Abstract
Free full text

Self-priming of Plk1 binding to BubR1 ensures accurate mitotic progression
Abstract
Plk1 is a key mitotic kinase that localizes to distinct subcellular structures to promote accurate mitotic progression. Plk1 recruitment depends on direct interaction between polo-box domain (PBD) on Plk1 and PBD binding motif (PBD BM) on the interactors. However, recent study showed that PBD BM alone is not enough for stable binding between CENP-U and Plk1 highlighting the complexity of the interaction which warrants further investigation. An important interactor for Plk1 during mitosis is the checkpoint protein BubR1. Plk1 bound to BubR1 via PBD interaction with pT620 phosphorylates BubR1 S676/T680 to promote BubR1-PP2A/B56 interaction. The BubR1-PP2A/B56 complex counteracts the destablizing effect on kinetochore-microtubule attachments by mitotic kinases to promote mitotic progression. Here we show that Plk1 phosphorylates T600/T608 on BubR1 and the double phosphorylation is critical for BubR1-Plk1 interaction. A similar mechanism for Plk1-Bub1 interaction also exists indicating a general principle for Plk1 kinetochore recruitment through self-priming. Mechanistically preventing BubR1 T600/T608 phosphorylation impairs chromosome congression and checkpoint silencing by reducing Plk1 and PP2A/B56 binding to BubR1. Increasing the binding affinity towards Plk1 and PP2A/B56 in BubR1 through protein engineering bypasses the requirement of T600/T608 phosphorylation for mitotic progression. These results reveal a new layer of regulation for accurate mitotic progression.
Introduction
Polo-like kinase 1 (Plk1) plays essential roles in mitosis by promoting mitotic entry, centrosome maturation, mitotic spindle assembly, chromosome arm resolution, chromosome alignment and cytokinesis1. Plk1 harbors a kinase domain in the N-terminal region and two polo-box domains (PBD) in the C-terminal region. Through direct interaction between the Plk1 PBD domains and the PBD binding motif (PBD BM hereafter) (S-[pS/pT]-[P/X], where p indicates phosphorylation and X means any amino acid) on interaction partners, Plk1 is recruited to different subcellular locations2. Many proteins have been reported as Plk1 receptors at these locations. For example, Plk1 kinetochore localization relies on the interaction with Bub1, CENP-U, INCENP, NudC, BubR1, Sgo1, CLASP2, Survivin, p27, NCAPG2, USP16, and RSF1 while recent studies identified Bub1 and CENP-U as the major Plk1 kinetochore receptors3–17.
Among the reported interactors, BubR1 and Bub1 are mitotic checkpoint proteins which play important roles at both monitoring and promoting kinetochore-microtubule attachment7,18–20. The two proteins are paralogues arising from the same ancestral gene. Sub-functionalization confers distinct roles to Bub1 and BubR1 in mitosis though the two proteins share some structural similarities. From the N-terminal to the C-terminal region, both proteins harbor a GLEBS domain, a PBD BM, a protein phosphatase PP2A/B56 binding motif and a kinase domain. Interestingly, the PP2A/B56 binding motif on Bub1 is dysfunctional which leaves BubR1 as the main PP2A/B56 receptor at outer kinetochores20. On the other hand, Bub1 contains an active kinase domain while BubR1 is a pseudokinase21. Such arrangement ensures a robust checkpoint and efficient attachment between kinetochores and microtubules20. Unlike Bub1, BubR1 marginally contributes to the Plk1 kinetochore recruitment though disruption of the BubR1-Plk1 interaction causes chromosome aligning defects6,7,17. Briefly, Plk1 bound with BubR1 PBD BM primed by Cdk1 further phosphorylates S676 and T680 close to the B56 binding motif (669-LxxIxE-675 or KARD motif) which significantly enhances the interaction between BubR1 and the B56 subunit. Thus, PP2A/B56 locates on outer kinetochores to counteract the destabilizing effect on kinetochore-microtubule interactions by mitotic kinases like Aurora B and Mps122–28. PP2A/B56 in turn reduces the phosphorylation on pT620 and associated Plk1. Thus, an intramolecular negative feedback loop forms on BubR129,30. Whether this working module is under additional regulation is not clear.
In this study, we show Plk1 recruited by BubR1 PBD BM phosphorylates BubR1 T600 and T608. The double phosphorylation is required for the stable BubR1-Plk1 interaction as prevention of the phosphorylation largely reduces Plk1 as well as PP2A/B56 bound to BubR1 and delays the mitotic progression due to inefficient kinetochore-microtubule attachment resulting in activation of the mitotic checkpoint. We show that a similar mechanism also operates on Bub1 indicating a general principle for Plk1 kinetochore recruitment. Grafting a strong PBD or PP2A/B56 binding motif onto BubR1 bypasses the requirement of the double phosphorylation for accurate mitosis. Interestingly, further analysis shows that the double phosphorylation on BubR1 does not generate a new Plk1 binding site. The enhanced binding to Plk1 after double phosphorylation may come from protein conformation changes which needs to be investigated in future.
Results
BubR1 T600/T608 phosphorylation is required for accurate mitotic progression
A previous phospho-proteomics study31 identified two phosphorylation sites on BubR1 (pT600 and pT608) that were highly regulated by PP2A/B56 during mitosis. We set out to investigate if these two phosphorylation sites have any biological function. To address this, we generated RNAi-resistant BubR1 T600A/T608A and BubR1 T600E/T608E (BubR1 2A and 2E hereafter) which mimic the unphosphorylated and phosphorylated BubR1 and tested their ability to support mitosis in mammalian cells. HeLa cells were co-transfected with RNAi oligos against BubR1 and RNAi-resistant plasmid expressing YFP-tagged BubR1. mCherry-Histone 3 expressing plasmid was also transfected as an indicator for chromosome movement. The mitotic progression was recorded by live cell imaging. Quantification of the mitotic timing from nuclear envelope breakdown (NEBD) to anaphase showed a significant shortening of mitotic timing in cells treated with BubR1 RNAi (Fig. 1a, b). Wild type BubR1 and BubR1 2E mutant fully restored the mitotic timing while BubR1 2A mutant caused a delayed mitosis (Fig. 1a, b). To find out the reason for the mitotic delay, cells were arrested in mitosis with the proteasome inhibitor MG132 before being fixed and examined by immunofluorescence microscopy. Indeed, cells complemented with BubR1 2A mutant showed a clear chromosome alignment defect (Fig. 1c, d). To determine if the chromosome alignment defects could cause chromosome segregation errors, we examined chromosome segregation at anaphase and found an increase in lagging chromosomes specifically in BubR1 2A complemented cells (Supplementary Fig. 1a, b). We also examined the effect of BubR1 2A mutation on the spindle assembly checkpoint (SAC). Low dose of nocodazole was applied to the cells which partially depolymerized microtubules and activated the checkpoint. The time cells spent in mitosis, a direct indicator for checkpoint strength, was recorded by live cell imaging. In cells complemented with BubR1 2A, the mitotic time was significantly elongated from 690 min in cells with wild type BubR1 to 805
min in cells with wild type BubR1 to 805 min showing an enhanced mitotic checkpoint. In contrast, the mitotic timing in BubR1 2E was reduced to 490
min showing an enhanced mitotic checkpoint. In contrast, the mitotic timing in BubR1 2E was reduced to 490 min (Fig. 1e). Consistently, the kinetochore localized checkpoint protein Mad2 was slightly enhanced in cells with BubR1 2A compared with cells with wild type or BubR1 2E (Supplementary Fig. 1c, d).
min (Fig. 1e). Consistently, the kinetochore localized checkpoint protein Mad2 was slightly enhanced in cells with BubR1 2A compared with cells with wild type or BubR1 2E (Supplementary Fig. 1c, d).
