sci. U. S. A. 2023, 120 (20), e221630812010.1073/pnas.2216308120. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ishiguro A.; Kimura N.; Watanabe Y.; Watanabe S.; Ishihama A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes to cells: devoted to molecular & cellular mechanisms 2016, 21 (5), 466–81. 10.1111/gtc.12352. [Abstract] [CrossRef] [Google Scholar]
- Begeman A.; Son A.; Litberg T. J.; Wroblewski T. H.; Gehring T.; Huizar Cabral V.; Bourne J.; Xuan Z.; Horowitz S. G-Quadruplexes act as sequence-dependent protein chaperones. EMBO Rep. 2020, 21 (10), e4973510.15252/embr.201949735. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cooper-Knock J.; Walsh M. J.; Higginbottom A.; Robin Highley J.; Dickman M. J.; Edbauer D.; Ince P. G.; Wharton S. B.; Wilson S. A.; Kirby J.; Hautbergue G. M.; Shaw P. J. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 2014, 137 (Pt 7), 2040–2051. 10.1093/brain/awu120. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Haeusler A. R.; Donnelly C. J.; Periz G.; Simko E. A.; Shaw P. G.; Kim M. S.; Maragakis N. J.; Troncoso J. C.; Pandey A.; Sattler R.; Rothstein J. D.; Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507 (7491), 195–200. 10.1038/nature13124. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Nagata K.; Kinjo M. Conformational analysis of misfolded protein aggregation by FRET and live-cell imaging techniques. International journal of molecular sciences 2015, 16 (3), 6076–92. 10.3390/ijms16036076. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bacia K.; Kim S. A.; Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods 2006, 3 (2), 83–9. 10.1038/nmeth822. [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Tornmalm J.; Demirbay B.; Piguet J.; Kinjo M.; Widengren J. Trans-cis isomerization kinetics of cyanine dyes reports on the folding states of exogeneous RNA G-quadruplexes in live cells. Nucleic acids research 2023, 51 (5), e2710.1093/nar/gkac1255. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sasaki A.; Yamamoto J.; Jin T.; Kinjo M. Raster image cross-correlation analysis for spatiotemporal visualization of intracellular degradation activities against exogenous DNAs. Sci. Rep. 2015, 5, 14428.10.1038/srep14428. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gitler A. D.; Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion 2011, 5 (3), 179–87. 10.4161/pri.5.3.17230. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kishor A.; White E. J. F.; Matsangos A. E.; Yan Z.; Tandukar B.; Wilson G. M. Hsp70’s RNA-binding and mRNA-stabilizing activities are independent of its protein chaperone functions. J. Biol. Chem. 2017, 292 (34), 14122–14133. 10.1074/jbc.M117.785394. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kerr A. G.; Wang Z.; Wang N.; Kwok K. H. M.; Jalkanen J.; Ludzki A.; Lecoutre S.; Langin D.; Bergo M. O.; Dahlman I.; Mim C.; Arner P.; Gao H. The long noncoding RNA ADIPINT regulates human adipocyte metabolism via pyruvate carboxylase. Nat. Commun. 2022, 13 (1), 2958.10.1038/s41467-022-30620-0. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Fujimoto A.; Kawashima R.; Lyu Y.; Sasaki K.; Hamada Y.; Moriya K.; Kurata A.; Takahashi K.; Brielmann R.; Bott L. C.; Morimoto R. I.; Kinjo M. Hetero-oligomerization of TDP-43 carboxy-terminal fragments with cellular proteins contributes to proteotoxicity. Commun. Biol. 2024, 7 (1), 743.10.1038/s42003-024-06410-3. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Iwasaki N.; Kinjo M. Molecular chaperone HSP70 prevents formation of inclusion bodies of the 25-kDa C-terminal fragment of TDP-43 by preventing aggregate accumulation. Cell stress & chaperones 2018, 23 (6), 1177–1183. 10.1007/s12192-018-0930-1. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Calabrese J. M.; Sharp P. A. Characterization of the short RNAs bound by the P19 suppressor of RNA silencing in mouse embryonic stem cells. RNA 2006, 12 (12), 2092–102. 10.1261/rna.224606. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitamura A.; Kubota H.; Pack C. G.; Matsumoto G.; Hirayama S.; Takahashi Y.; Kimura H.; Kinjo M.; Morimoto R. I.; Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nature cell biology 2006, 8 (10), 1163–70. 10.1038/ncb1478. [Abstract] [CrossRef] [Google Scholar]
