Abstract
Free full text

Identification of reproduction-related genes in the hypothalamus of sheep (Ovis aries) using the nanopore full-length transcriptome sequencing technology
Abstract
The hypothalamus is the coordination center of the sheep (Ovis aries) endocrine system and plays an important role in the reproductive processes of sheep. However, the specific mechanism by which the hypothalamus affects sheep reproductive performance remains unclear. In this study, the hypothalamus tissues of high-reproduction small-tailed Han sheep and low-reproduction Wadi sheep were collected, and full-length transcriptome sequencing by Oxford Nanopore Technologies (ONT) was performed to explore the key functional genes associated with sheep fecundity. The differentially expressed genes (DEGs) were screened and enriched using DESeq2 software through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Approximately 41.75 million clean reads were obtained from the hypothalamus tissues of high- and low-reproduction sheep, after quality control, 32,194,872 high-quality full-length sequences and 2,114 DEGs were obtained, including 1,247 upregulated genes and 867 downregulated genes (P adjust <
< 0.05, |log2FC|>1). Some DEGs were enriched in oocyte meiosis, progesterone-mediated oocyte maturation, estrogen signaling pathway, GnRH signaling pathway and other development-related signaling pathways. The constructed protein-protein interaction (PPI) networks identified the reproduction-related genes, such as GSK3B, PPP2R1B, and PPP2CB. The results of this study will enrich and supplement the genomic information available for small-tailed Han sheep and Wadi sheep, as well as expand the understanding of the molecular mechanisms underlying the regulation of animal reproduction by the hypothalamus, and they also provided reference data for further investigations on the mechanism of high reproduction in sheep.
0.05, |log2FC|>1). Some DEGs were enriched in oocyte meiosis, progesterone-mediated oocyte maturation, estrogen signaling pathway, GnRH signaling pathway and other development-related signaling pathways. The constructed protein-protein interaction (PPI) networks identified the reproduction-related genes, such as GSK3B, PPP2R1B, and PPP2CB. The results of this study will enrich and supplement the genomic information available for small-tailed Han sheep and Wadi sheep, as well as expand the understanding of the molecular mechanisms underlying the regulation of animal reproduction by the hypothalamus, and they also provided reference data for further investigations on the mechanism of high reproduction in sheep.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79140-5.
Author information:
Introduction
Sheep (Ovis aries) is an important economic animal in China, its meat, milk and furs greatly enrich human life, and its reproductive performance is an important indicator of its economic traits1. Reproduction is a complex and important physiological process that depends on the coordination, synthesis and release of gonadal axis hormones2. The hypothalamus-pituitary-gonadal (HPG) axis underlies the endocrine control of reproduction in mammals, and together, they regulate the development and reproductive function in mammals3. As the starting organ of the HPG axis, the hypothalamus can generate a GnRH signal to regulate the secretion of downstream hormones, including follicle-stimulating hormone (FSH) and luteinizing hormone (LH)4. These two hormones are regulatory factors of the ovary, by maintaining the function of the corpus luteum, they drive the follicle to produce progesterone (P4) and estrogen (E2)5. GnRH (gonadotropin releasing hormone) secreted by the hypothalamus can serve as an initiator of reproductive activities and plays an important role in reproductive regulation6, and changes in hypothalamus function affect reproductive activities7,8, such as follicle development, ovulation and egg laying9. Therefore, the hypothalamus is involved in the regulation of reproductive activities in mammals, and the hypothalamus is a key region in the brain that initiates reproductive activities.