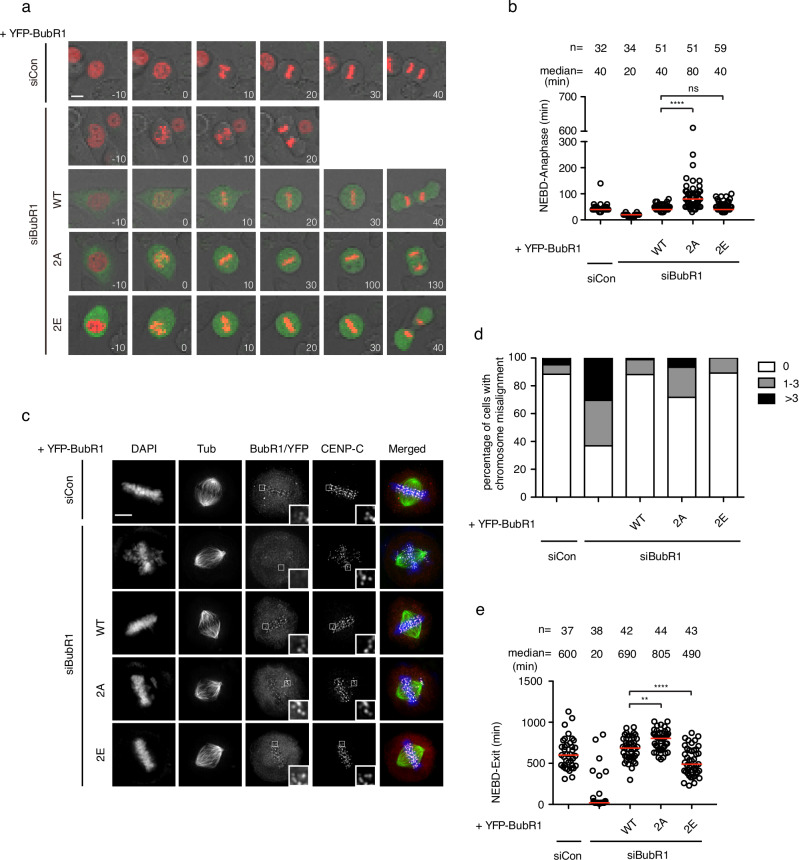
a Representative stills of live cell imaging of cells transfected with siRNA oligos against luciferase as control or with siRNA oligos against BubR1 with RNAi-resistant constructs expressing YFP-BubR1. mCherry-H3 construct was co-transfected for chromosome indication. Scale bar is 10 μm. b The mitotic timing of unperturbed mitosis recorded in (a). Each circle represents the time from NEBD to anaphase of a single cell. Red line indicates the median time. The number of cells analyzed per condition is indicated above (n = X). Mann–Whitney U-test was applied. ns means not significant; ****P <
< 0.0001. c Representative images of mitotic cells transfected with the same procedures as in (a). The cells were released from RO3306 into the medium with MG132 for 105
0.0001. c Representative images of mitotic cells transfected with the same procedures as in (a). The cells were released from RO3306 into the medium with MG132 for 105 min before fixation and stained with corresponding antibodies. Scale bar is 10
min before fixation and stained with corresponding antibodies. Scale bar is 10 µm. d Quantification of chromosome aligning defects from (c). 0 means all the chromosomes were aligned at the metaphase plate; 1–3 means there were 1 to 3 unaligned chromosomes outside of the metaphase plate. >3 means there were more than 3 chromosomes outside of the metaphase plate. More than 100 mitotic cells quantified in each condition. e Plot showing the time from NEBD to mitotic exit of the cells transfected with the same procedures as in (a) and (c). Low dose of nocodazole (100
µm. d Quantification of chromosome aligning defects from (c). 0 means all the chromosomes were aligned at the metaphase plate; 1–3 means there were 1 to 3 unaligned chromosomes outside of the metaphase plate. >3 means there were more than 3 chromosomes outside of the metaphase plate. More than 100 mitotic cells quantified in each condition. e Plot showing the time from NEBD to mitotic exit of the cells transfected with the same procedures as in (a) and (c). Low dose of nocodazole (100 ng/ml) was applied into the medium before live cell imaging conducted. Each circle represents the time spent in mitosis of a single cell. Red line indicates the median time. The number of cells analyzed per condition is indicated above (n = X). Mann–Whitney U-test was applied. **P
ng/ml) was applied into the medium before live cell imaging conducted. Each circle represents the time spent in mitosis of a single cell. Red line indicates the median time. The number of cells analyzed per condition is indicated above (n = X). Mann–Whitney U-test was applied. **P <
< 0.01; ****P
0.01; ****P <
< 0.0001.
0.0001.
The above results reveal an important role of BubR1 T600/T608 phosphorylation for mitotic progression.
BubR1 T600/T608 phosphorylation promotes BubR1-B56 interaction
It is well characterized that PP2A/B56 bound to BubR1 plays important roles for both chromosome alignment and checkpoint silencing which resembles the phenotype observed in BubR1 2A22–24. To test whether the observed mitotic defects caused by BubR1 2A is related to PP2A/B56, we first examined the kinetochore signals of the PP2A/B56 substrate Mps1 pT3320,28,32 by quantitative immunofluorescence since B56 on kinetochores is beyond detection by fluorescence microscopy22,23. The kinetochore signals of Mps1 pT33 was increased more than 30% in cells expressing BubR1 2A while reduced around 30% in cells with BubR1 2E compared with the cells with wild type BubR1 (Supplementary Fig. 1e, f) indicating PP2A/B56 could not be efficiently recruited to BubR1 2A. We then examined the kinetochore signals of BubR1 pT680 which is phosphorylated by Plk1 and required for efficient binding of PP2A/B56 to BubR1. Intriguingly, there was around 80% reduction of pT680 kinetochore signals in BubR1 2A cells compared with wild-type cells while in cells expressing BubR1 2E, the kinetochore signals of pT680 were partially rescued (Fig. 2a, b).
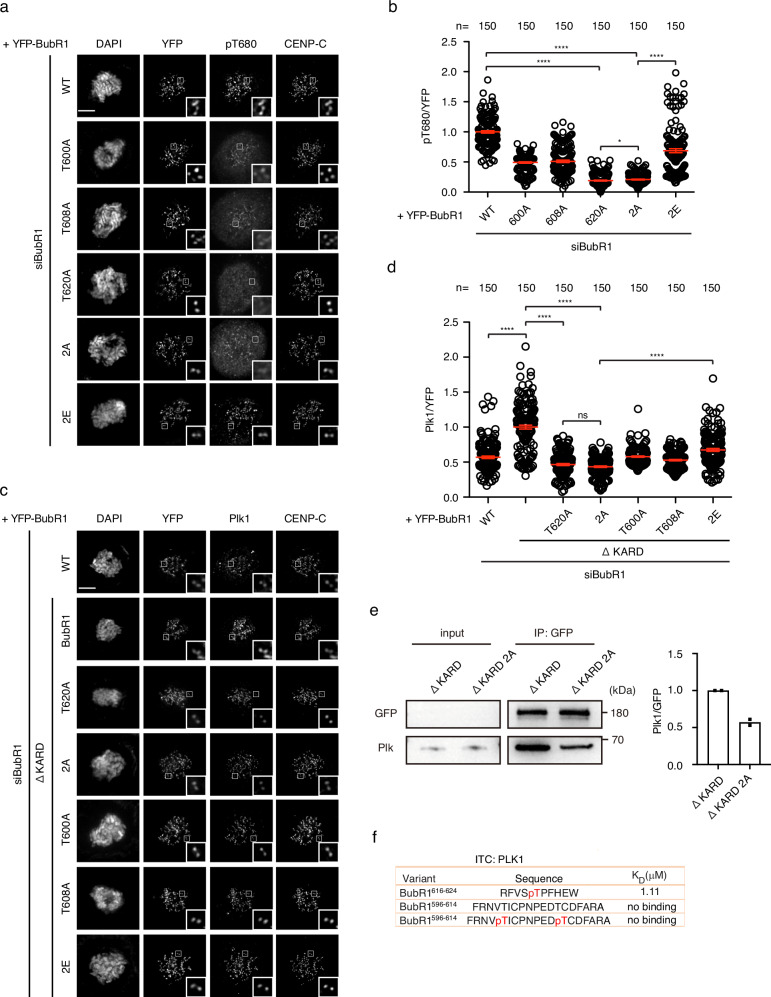
a Representative images of mitotic cells transfected with siRNA oligos against BubR1 and RNAi-resistant constructs expressing YFP-BubR1. The cells were released from RO3306 into the medium with nocodazole (200 ng/ml) for 45
ng/ml) for 45 min before fixation and staining with corresponding antibodies. Scale bar is 10
min before fixation and staining with corresponding antibodies. Scale bar is 10 µm. b Quantification of of kinetochore signals of BubR1 pT680 against YFP-BubR1 from (a). The values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for wild type BubR1 sample (WT) and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. *P
µm. b Quantification of of kinetochore signals of BubR1 pT680 against YFP-BubR1 from (a). The values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for wild type BubR1 sample (WT) and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. *P <
< 0.1; ****P
0.1; ****P <
< 0.0001. c Similar treatment as (a). d Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (c). The values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P
0.0001. c Similar treatment as (a). d Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (c). The values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P <
< 0.0001. e YFP-tagged BubR1 was expressed in HeLa cells and immunoprecipitated by GFP-trap beads. Plk1 were detected by western blot from the immunoprecipitate. Quantification of Plk1 intensity against YFP intensity was plotted on the right from two repeats. The column represents median value. The ratio from ΔKARD 2A was normalized against the ratio from ΔKARD which was set to 1. f ITC showing the binding between BubR1 peptides and recombinant Plk1.