- Fang Y. S.; Tsai K. J.; Chang Y. J.; Kao P.; Woods R.; Kuo P. H.; Wu C. C.; Liao J. Y.; Chou S. C.; Lin V.; Jin L. W.; Yuan H. S.; Cheng I. H.; Tu P. H.; Chen Y. R. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat. Commun. 2014, 5, 4824.10.1038/ncomms5824. [Abstract] [CrossRef] [Google Scholar]
- Carey J. L.; Guo L. Liquid-Liquid Phase Separation of TDP-43 and FUS in Physiology and Pathology of Neurodegenerative Diseases. Front Mol. Biosci 2022, 9, 82671910.3389/fmolb.2022.826719. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Shodai A.; Morimura T.; Ido A.; Uchida T.; Ayaki T.; Takahashi R.; Kitazawa S.; Suzuki S.; Shirouzu M.; Kigawa T.; Muto Y.; Yokoyama S.; Takahashi R.; Kitahara R.; Ito H.; Fujiwara N.; Urushitani M. Aberrant assembly of RNA recognition motif 1 links to pathogenic conversion of TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 2013, 288 (21), 14886–905. 10.1074/jbc.M113.451849. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gruber A. R.; Lorenz R.; Bernhart S. H.; Neubock R.; Hofacker I. L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36 (suppl_2), W70–W74. 10.1093/nar/gkn188. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cammas A.; Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017, 45 (4), 1584–1595. 10.1093/nar/gkw1280. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fay M. M.; Anderson P. J.; Ivanov P. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell reports 2017, 21 (12), 3573–3584. 10.1016/j.celrep.2017.11.093. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Breevoort S.; Gibson S.; Figueroa K.; Bromberg M.; Pulst S. Expanding Clinical Spectrum of C9ORF72-Related Disorders and Promising Therapeutic Strategies: A Review. Neurol.: Genet. 2022, 8 (3), e67010.1212/NXG.0000000000000670. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gijselinck I.; Cruts M.; Van Broeckhoven C. The Genetics of C9orf72 Expansions. Cold Spring Harb Perspect Med. 2018, 8 (4), a02675710.1101/cshperspect.a026757. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Oyama R.; Takashima H.; Yonezawa M.; Doi N.; Miyamoto-Sato E.; Kinjo M.; Yanagawa H. Protein-protein interaction analysis by C-terminally specific fluorescence labeling and fluorescence cross-correlation spectroscopy. Nucleic acids research 2006, 34 (14), e10210.1093/nar/gkl477. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from JACS Au are provided here courtesy of American Chemical Society
Citations & impact
This article has not been cited yet.
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/168260136
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[The molecular mechanisms of intracellular TDP-43 aggregates].
Brain Nerve, 61(11):1292-1300, 01 Nov 2009
Cited by: 1 article | PMID: 19938686
Review
Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1.
J Neurochem, 116(2):248-259, 02 Dec 2010
Cited by: 135 articles | PMID: 21062285
Mitigation of TDP-43 toxic phenotype by an RGNEF fragment in amyotrophic lateral sclerosis models.
Brain, 147(6):2053-2068, 01 Jun 2024
Cited by: 1 article | PMID: 38739752 | PMCID: PMC11146434
Aberrant assembly of RNA recognition motif 1 links to pathogenic conversion of TAR DNA-binding protein of 43 kDa (TDP-43).
J Biol Chem, 288(21):14886-14905, 04 Apr 2013
Cited by: 44 articles | PMID: 23558684 | PMCID: PMC3663511