With the development of high-throughput sequencing technology, breed selection has entered the era of molecular breeding. Candidate gene analysis can be used to identify individual genes responsible for economically important traits, and many candidate genes associated with animal fecundity have been continuously mined10. Currently, many signaling pathways related to sheep reproductive performance have been identified through molecular biology techniques11, but the genetic and physiological mechanisms involved remain to be further explored. Small-tailed Han sheep is a predominant indigenous breed in China, mainly distributed in provinces such as Shandong, Henan, Anhui, etc12. Small-tailed Han sheep can be in estrus for four seasons and can be bred all year round, the average annual lambing rate is approximately 267.1%, which are known for their high fecundity. Wadi sheep is a local breed in Shandong Province that can be used for both skin and meat purposes, which are often in estrus in spring and fall, and the lambing rate of primiparous ewes is only 178%13. The fecundity of Wadi sheep is lower than that of small-tailed Han sheep. The differences in fertility between small-tailed Han sheep and Wadi sheep as well as the molecular mechanisms underlying the regulation of animal reproduction by the hypothalamus have not been studied.
Nanopore full-length transcriptome sequencing based on the Oxford Nanopore Technologies (ONT) platform, is a recently developed third-generation sequencing technology14. Using nanopore sequencing, a single molecule of DNA or RNA can be sequenced without the need for PCR amplification or chemical labeling of the sample. ONT is having the advantages of long sequencing reads, high mobility for testing, and short processing time with the ability to display results in real-time. And which has unique advantages in alternative splicing, gene fusion, and the identification of complicated transcripts, as well as higher accuracy in identifying transcript expression levels. In this study, we used hypothalamus tissues from high-reproduction small-tailed Han sheep ewes and low-reproduction Wadi sheep ewes as the research objects. Meanwhile, Nanopore full-length transcriptome sequencing technology by ONT was used to screen and mine related genes involved in the reproduction regulation of sheep. This is of great significance for the comprehensive understanding of the mechanism of the hypothalamus in regulating animal reproduction, and may provide a more theoretical basis for the future elucidation of the molecular mechanism of high reproduction in sheep.
Methods
Sample collection and ethics declarations
Based on pedigree and lambing records, three one-year-old small-tailed Han sheep ewes from the same family line, with no significant differences in weight, height, body shape or body condition, and with the same age, which were feed and managed in small-tailed Han sheep breeding farm (Jiaxiang County, Shandong Province, China, National-level Mutton Sheep Core Breeding Farm), were selected and named as High Fecundity Group (XH). Three Wadi sheep ewes, from Binzhou Animal Husbandry and Veterinary Research Institute of Shandong Province, China (Wadi Sheep Breeding Farm of Shandong Province), were also selected with the same selection criteria as small-tailed Han sheep, and named as Low Fecundity Group (XL). All these experimental sheep were in estrus after synchronous estrus treatment with sponge plugs, and their basic traits were also listed in Table S1. All sheep were sacrificed without pain after general anesthesia by intramuscular injection of Xylazine Hydrochloride injection solution (0.3 milligrams per kilogram of body weight), and every effort was made to reduce the suffering of animals in the experiments. The hypothalamus tissues were removed and placed into sterile cryopreservation tubes, numbered and quickly placed in the nitrogen tank for extraction of total RNA and subsequent experiments. All experimental protocols were approved by Experimental Animal Management Committee of Shandong Agricultural University (SDAUA-2023-237), all methods were carried out in accordance with relevant guidelines and regulations, all methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Construction and sequencing of the ONT library
Total RNA was extracted respectively from each hypothalamus tissue of six experimental sheep using TRIzol reagent (Thermo Fisher Scientific, USA), the RNA purity (D260/D280 value) was determined using a NanoDrop ND-2000 (Thermo Science, USA)15, the RNA concentration was precisely quantified using a Qubit 3.0 fluorometer16, and the integrity of the RNA was examined by agarose gel electrophoresis. After passing the quality inspection, 500 ng of total RNA is required for each experimental sample and used for for connecting reverse transcription primers with 1 µL VN primers (VNP), and complementary DNA (cDNA) was obtained by low-cycle polymerase chain reaction (PCR) with Strand-Switching Primer (2 µL) and Reverse Transcriptase (1 µL). Subsequently, the amplification reaction was performed with the following system, Reverse-transcribed RNA sample 5 µL, PCR Barcode 1.5 µL, 2x LongAmp Taq Master Mix 25 µL, and added to 50 µL with Nuclease-free water 18.5 µL, the PCR products were purified using AMPure beads. After adding sequencing adapters (including motor proteins) with Rapid Adapter (RAP, 2 µL), the libraries were configured with Squencing Buffer(SQB) 75 µL, and Loading Beads(LB) 51 µL, and loaded on FLO-PRO002 microarray (R9.4)17. The sequencing was performed for each library using the PromethION sequencer (Oxford Nanopore Technologies, Oxford, UK). ONT library construction and all sequencing work were completed by Bena (Wuhan) Information Technology Co., Ltd. (Wuhan, China).