0.0001. e YFP-tagged BubR1 was expressed in HeLa cells and immunoprecipitated by GFP-trap beads. Plk1 were detected by western blot from the immunoprecipitate. Quantification of Plk1 intensity against YFP intensity was plotted on the right from two repeats. The column represents median value. The ratio from ΔKARD 2A was normalized against the ratio from ΔKARD which was set to 1. f ITC showing the binding between BubR1 peptides and recombinant Plk1.
The above results clearly show that BubR1 pT600/pT608 is required for efficient phosphorylation of BubR1 T680 and its interaction with PP2A/B56 as a result.
Phosphorylation of T600/T608 is required for stable interaction of Plk1 with BubR1
How could the phosphorylation on BubR1 T600/T608 enhances the phosphorylation of BubR1 T680? Since T600 and T608 are close to the cannonical Plk1 binding motif (619-SpTP-621), we reasoned that the double phosphorylation might be involved in the BubR1-Plk1 interaction. We decided to test whether BubR1 pT600/pT608 is required for kinetochore localization of Plk1. Based on the fact that BubR1 has only minor contribution to kinetochore recruited Plk1, it is difficult to precisely measure the signal variations of BubR1-associated Plk1 within such a small window. On the other hand, BubR1 phosphorylation is negatively regulated by PP2A/B5629–31. Thus, it is possible to increase the kinetochore signals of BubR1-associated Plk1 by disrupting BubR1 binding to PP2A/B56 which allows more accurate quantification of the Plk1 signals. We first examined the kinetochore localization of Plk1 in BubR1ΔKARD background which is not able to bind B56 due to two alanine mutations within the core binding motif (LxxIxE into AxxAxE). Indeed, loss of PP2A/B56 binding increased ~2-fold the kinetochore levels of Plk1 (Fig. 2c, d). In this background, we examined the kinetochore localization of Plk1 in cells expressing BubR1 2A and found the Plk1 signals was significantly reduced to similar extent as T620A mutant while single alanine mutation T600A or T608A caused mild Plk1 reduction (Fig. 2c, d). Similar result was achieved by immunoprecipitating YFP-tagged BubR1 in cells expressing corresponding constructs. BubR1-associated Plk1 was strongly reduced in YFP-BubR1 2A sample compared with wild type BubR1 sample in ΔKARD background (Fig. 2e). To investigate if the phosphorylation of T620 was affected by the T600A/T608A mutation we analyzed the phosphorylation pattern of purified YFP-BubR1 and BubR1 2A by quantitative mass spectrometry. We did not directly identify the phosphorylated pT620 peptide but measured the unphosphorylated FVSTPFHEIMSLK peptide levels which was strongly reduced in BubR1 2A purifications suggesting increased T620 phosphorylation (Supplementary Fig. 1g). We also detected pS670 which was similar in both BubR1 WT and 2A (Supplementary Fig. 1h).
We then tested if double phosphorylation of T600/608 creates a second Plk1 binding site besides the canonical one. We synthesized peptide BubR1596-614 with T600/T608 phosphorylated. As a control, we also synthesized a peptide BubR1616-624 with T620 phosphorylated. Using isothermal titration calorimetry (ITC), we measured the binding affinity between recombinant Plk1 PBD and the peptides. The assay clearly showed binding of BuBR1616-624, but not of BubR1596-614 to the recombinant protein indicating no extra binding site generated on BubR1 by the double phosphorylation (Fig. 2f; Supplementary Fig. 2a–c).
Both the T600 and T608 phosphorylation sites fit the consensus recognized by Plk1, [D/N/E/Y]-X-S/T-[F/Φ; no P]-[Φ/X], where Φ is hydrophobic amino acid and X indicates any amino acid17. To test whether these two phosphorylation relies on Plk1 kinase activity, we generated a BubR1 pT600 phosphor antibody and characterized it by quantitative immunofluorescence. This antibody strongly decorated kinetochores of the cells treated with nocodazole and this decoration was fully abolished after lambda phosphatase treatment (Supplementary Fig. 3a). In cells depleted of BubR1 or treated with Plk1 inhibitor BI2536, the kinetochore signals recognized by the antibody were largely reduced (Supplementary Fig. 3b–d). There was around 30% of the kinetochore signals remained in both cases indicating this antibody cross-reacts with other phosphorylation modification on kinetochores. We further characterized this antibody in cells complemented with RNAi-resistant constructs expressing YFP-tagged wild type or mutant BubR1 with endogenous BubR1 depleted by RNAi. To eliminate the negative regulation by PP2A/B56, all these BubR1 proteins harbor defective B56 binding motif. The quantification showed a significant reduction of pT600 kinetochore signals in either T600A or T620A mutant to the same extent compared with BubR1ΔKARD indicating the phosphorylation of T600 requires the phosphorylation of T620 (Supplementary Fig. 3e). Using this antibody, we also examined the occurance of BubR1 T600 phosphorylation at distinct phases of mitosis. The phosphorylations happens as early as in prophase when BubR1 starts localizing onto kinetochores. The signals peak at prometaphase and declines continuously till anaphase (Supplementary Fig. 3f).
These data suggest that BubR1 binds Plk1 through the cannonical STP motif after T620 gets phosphorylated by Cdk1. Plk1 in turn, mediates the phosphorylation of T600 and T608 to enhance its binding to BubR1. Once Plk1 stably binds BubR1, it further phosphorylates amino acids S676 and T680 to promote the binding of PP2A/B56 with BubR1. Thus, kinetochore localized PP2A/B56 ensures accurate chromosome segregation.
The double phosphorylation can be bypassed by enhancing PBD binding or B56 binding to support accurate mitotic progression
According to the above model, we predict that enhancing the binding of Plk1 with BubR1 may bypass the requirement of the double phosphorylation on T600 and T608 for accurate mitosis. To test this, we engineered BubR1 2A mutant by replacing the BubR1 PBD BM (RFVSTPFHE) with WDR47 PBD BM (IHTSTPRNP) whose affinity towards Plk1 PBD is almost 25 times stronger (BubR1 2A +
+ WDR47 hereafter) (Fig. 3a; Supplementary Figs. 2d, e and 4a). We first examined the ability of the engineered protein BubR1 2A
WDR47 hereafter) (Fig. 3a; Supplementary Figs. 2d, e and 4a). We first examined the ability of the engineered protein BubR1 2A +
+ WDR47 to recruit Plk1 onto kinetochores by quantitative immunofluorescence. As expected, BubR1 2A
WDR47 to recruit Plk1 onto kinetochores by quantitative immunofluorescence. As expected, BubR1 2A +
+ WDR47 efficiently recruited Plk1 to the same extent as BubR1 itself (Fig. 3b, c). We then tested the ability of the engineered protein to support accurate mitotic progression by live cell imaging. The results showed the mitotic delay caused by mutating T600/T608 to alanine (80
WDR47 efficiently recruited Plk1 to the same extent as BubR1 itself (Fig. 3b, c). We then tested the ability of the engineered protein to support accurate mitotic progression by live cell imaging. The results showed the mitotic delay caused by mutating T600/T608 to alanine (80 min) was fully restored to normal (40
min) was fully restored to normal (40 min) in the presence of WDR47 PBD BM (Fig. 3d, e). Live cell imaging testing the SAC activity also showed efficient silencing of the SAC in cells complemented with BubR1 2A
min) in the presence of WDR47 PBD BM (Fig. 3d, e). Live cell imaging testing the SAC activity also showed efficient silencing of the SAC in cells complemented with BubR1 2A +
+ WDR47 (Supplementary Fig. 4b).