Quality control and statistics of the sequencing data
The obtained raw sequencing data were subjected to base calling using GUPPY (version: 5.0.16) software, converted to fastq format to remove low-quality sequences and adaptor sequences, and then processed using NanoFilt 50 (version: 2.8.0; parameters: -q 7 -l 50) software18 to filter out sequences with a quality value less than 7 and a length less than 50 bp to obtain effective clean reads. The SeqKit (version: 0.12.0; parameter: default) software19 was used to count the effective clean reads. The full-length sequences in the valid sequencing data were identified to direct and trim the full-length Nanopore cDNA sequences using Pychopper software (version: 2.4.0; parameters: -Q 7 -z 50) and NanoFilt 50 (version: 2.8.0; parameters: -q 7 -l 50) software18 was further used to filter the obtained full-length sequences. minimap2 (version: 2.17- r941; parameters: -ax splice -uf -k14./reference/ref-sheep-ont.mmi $i/out.pass.fq -t 10 −
− 0 $i/aln.sam.)20 was used to align the filtered full-length sequence with the sheep reference genome (ARS-UI_Ramb_v2.0). And SAMtools software (version: 1.11; parameter: flagstat)21 was used to analyze the genome alignment results.
0 $i/aln.sam.)20 was used to align the filtered full-length sequence with the sheep reference genome (ARS-UI_Ramb_v2.0). And SAMtools software (version: 1.11; parameter: flagstat)21 was used to analyze the genome alignment results.
Analysis of DEGs between the high- and low- fecundity groups
Transcripts per kilobase million (TPM) was used to measure gene expression levels, and gene expression quantification was performed using Salmon software (version: 1.4.0)22. DESeq2 software (version: 1.26.0)23 was used to analyze the read count data in each sample for differential expression analysis, and the screening thresholds were P adjust <
< 0.05 and |log2FoldChange| > 1. For the calculation of TPM, for each gene, the read counts were divided by the length (in kilobases) of the gene to obtain the reads per kilobase (RPK), and all RPK values in a sample were calculated and divided by 1,000,000 to obtain the “per million” scaling factor. Then, the RPK value was divided by the “per million” scaling factor to obtain the TPM value24.
0.05 and |log2FoldChange| > 1. For the calculation of TPM, for each gene, the read counts were divided by the length (in kilobases) of the gene to obtain the reads per kilobase (RPK), and all RPK values in a sample were calculated and divided by 1,000,000 to obtain the “per million” scaling factor. Then, the RPK value was divided by the “per million” scaling factor to obtain the TPM value24.
GO and KEGG enrichment analysis of DEGs
The Gene Ontology (GO) database (http://www.geneontology.org/)25 was used for the GO annotation of the DEGs, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/)26 was used to perform pathway analysis on the DEGs. GO analysis included mainly cellular component (CC), molecular function (MF) and biological process (BP). The GO items and KEGG pathways that were significantly enriched in DEGs were calculated using hypergeometric distribution, and the GO items and KEGG pathways with P ≤
≤ 0.05 were considered significantly enriched. The enrichment analysis software used for DEGs was ClusterProfiler (version 3.14.3)27.
0.05 were considered significantly enriched. The enrichment analysis software used for DEGs was ClusterProfiler (version 3.14.3)27.
Construction of the regulatory network
Based on the GO and KEGG enrichment results of the DEGs, Cytoscape software (version 3.10.0, http://www.cytoscape.org/)28 and the STRING database (version 11.5, https://cn.string-db.org)29 was used to construct a gene regulatory network.
Validation of DEGs by quantitative real-time PCR (qRT-PCR)
To verify the reliability of the sequencing results, eight DEGs were randomly selected from the transcriptome sequencing data. The GAPDH (Ovis aries) gene was used as an internal reference gene for the qRT-PCR. The primers were designed using Primer 5 software (Table S2). Quantification was performed according to the instruction manual of the SYBR Green Premix Pro Taq HS qPCR Kit (Code: AG11701), and PCR amplification was performed on a LightCycler 480 system (Roche). The specific reaction conditions were as follows: denaturation at 95 °C for 30 s, followed by for 40 cycles of amplification at 95 °C for 5 s and 40 cycles of amplification at 60 °C for 30 s. The relative expression levels of genes were calculated by the 2−ΔΔCt method, and the significance standard was P <
< 0.0530. The results were visualized using GraphPad 8.0 (GraphPad Software, USA).
0.0530. The results were visualized using GraphPad 8.0 (GraphPad Software, USA).
Results
Quality assessment and reference genome comparison of sequencing data
In this study, the hypothalamus tissues of high-reproduction small-tailed Han sheep (XH) and low-reproduction Wadi sheep (XL) were selected for ONT sequencing. For the XH group (X1-X3), 9,530,456, 9,962,624, and 9,855,014 raw reads were obtained, of which 8,998,836, 9,496,791, and 9,415,631 were clean reads, accounting for 94.4%, 95.3% and 95.5% of the raw reads, respectively (Table 1; Fig. 1A). For the XL (X4-X6) group, 4,498,641, 4,965,510 and 4,958,213 raw reads were obtained, of which 4,313,447, 4,776,002 and 4,754,922 were clean reads, accounting for 95.9%, 96.2% and 95.9% of the raw reads, respectively (Table 1; Fig. 1A). For the XH group (X1-X3), there were 6,840,076, 7,329,826, and 7,397,089 full-length sequences, respectively, and 3,291,592, 3,700,877, and 3,635,412 full-length sequences were also identified for the XL group (X4-X6), respectively. The full-length sequences were aligned to the reference genome, and the alignment rate was greater than 80% (Table 1). In addition, the correlation analysis of RNA-Seq data based on the gene expression levels showed that the Pearson correlation coefficient (R) between the two groups was greater than 0.96 (Fig. 1B), indicating that the similarity in the expression patterns between the two groups of samples was high, which increased the reliability of the results. These results indicated that high-quality full-length transcriptome sequencing results were obtained.
Table 1
Sequencing data information and alignment statistics with reference genome for six samples.
| Sample | Raw reads | Clean reads | Total reads | Mapped reads | Map_rate |
|---|---|---|---|---|---|
| X1 | 9,530,456 | 8,998,836 | 6,840,076 | 5,503,728 | 80.46% |
| X2 | 9,962,624 | 9,496,791 | 7,329,826 | 5,935,547 | 80.98% |
| X3 | 9,855,014 | 9,415,631 | 7,397,089 | 6,226,032 | 84.17% |
| X4 | 4,498,641 | 4,313,447 | 3,291,592 | 3,209,994 | 97.52% |
| X5 | 4,965,510 | 4,776,002 | 3,700,877 | 3,617,334 | 97.74% |
| X6 | 4,958,213 | 4,754,922 | 3,635,412 | 3,573,424 | 98.29% |
X1, X2 and X3 represent the different library in small-tailed Han sheep group (high-reproduction, XH); X4, X5 and X6 represent the different library in Wadi sheep group (low-reproduction, XL).