WDR47 (Supplementary Fig. 4b).
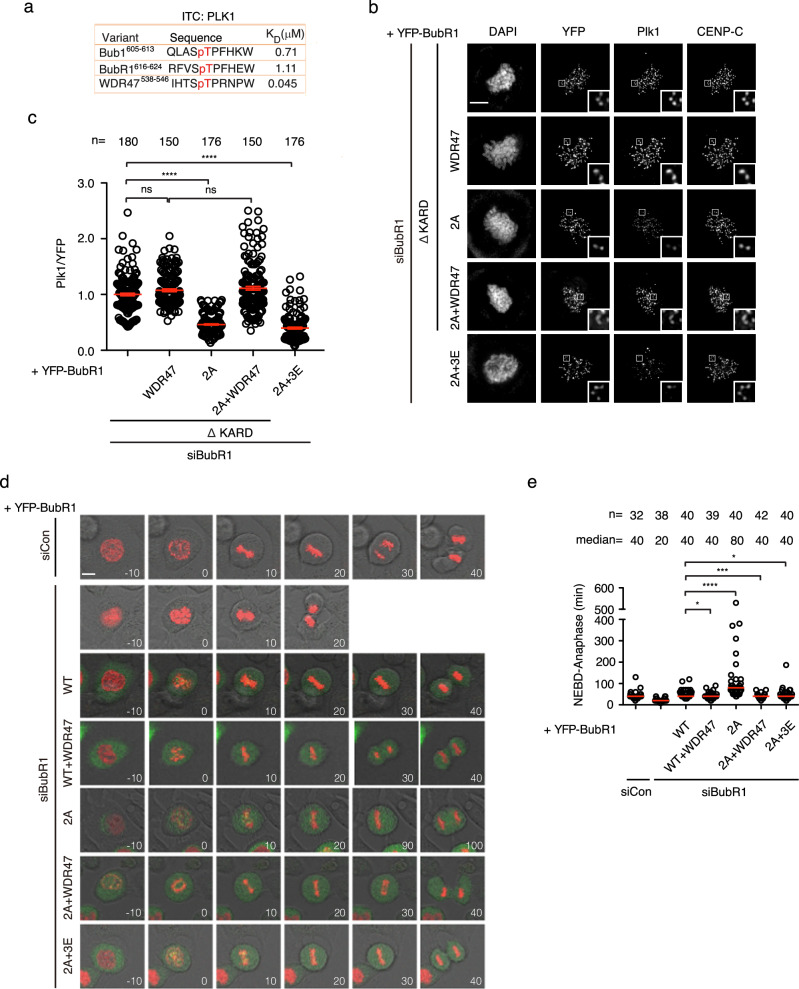
a KD measured by ITC of recombinant Plk1 with phospho-modified peptides. b Representative images of mitotic cells transfected with siRNA oligos against BubR1 and RNAi-resistant constructs expressing YFP-BubR1. The cells were released from RO3306 into the medium with nocodazole (200 ng/ml) for 45
ng/ml) for 45 min before fixation and staining with corresponding antibodies. Scale bar is 10
min before fixation and staining with corresponding antibodies. Scale bar is 10 µm. c Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (b). Mean values of at least 150 kintochores from at least 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P
µm. c Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (b). Mean values of at least 150 kintochores from at least 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P <
< 0.0001. d Representative stills of unperturbed mitosis recorded by live cell imaging. The cells were transfected with siRNA oligos against luciferase as control or siRNA oligos against BubR1 with RNAi-resistant constructs expressing YFP-BubR1. mCherry-H3 expressing plasmid was co-transfected for chromosome indication. Scale bar is 10
0.0001. d Representative stills of unperturbed mitosis recorded by live cell imaging. The cells were transfected with siRNA oligos against luciferase as control or siRNA oligos against BubR1 with RNAi-resistant constructs expressing YFP-BubR1. mCherry-H3 expressing plasmid was co-transfected for chromosome indication. Scale bar is 10 µm. e Quantification of the mitotic time from NEBD to anaphase from (d). Each circle represents the time from NEBD to anaphase of a single cell. Red line indicates the median time. The number of cells analyzed per condition is indicated above (n = X). Mann–Whitney U-test was applied. *P
µm. e Quantification of the mitotic time from NEBD to anaphase from (d). Each circle represents the time from NEBD to anaphase of a single cell. Red line indicates the median time. The number of cells analyzed per condition is indicated above (n = X). Mann–Whitney U-test was applied. *P <
< 0.1; ***P
0.1; ***P <
< 0.001; ****P
0.001; ****P <
< 0.0001.
0.0001.
It is known that BubR1-bound Plk1 promotes BubR1-PP2A/B56 interaction. We wondered whether enhancing the binding of PP2A/B56 with BubR1 could bypass the requirement of the pT600/pT608. To test the idea, we engineered a BubR1 2A mutant with D675E/S676E/R677E (BubR1 2A +
+ 3E hereafter) which efficiently binds B56 as described before33 (Supplementary Fig. 4a). Quantitative immunofluorescence showed the engineered protein could not efficiently recruit Plk1 onto kinetochores similar as BubR1 2A mutant (Fig. 3b, c). However, live cell imaging showed BubR1 2A
3E hereafter) which efficiently binds B56 as described before33 (Supplementary Fig. 4a). Quantitative immunofluorescence showed the engineered protein could not efficiently recruit Plk1 onto kinetochores similar as BubR1 2A mutant (Fig. 3b, c). However, live cell imaging showed BubR1 2A +
+ 3E was as efficient as wild type BubR1 in promoting accurate mitotic progression as well as SAC silencing (Fig. 3d, e; Supplementary Fig. 4b).
3E was as efficient as wild type BubR1 in promoting accurate mitotic progression as well as SAC silencing (Fig. 3d, e; Supplementary Fig. 4b).
The above data further demonstrates BubR1-Plk1 interaction requires both PBD BM and the double phosphorylation in front. However, for BubR1 with optimal PBD BM, the phosphorylation is not necessary anymore for accurate mitosis. The results also show the BubR1-Plk1 interaction is largely dispensible for accurate mitosis in the presence of enhanced binding between BubR1 and PP2A/B56.
Efficient Bub1-Plk1 binding also requires extra phosphorylation besides the PBD BM
Bub1, the paralogue of BubR1, is one of the two main Plk1 receptors on outer kinetochores. Whether Bub1 shares similar principle for Plk1 interaction as BubR1 is not clear. At least five amino acids (T589, S593, S596, T601, and S602) N terminal to the canonical Plk1 binding site (608-SpTP-610) of Bub1 are phosphorylated (phosphosite.org). Two of them, T589 and T601 fit the consensus motif for Plk1. Another two S593 and S596 are likely phosphorylated by Cdk1. We mutated all the four residues into alanine (Bub1 4A) or into glutamic acid (Bub1 4E) to mimic the unphosphorylated and phosphorylated Bub1. To avoid the signal interference from BubR1-associated Plk1, the BubR1 binding domain (amino acids 266-311)34 was removed from all the above mutants. We measured the kinetochore signals of Plk1 in HeLa cells depleted of endogenous Bub1 and complemented RNAi-resistant YFP-tagged Bub1 constructs. Bub1 4A reduced the kinetochore Plk1 level to less than 50% while Bub1 4E maintained Plk1 around 80% of Bub1ΔBubR1 (Fig. 4a, b).
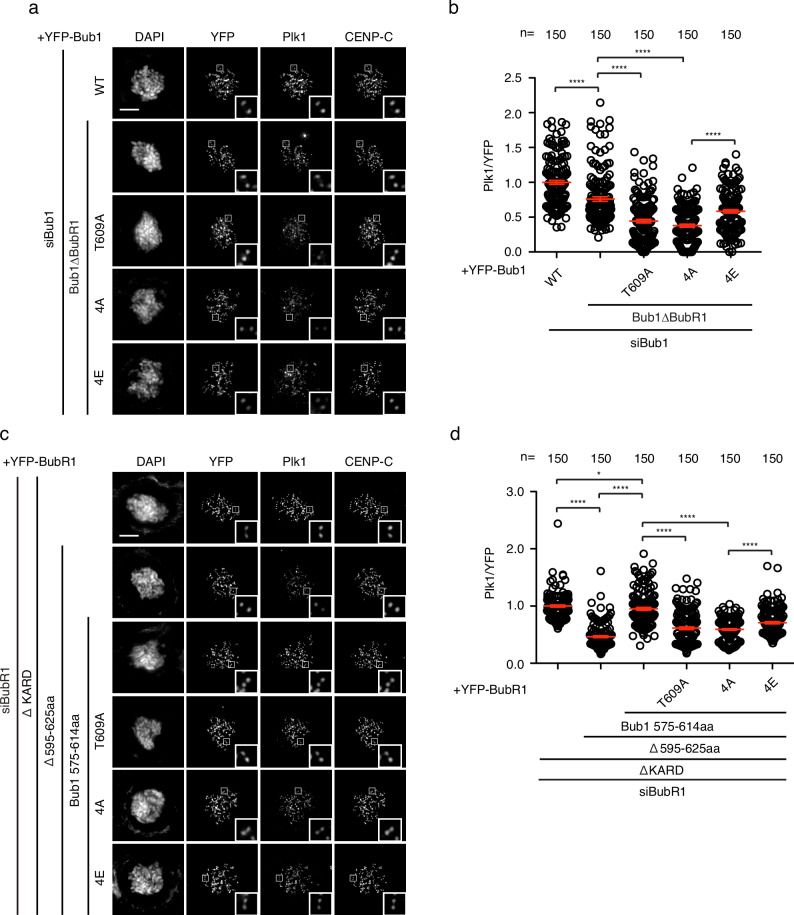
a Representative images of mitotic cells transfected with siRNA oligos against Bub1 and RNAi-resistant constructs expressing YFP-Bub1. The cells were released from RO3306 into the medium with nocodazole (200 ng/ml) for 45
ng/ml) for 45 min before fixation and staining with corresponding antibodies. Scale bar is 10
min before fixation and staining with corresponding antibodies. Scale bar is 10 µm. b Quantification of kinetochore signals of Plk1 against YFP-Bub1 from (a). Mean values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for wild type Bub1 (WT) sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ****P
µm. b Quantification of kinetochore signals of Plk1 against YFP-Bub1 from (a). Mean values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for wild type Bub1 (WT) sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ****P <
< 0.0001. c Representative images of mitotic cells transfected with siRNA oligos against BubR1 and RNAi-resistant constructs expressing YFP-BubR1. The cells were released from RO3306 into the medium with nocodazole (200
0.0001. c Representative images of mitotic cells transfected with siRNA oligos against BubR1 and RNAi-resistant constructs expressing YFP-BubR1. The cells were released from RO3306 into the medium with nocodazole (200 ng/ml) for 45
ng/ml) for 45 min before fixation and staining with corresponding antibodies. Scale bar is 10
min before fixation and staining with corresponding antibodies. Scale bar is 10 µm. d Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (c). Mean values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. *P
µm. d Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (c). Mean values of 150 kintochores from 10 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. *P <
< 0.1; ****P
0.1; ****P <
< 0.0001.
0.0001.
Since the Bub1 Plk1 binding module (the region containing the phosphorylation sites and PBD BM) could efficiently recruit Plk1 to kinetochores, we wonder whether this module could replace the BubR1 Plk1 binding module for efficient Plk1 binding. We grafted Bub1 Plk1 binding module (575–614aa) to the corresponding region on BubR1 (595–625aa) and measured the kinetochore signals of Plk1 in HeLa cells expressing the engineered protein after depletion of endogenous BubR1. The results showed BubR1 containing Bub1 Plk1 binding module but not the 4A mutant could recruit Plk1 as efficiently as BubR1 itself (Fig. 4c, d).
These data suggest self-priming may be a general principle for Plk1 interaction with PBD BM-containing proteins.
Two neighboring PBD BMs on BubR1 cannot efficiently recruit Plk1
If a single BubR1 PBD BM does not efficiently recruit Plk1, can tandem PBD BMs do this as revealed recently on CENP-U17? We generated a BubR1 mutant with two copies of the PBD BMs separated by 18 amino acids (2 x STP) and tested its ability to recruit Plk1 (Fig. 5a). Surprisingly, the engineered protein recruited Plk1 as low as 2A mutant (Fig. 5b, c). We reasoned probably one of the two PBD BM needs to have a high affinity towards Plk1 molecule to initiate the binding, then through dimerziation, it stabilizes the interaction of the second Plk1 with a PBD BM with low affinity. We then grafted WDR47 PBD BM in front of BubR1 PBD BM (WDR47 +
+ STP) and tested again the Plk1 recruitment. As a control, another BubR1 construct containing two WDR47 PBD BMs (2 x WDR47) was also generated (Fig. 5a). Quantitative immunofluorescence showed both engineered proteins with two PBD BMs recruited much less Plk1 onto kinetochores than the one with a single WDR47 PBD BM (2A
STP) and tested again the Plk1 recruitment. As a control, another BubR1 construct containing two WDR47 PBD BMs (2 x WDR47) was also generated (Fig. 5a). Quantitative immunofluorescence showed both engineered proteins with two PBD BMs recruited much less Plk1 onto kinetochores than the one with a single WDR47 PBD BM (2A +
+ WDR47) indicating an interference between the two PBD BMs in such a short distance (Fig. 5b, c).
WDR47) indicating an interference between the two PBD BMs in such a short distance (Fig. 5b, c).
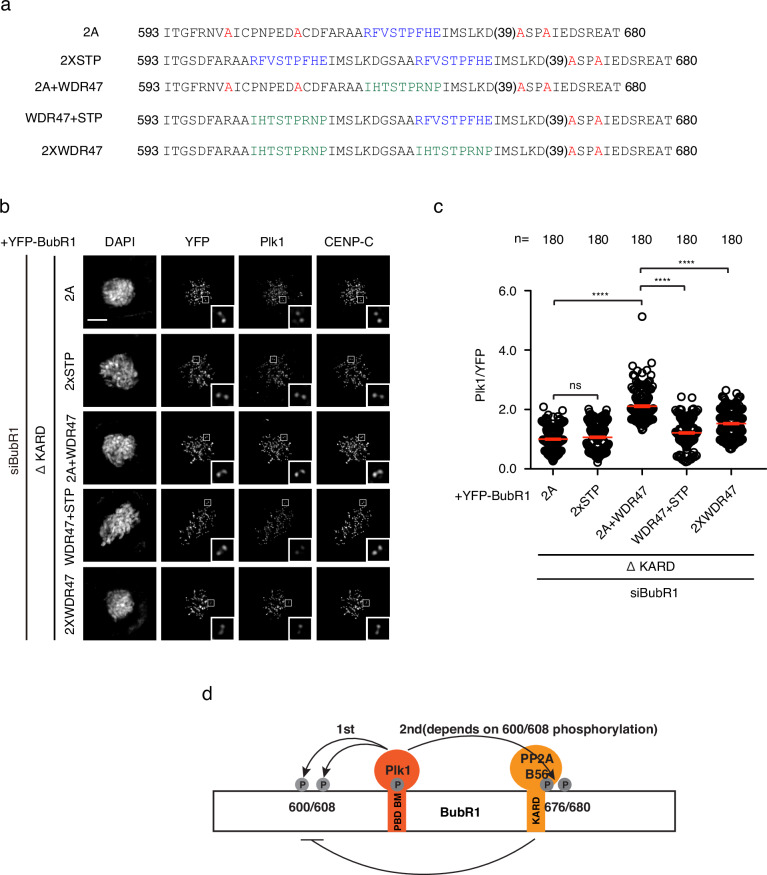
a Amino acid sequences of the engineered proteins with one PBD BM or two PBD BMs. b Representative images of mitotic cells transfected with siRNA oligos against BubR1 and RNAi-resistant constructs expressing YFP-BubR1. The cells were released from RO3306 into the medium with nocodazole (200 ng/ml) for 45
ng/ml) for 45 min before fixation and staining with corresponding antibodies. Scale bar is 10
min before fixation and staining with corresponding antibodies. Scale bar is 10 µm. c Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (b). Mean values of 180 kintochores from 12 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 T600A/T608A ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P
µm. c Quantification of kinetochore signals of Plk1 against YFP-BubR1 from (b). Mean values of 180 kintochores from 12 cells for each condition were presented. The red line indicates the mean value which was set to 1 for BubR1 T600A/T608A ΔKARD sample and the rest was normalized to it. Bar is standard error of the mean. Mann–Whitney U-test was applied. ns means not significant; ****P <
< 0.0001. d Working model showing self-priming of Plk1 on BubR1 enhances BubR1-Plk1 interaction as well as BubR1-PP2A/B56 as a result.