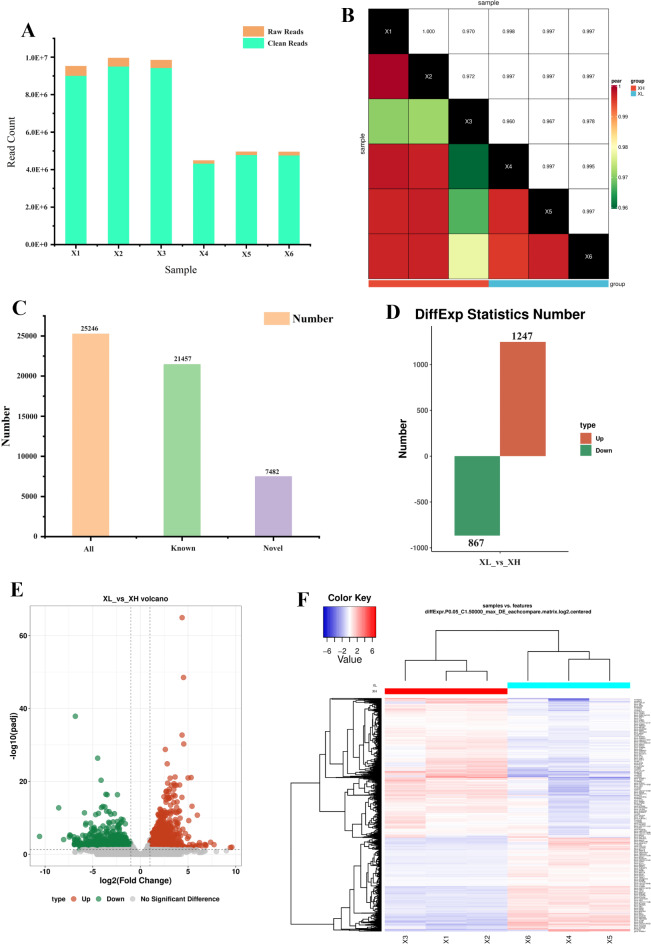
Sequencing data statistics and differential gene identification analysis. (A) Statistics of reads filtering information; (B) Pearson correlation between different samples; (C) Statistics of the gene numbers obtained from sequencing; (D) Number of differentially expressed genes between different groups; (E) Volcano plot of differentially expressed genes; (F) Clustering analysis heatmap of differentially expressed genes.
Identification and expression analysis of DEGs
The hypothalamus is an important organ of the hypothalamic-pituitary-ovarian (HPO) axis in female mammals, and the differential expression of genes can affect the fecundity of sheep. To further explore important regulatory factors in the hypothalamus associated with sheep reproductive capacity, the DEGs between the XL and XH groups were screened according to the criteria of padj <
< 0.05 and |log2FC|>1. A total of 25,246 genes, including 21,457 known genes and 7,482 novel genes (genes with new transcripts were counted as novel genes), were identified between the two groups (Fig. 1C, Table S3). A total of 2,114 DEGs were obtained in these two groups, with 1,247 upregulated genes and 867 downregulated genes (Fig. 1D, Table S4). The volcano map shows the basic distribution of the DEGs obtained by sequencing (Fig. 1E). Hierarchical cluster analysis of DEGs in different samples revealed significant segregation between the XH group (X1-X3) and the XL group (X4-X6), indicating significant differences in gene expression between the two groups. In the hypothalamus, the three samples (X1-X3) of the XH group were clustered in the same cluster, and the three samples (X4-X6) of the XL group were clustered in the same cluster, indicating that the gene expression levels and patterns of the three samples within the same group were similar, and differences were also existed between different groups (Fig. 1F).
0.05 and |log2FC|>1. A total of 25,246 genes, including 21,457 known genes and 7,482 novel genes (genes with new transcripts were counted as novel genes), were identified between the two groups (Fig. 1C, Table S3). A total of 2,114 DEGs were obtained in these two groups, with 1,247 upregulated genes and 867 downregulated genes (Fig. 1D, Table S4). The volcano map shows the basic distribution of the DEGs obtained by sequencing (Fig. 1E). Hierarchical cluster analysis of DEGs in different samples revealed significant segregation between the XH group (X1-X3) and the XL group (X4-X6), indicating significant differences in gene expression between the two groups. In the hypothalamus, the three samples (X1-X3) of the XH group were clustered in the same cluster, and the three samples (X4-X6) of the XL group were clustered in the same cluster, indicating that the gene expression levels and patterns of the three samples within the same group were similar, and differences were also existed between different groups (Fig. 1F).