0.0001. d Working model showing self-priming of Plk1 on BubR1 enhances BubR1-Plk1 interaction as well as BubR1-PP2A/B56 as a result.
In the end, we propose a working model based on our observations. Cdk1 phosphorylates BubR1 T620 to generate the docking site for Plk1. In turn, Plk1 transiently bound to pT620 phosphorylates two BubR1 residues T600 and T608 in front which enhances the binding between Plk1 and BubR1. Now, stably bound Plk1 on BubR1 could efficiently phosphorylate BubR1 on T676 and T680 to promote the interaction between BubR1 and PP2A/B56 which is critical for the proper attachment between kinetochores and microtubules as well as the timely silencing of SAC (Fig. 5d).
Discussion
Numerous proteins containing the general PBD BM (S-[pS/pT]-[P/X]) have been reported to interact with Plk1 and recruit it to subcellular apparatus for proper functioning in mitosis. However, a recent study indicates that in the case of CENP-U, the PBD BM is not able to efficiently bind Plk117. The phosphorylation of neighboring amino acids in front of the PBD BM generates a second Plk1 binding site and the two Plk1 molecules dimerize to form stable interaction with CENP-U17. In this study, we found the binding between Plk1 and BubR1 as well as Plk1 and Bub1 also relies on the phosphorylation of residues in front of the PBD BM.
Does the double phosphorylation promote the interaction between Plk1 and BubR1 via dimerization of two Plk1 molecules like what happens on CENP-U? Due to the following reasons, we do not prefer the dimerization mechanism for BubR1-Plk1 interaction. First, neither pT600 nor pT608 fits Plk1 PBD BM consensus. Second, there are three phosphorylation sites (T600, T608, T620) in close approximity on BubR1 involved in Plk1 binding which makes the recruitment of another two Plk1 molecules very difficult due to spatial hindrance. Third, the peptide binding assay could not detect any interaction between the pT600/pT608 peptide with the recombinant Plk1. Fourth, engineering two PBD BMs on BubR1 did not recruit additional Plk1 onto kinetochores than a single PBD BM. So far the mechanism of the double phosphorylation on T600 and T608 enhancing BubR1-Plk1 interaction is not clear yet. One possibility is that double phosphorylation causes BubR1 conformation changes which exposes pT620 more efficiently for Plk1 binding. Future work is needed to fully understand the enhancement.
The binding affinity of Bub1 and BubR1 towards Plk1 is very similar and both have amino acids phosphorylated in front of the PBD BM. Then, what makes Bub1 but not BubR1 as the main kinetochore receptor for Plk115–17? From this and previous studies20,29–31, we belive that direct binding of PP2A/B56 to BubR1 keeps the phosphorylation and the associated Plk1 at low levels. On the other hand, the putative B56 binding site on Bub1 loses the ability to bind B56 allowing higher phosphorylation and more Plk1 associated. The presence of BubR1-PP2A/B56 interaction may also explain the fact that the phosphorylation of T680 requires more stably BubR1-Plk1 binding than the phosphorylation of T600 and T608 as the latter are closer to the kinase but further away from the phosphatase while T680 is vice versa.
Based on the above results, we propose that efficient Plk1 binding with the PBD BM not only depends on the binding affinity between them but also the phosphorylation of the amino acids in front of PBD BM. Plk1 transiently binds the canonical PBD BM and phosphorylates amino acids in front of PBD BM to enhance the interaction. Two distinct mechanisms may exist to enhance the interaction between Plk1 and interactors. One involves two Plk1 molecules dimerization on two neighboring PBD BMs while the other may rely on protein conformation changes after being phosphorylated at the amino acids in front of PBD BM. This working model applies to the interaction of Plk1 with two major and one marginal kinetochore receptors. Does this model also apply to Plk1 recruitment to other subcellular structures awaits for futher investigation.
Methods
Cell culture and transfection
HeLa cells were cultivated in DMEM medium (Gibco) supplemented with 10% FBS and Pen/Strep. Double thymidine block was applied to cells for synchronization. siRNA oligo with RNAi-resistant constructs were transfected into cells in-between the two thymidine block using Lipofectamine 2000 (Thermo Fisher Scientific). For live cell imaging, the cells were released from the second thymidine block and recorded by microscopy 6 h later. For immunofluorescence assay, cells were released from the second thymidine block into RO3306 (5
h later. For immunofluorescence assay, cells were released from the second thymidine block into RO3306 (5 μM, Selleck) containing DMEM medium. 12
μM, Selleck) containing DMEM medium. 12 h later, the cells were released from RO3306 into DMEM mediuim with nocodazole (200
h later, the cells were released from RO3306 into DMEM mediuim with nocodazole (200 ng/ml, Selleck) for 45
ng/ml, Selleck) for 45 min or MG132 (10
min or MG132 (10 μM, Selleck) for 105
μM, Selleck) for 105 min before fixation. RNAi oligos targeting Bub1 (5’GAGUGAUCACGAUUUCUAAdTdT3’), BubR1 (5’GAUGGUGAAUUGUGGAAUAdTdT3’), and luciferase (5’ CGUACGCGGAAUACUUCGAdTdT3’) were synthesized from Genepharma.
min before fixation. RNAi oligos targeting Bub1 (5’GAGUGAUCACGAUUUCUAAdTdT3’), BubR1 (5’GAUGGUGAAUUGUGGAAUAdTdT3’), and luciferase (5’ CGUACGCGGAAUACUUCGAdTdT3’) were synthesized from Genepharma.
Molecular cloning
The constructs used in this study were generated by standard cloning method and mutagenesis was conducted by mutation PCR. Briefly, wild-type Bub1 or BubR1 was cloned into pcDNA5/FRT/TO N-YFP vector by KpnI and NotI. For BubR1 engineered protein containing distinct PBD BM, BamHI site was first introduced by mutation PCR to replace the DNA sequence at designed positions. Afterward, the DNA encoding distinct PBD BM was amplified and inserted by BamHI restrictive enzyme and T4 DNA ligase. Gene amplification or mutation PCR was performed with KOD DNA polymerase (Toyobo). All the restriction enzymes and T4 DNA ligase were purchased from Thermo Fisher Scientific.
Live cell imaging
HeLa cells were first seeded in 6-well plate and synchronized with thymidine. RNA interference and plasmid transfection was performed as described in the above section. 900 ng of RNAi-resistant YFP-BubR1 or YFP-Bub1 construct and 30
ng of RNAi-resistant YFP-BubR1 or YFP-Bub1 construct and 30 ng of mCherry-H3 were co-transfected together with 50
ng of mCherry-H3 were co-transfected together with 50 nM of RNAi oligos in each well for undisburbed mitosis imaging. For SAC strength assays, only YFP-BubR1 and RNAi oligos were co-transfected. 24
nM of RNAi oligos in each well for undisburbed mitosis imaging. For SAC strength assays, only YFP-BubR1 and RNAi oligos were co-transfected. 24 h after transfection, the cells were re-seeded into an 8-well chamber slide (Ibidi) during the second thymidine block. 18
h after transfection, the cells were re-seeded into an 8-well chamber slide (Ibidi) during the second thymidine block. 18 h later, cells were released from the second thymidine block and recorded by live cell imaging 6
h later, cells were released from the second thymidine block and recorded by live cell imaging 6 h later. Leibovitz’s L-15 medium (Thermo Fisher Scientific) supplemented with 10% FBS was applied into each chamber before the slide was mounted onto microscopy. For SAC assays, nocodazole (30
h later. Leibovitz’s L-15 medium (Thermo Fisher Scientific) supplemented with 10% FBS was applied into each chamber before the slide was mounted onto microscopy. For SAC assays, nocodazole (30 ng/ml or 100
ng/ml or 100 ng/ml as specified in figure legends, Selleck) was added into the L15 medium. Nikon A1HD25 imaging system (Nikon) was used for live cell imaging. DIC, YFP, and RFP signals were collected every 10
ng/ml as specified in figure legends, Selleck) was added into the L15 medium. Nikon A1HD25 imaging system (Nikon) was used for live cell imaging. DIC, YFP, and RFP signals were collected every 10 min for a total of 18
min for a total of 18 h. NIS-Elements AR Analysis (Nikon) was used for data analysis. At least two repeats for each assay were performed with more than 30 cells in each condition quantified.
h. NIS-Elements AR Analysis (Nikon) was used for data analysis. At least two repeats for each assay were performed with more than 30 cells in each condition quantified.
Immunoprecipitation and western blot
HeLa cells were synchronized by double thymidine and transfected with YFP-BubR1 constructs after the first thymidine arrest. 36 h later, the cells were released from the second thymidine arrest into medium containing nocodazole (200
h later, the cells were released from the second thymidine arrest into medium containing nocodazole (200 ng/ml) for an additional 12
ng/ml) for an additional 12 h. Mitotic cells were collected by shake off and lysed in buffer containing 10
h. Mitotic cells were collected by shake off and lysed in buffer containing 10 mM Tris pH 7.5, 150
mM Tris pH 7.5, 150 mM NaCl, 0.5
mM NaCl, 0.5 mM EDTA, and 0.5% NP40. After centrifugation at 16,000
mM EDTA, and 0.5% NP40. After centrifugation at 16,000 ×
× g for 15
g for 15 min, the supernatant was incubated with GFP-Trap agarose beads (Proteintech) and shaken at 1100
min, the supernatant was incubated with GFP-Trap agarose beads (Proteintech) and shaken at 1100 rpm for 120
rpm for 120 min at 4
min at 4 °C on thermomixer (Eppendorf). After three washes, the bound protein was eluted in 2 x SDS sample buffer and separated by SDS-PAGE. Target proteins were detected by western blot. Antibodies used in this study include YFP antibody (homemade in JN lab, 1:2000), Plk1 (Santa Cruz sc-17783, 1:500).
°C on thermomixer (Eppendorf). After three washes, the bound protein was eluted in 2 x SDS sample buffer and separated by SDS-PAGE. Target proteins were detected by western blot. Antibodies used in this study include YFP antibody (homemade in JN lab, 1:2000), Plk1 (Santa Cruz sc-17783, 1:500).
Immunofluorescence
Cells growing on coverslips in 6-well plate were treated as described in the above. Cells were fixed by 4% paraformaldehyde in PHEM buffer (60 mM PIPES, 25
mM PIPES, 25 mM HEPES, pH 6.9, 10
mM HEPES, pH 6.9, 10 mM EGTA, and 4
mM EGTA, and 4 mM MgSO4) at room temperature for 20
mM MgSO4) at room temperature for 20 min. The fixed cells were permeabilized by 0.5% Triton X-100 in PHEM for 10
min. The fixed cells were permeabilized by 0.5% Triton X-100 in PHEM for 10 min at room temperature before being stained by corresponding antibodies. The antibodies used in this study include Bub1 (Abcam, ab54893, 1:200), BubR1 (home made in JN lab, 1:200), CENP-C (MBL, PD030, 1:800), BubR1 pT680 (Abcam200061, 1:500), BubR1 pT600 (Abclonal, 1:200), α tubulin (Sigma, F2168, 1:400), Mad2 (home made in JN lab, 1:200), Mps1 pT33 (home made in JN lab, 1:200) and Plk1 (Santa Cruz, sc-17783, 1:100). The BubR1 phospho antibody was raised in rabbit against C-RNV[pT]ISPNPE-amide peptide coupled to carrier and the antibody was affinity purified (Abclonal). To avoid interferring the coupling, the original C at 602 was replaced by S. Fluorescent secondary antibodies are Alexa Fluor Dyes (Invitrogen, 1:1000) except GFP booster Alexa Fluor 488 was used for YFP detection (Proteintech, gb2AF488-10, 1:500). Z-stacks with 200
min at room temperature before being stained by corresponding antibodies. The antibodies used in this study include Bub1 (Abcam, ab54893, 1:200), BubR1 (home made in JN lab, 1:200), CENP-C (MBL, PD030, 1:800), BubR1 pT680 (Abcam200061, 1:500), BubR1 pT600 (Abclonal, 1:200), α tubulin (Sigma, F2168, 1:400), Mad2 (home made in JN lab, 1:200), Mps1 pT33 (home made in JN lab, 1:200) and Plk1 (Santa Cruz, sc-17783, 1:100). The BubR1 phospho antibody was raised in rabbit against C-RNV[pT]ISPNPE-amide peptide coupled to carrier and the antibody was affinity purified (Abclonal). To avoid interferring the coupling, the original C at 602 was replaced by S. Fluorescent secondary antibodies are Alexa Fluor Dyes (Invitrogen, 1:1000) except GFP booster Alexa Fluor 488 was used for YFP detection (Proteintech, gb2AF488-10, 1:500). Z-stacks with 200 nm interval were taken by DMi8 fluorescent microscopy (Leica) using a 100× oil objective followed by deconvolution with Thunder cleaning function. Signal quantification was performed by drawing a circle around each kinetochore marked by CENP-C staining. The three continuous peak values within the circle on the interested channel were averaged and subtracted of the background values from a neighboring circle. Two repeats were performed for each immunofluorescence assay with at least 150 kinetochores from 10 cells in one repeat quantified.
nm interval were taken by DMi8 fluorescent microscopy (Leica) using a 100× oil objective followed by deconvolution with Thunder cleaning function. Signal quantification was performed by drawing a circle around each kinetochore marked by CENP-C staining. The three continuous peak values within the circle on the interested channel were averaged and subtracted of the background values from a neighboring circle. Two repeats were performed for each immunofluorescence assay with at least 150 kinetochores from 10 cells in one repeat quantified.
Recombinant protein production
Plk1 367-603aa was expressed in E. coli at 18 °C overnight. Cells were collected by centrifugation and were resuspended in lysis buffer (50
°C overnight. Cells were collected by centrifugation and were resuspended in lysis buffer (50 mM NaP pH 7.5, 300
mM NaP pH 7.5, 300 mM NaCl, 10
mM NaCl, 10 mM imidazole, 10% glycerol, 0.5
mM imidazole, 10% glycerol, 0.5 mM TCEP, protease inhibitors). After sonication, cell lysate was centrifuged at 17,000
mM TCEP, protease inhibitors). After sonication, cell lysate was centrifuged at 17,000 ×
× g at 4
g at 4 °C for 30
°C for 30 min to get rid of cell debris. The lysate was loaded onto a 5
min to get rid of cell debris. The lysate was loaded onto a 5 ml HiTrap column and washed with IMAC binding buffer (50
ml HiTrap column and washed with IMAC binding buffer (50 mM NaP pH 7.5, 300
mM NaP pH 7.5, 300 mM NaCl, 10
mM NaCl, 10 mM imidazole, 10% glycerol, 0.5
mM imidazole, 10% glycerol, 0.5 mM TCEP) and eluted with a gradient of IMAC elution buffer (50
mM TCEP) and eluted with a gradient of IMAC elution buffer (50 mM NaP 7.5, 300
mM NaP 7.5, 300 mM NaCl, 500
mM NaCl, 500 mM imidazole, 10% glycerol, 0.5
mM imidazole, 10% glycerol, 0.5 mM TCEP). The tag was removed by TEV cleavage and following dialysis the tag and TEV was removed by running the sample on a 5
mM TCEP). The tag was removed by TEV cleavage and following dialysis the tag and TEV was removed by running the sample on a 5 ml HiTrap column. The flow through was collected. The protein was concentrated and run on a Superdex 200 column equilibrated with GF buffer (50
ml HiTrap column. The flow through was collected. The protein was concentrated and run on a Superdex 200 column equilibrated with GF buffer (50 mM NaP pH 7.5, 150
mM NaP pH 7.5, 150 mM NaCl, 10% glycerol, 0.5
mM NaCl, 10% glycerol, 0.5 mM TCEP). The identity of the protein was confirmed by MS analysis.
mM TCEP). The identity of the protein was confirmed by MS analysis.