GO functional annotation and classification of DEGs
Among the 2,114 DEGs obtained from the hypothalamus, 1,456 DEGs were enriched in 2,968 GO terms, including 1,717 in BP, 517 in CC and 734 in MF (Table S5). In BP, DEGs were enriched mainly in vesicle-mediated transport, translation, and protein localization to the plasma membrane; in CC, DEGs were enriched mainly in Golgi apparatus, Ribosom, and Mitochondrial respiratory chain complex IV; and in MF, DEGs were enriched mainly in structural constituent of ribosome, GTPase activity, and ubiquitin protein ligase binding. Based on P ≤
≤ 0.05, among the significant DEGs between the XH and XL groups, the top 20 GO terms with significant enrichment were screened, of which 10 GO terms belonging to CC, 5 GO terms belonging to BP, and 5 GO terms belonging to MF (Fig. 2A, B). Among the different GO categories, the top 20 GO terms in each of BP, CC, and MF are shown in Fig. 2C.
0.05, among the significant DEGs between the XH and XL groups, the top 20 GO terms with significant enrichment were screened, of which 10 GO terms belonging to CC, 5 GO terms belonging to BP, and 5 GO terms belonging to MF (Fig. 2A, B). Among the different GO categories, the top 20 GO terms in each of BP, CC, and MF are shown in Fig. 2C.
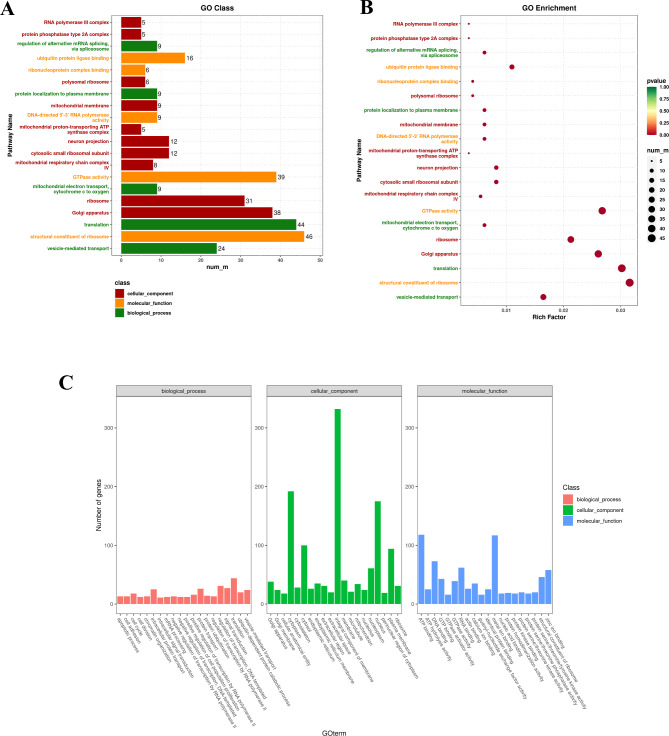
GO Annotation and enrichment analysis of differentially expressed genes in the hypothalamus. (A) Distribution of the top 20 GO terms in different GO Classes; (B) The top 20 significantly enriched GO terms for differentially expressed genes; (C) Distribution of the top 20 enriched GO terms for differentially expressed genes in different GO Classes.