Peptide binding assay
Peptides were ordered from Peptide 2.0 Inc (Chantilly, VA, USA). The purity was 95–98% as determined by high performance liquid chromatography (HPLC) and by mass spectrometry. The protein and the peptides were extensively dialyzed against 50 mM NaP, 150
mM NaP, 150 mM NaCl, 0.5
mM NaCl, 0.5 mM TCEP, pH 7.5. All ITC experiments were performed on Auto-iTC200 (Microcal, Malvern Instruments Ltd.) at 25
mM TCEP, pH 7.5. All ITC experiments were performed on Auto-iTC200 (Microcal, Malvern Instruments Ltd.) at 25 °C. The concentrations of peptide and protein were determined by measuring the absorbance at 280
°C. The concentrations of peptide and protein were determined by measuring the absorbance at 280 nm using a spectrometer and applying values for the extinction coefficients computed for the corresponding sequences by the ProtParam program (http://web.expasy.org/protparam/). The peptides at ~450
nm using a spectrometer and applying values for the extinction coefficients computed for the corresponding sequences by the ProtParam program (http://web.expasy.org/protparam/). The peptides at ~450 μM or 120
μM or 120 μM (for submicromolar affinities) were loaded into the syringe and titrated into the calorimetric cell containing the recombinant Plk1, respectively, at 35
μM (for submicromolar affinities) were loaded into the syringe and titrated into the calorimetric cell containing the recombinant Plk1, respectively, at 35 μM or 10
μM or 10 μM. The titration sequence consisted of a single 0.4
μM. The titration sequence consisted of a single 0.4 μl injection followed by 19 injections, 2
μl injection followed by 19 injections, 2 μl each, with 150
μl each, with 150 s spacing between injections to ensure that the thermal power returns to the baseline before the next injection. The stirring speed was 750
s spacing between injections to ensure that the thermal power returns to the baseline before the next injection. The stirring speed was 750 rpm. Control experiments with the peptides injected in the sample cell filled with buffer were carried out under the same conditions. These control experiments showed heats of dilution negligible in all cases. The heats per injection normalized per mole of injectant versus the molar ratio [peptide]/[protein] were fitted to a single-site model. A 1:1 stoichiometry complex was assumed for the fitting of the ITC binding isotherms in the case of the low c-assays. Data were analyzed with MicroCal PEAQ-ITC (version 1.1.0.1262) analysis software (Malvern Instruments Ltd.).
rpm. Control experiments with the peptides injected in the sample cell filled with buffer were carried out under the same conditions. These control experiments showed heats of dilution negligible in all cases. The heats per injection normalized per mole of injectant versus the molar ratio [peptide]/[protein] were fitted to a single-site model. A 1:1 stoichiometry complex was assumed for the fitting of the ITC binding isotherms in the case of the low c-assays. Data were analyzed with MicroCal PEAQ-ITC (version 1.1.0.1262) analysis software (Malvern Instruments Ltd.).
Mass spectrometry
Eluates were reduced with 5 mM DTT at 55
mM DTT at 55 °C for 30
°C for 30 min, cooled to room temperature, and alkylated with 15
min, cooled to room temperature, and alkylated with 15 mM iodoacetamide at room temperature for 45
mM iodoacetamide at room temperature for 45 min in the dark. Alkylation reactions were quenched with an additional 5
min in the dark. Alkylation reactions were quenched with an additional 5 mM of DTT. Proteins were enriched from eluates using the SP3 method35 and digested overnight in 25
mM of DTT. Proteins were enriched from eluates using the SP3 method35 and digested overnight in 25 mM ammonium bicarbonate with trypsin for mass spectrometric analysis. Peptides were analyzed on an Orbitrap Lumos mass spectrometer (ThermoFisher Scientific) equipped with a Vanquish Neo UHPLC system (ThermoFisher Scientific). Raw data were searched using COMET in high-resolution mode36 against a target-decoy (reversed)37 version of the human proteome sequence database (UniProt; downloaded 2/2020, 40704 entries of forward and reverse protein sequences), requiring fully tryptic peptides (K, R; not preceding P) with up to three mis-cleavages. Static modifications included carbamidomethylcysteine and variable modifications included oxidized methionine and S/T/Y phosphorylation. Searches were filtered using orthogonal measures, and quantification of LC-MS/MS spectra was performed using MassChroQ38.
mM ammonium bicarbonate with trypsin for mass spectrometric analysis. Peptides were analyzed on an Orbitrap Lumos mass spectrometer (ThermoFisher Scientific) equipped with a Vanquish Neo UHPLC system (ThermoFisher Scientific). Raw data were searched using COMET in high-resolution mode36 against a target-decoy (reversed)37 version of the human proteome sequence database (UniProt; downloaded 2/2020, 40704 entries of forward and reverse protein sequences), requiring fully tryptic peptides (K, R; not preceding P) with up to three mis-cleavages. Static modifications included carbamidomethylcysteine and variable modifications included oxidized methionine and S/T/Y phosphorylation. Searches were filtered using orthogonal measures, and quantification of LC-MS/MS spectra was performed using MassChroQ38.
Statistics and reproducibility
Mann–Whitney U-test was applied for all the statistic analysis for live imaging results, immunofluorescence quantification and peptide intensity which have been repeated at least twice. ns means not significant; *P <
< 0.1; **P
0.1; **P <
< 0.01; ***P
0.01; ***P <
< 0.001; ****P
0.001; ****P <
< 0.0001.
0.0001.
Supplementary information
Acknowledgements
We thank the Protein Production and Characterization Platform at the Novo Nordisk Foundation Center for Protein Research for the production of Plk1 protein. We also thank Dr. Adrian Saurin at the University of Dundee for constructive communications. This work was supported by the National Natural Science Foundation of China (31970666) and Taishan Scholar Project (tsqn201812054) from Shandong, China. Work at the Novo Nordisk Foundation Center for Protein Research was supported by grant NNF14CC0001 and J.N. is supported by grants from the Danish Cancer Society (R269-A15586-B17), Independent Research Fund Denmark (8021-00101B and 0134-00199B) and Novo Nordisk Foundation (NNF20OC0065098). A.N.K. is supported by grant from NIH (R35GM119455).
Author contributions
C.S. did all the cloning, live cell imaging, immunofluorescence with the help from M.Z., Y.Z. (Yuqing Zhang), Y.Z. (Yujing Zhai), Y.W., and T.L. B.L.M. did ITC analysis. M.Z. conducted immunoprecipitation. M.H.M. did the initial phosphorylation mutant characterization. T.K. prepared samples for mass spectrometry and A.N.K. performed mass spectrometry analysis. G.Z. supervised the whole project and wrote the manuscript with J.N.
Peer review
Peer review information
Communications Biology thanks Eleni Petsalaki and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Patrick Meraldi and Kaliya Georgieva. A peer review file is available.
Data availability
All data supporting the findings of this study are available within the paper and the supplementary information except the mass spectrometry proteomics data which have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the access code PXD057400. Source data can be found in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chunlin Song, Mingzhe Zhang.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07205-2.
References
Articles from Communications Biology are provided here courtesy of Nature Publishing Group
Funding
Funders who supported this work.
Taishan Scholar Project of Shandong Province (1)
Grant ID: tsqn201812054

 1
1