GO functional annotation and classification of the DEGs in small-tailed Han sheep (XH group)
To more clearly explain the functional role of the enriched GO terms in the reproductive process, GO annotation analysis on the upregulated and downregulated DEGs in the hypothalamus of small-tailed Han sheep was performed (Table S6). Among the 746 upregulated DEGs, 1,710 GO terms, including 937 in BP, 320 in CC and 453 in MF, were enriched, and the GO terms related to reproductive and physiological processes in sheep, such as ovarian follicle development and utero embryonic development were annotated and obtained. The top 20 GO terms distributed in each of BP, CC, and MF for upregulated genes are shown in Fig. 3A and based on P ≤
≤ 0.05, the top 20 GO terms with significant enrichment were screened (Fig. 3B), and the most enriched GO term was integral components of the membrane, with 178 enriched genes. The 710 downregulated DEGs were enriched in 1,920 GO terms, including 1,059 in BP, 377 in CC, and 484 in MF, and the annotation obtained GO terms, such as positive regulation of growth hormone secretion and estrogen metabolic process. The 20 GO terms distributed in BP, CC, and MF for downregulated genes are shown in Fig. 3C and the top 20 GO terms with significant enrichment for downregulated genes based on P
0.05, the top 20 GO terms with significant enrichment were screened (Fig. 3B), and the most enriched GO term was integral components of the membrane, with 178 enriched genes. The 710 downregulated DEGs were enriched in 1,920 GO terms, including 1,059 in BP, 377 in CC, and 484 in MF, and the annotation obtained GO terms, such as positive regulation of growth hormone secretion and estrogen metabolic process. The 20 GO terms distributed in BP, CC, and MF for downregulated genes are shown in Fig. 3C and the top 20 GO terms with significant enrichment for downregulated genes based on P ≤
≤ 0.05 are shown in Fig. 3D.
0.05 are shown in Fig. 3D.
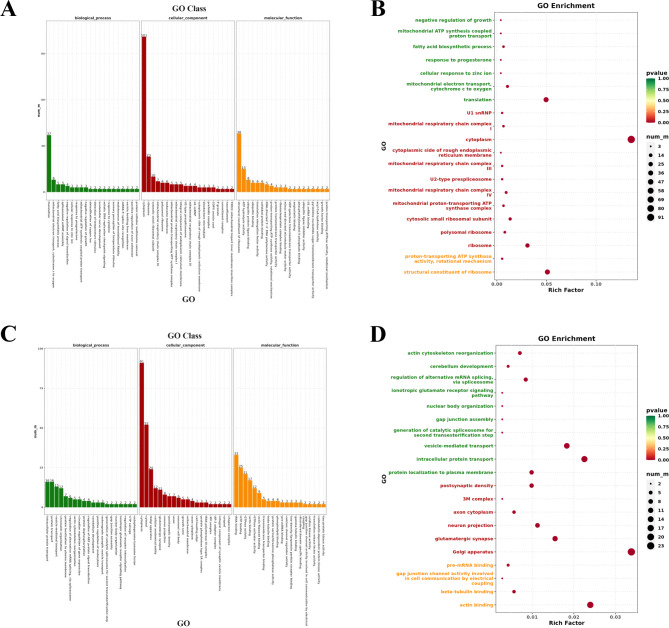
GO annotation and enrichment analysis of upregulated and downregulated genes in the hypothalamus of Small-tailed Han Sheep. (A) Distribution of the top 20 GO terms for upregulated genes in different GO Classes; (B) The top 20 significantly enriched GO terms for upregulated genes; (C) Distribution of the top 20 GO terms for downregulated genes in different GO Classes; (D) The top 20 significantly enriched GO terms for downregulated genes.
KEGG metabolic pathway analysis of DEGs
To further clarify the contribution of specific signaling pathways to sheep fecundity, KEGG enrichment analysis was performed on the DEGs of sheep with high and low fecundity (Table S7). A total of 826 DEGs were involved in 322 KEGG pathways (Fig. 4A), and 19 signaling pathways were closely related to sheep reproduction (Fig. 4B), involving oocyte meiosis (ko04114), progesterone-mediated oocyte maturation (ko04914), estrogen signaling pathway (ko04915), oxytocin signaling pathway (ko04921), and GnRH signaling pathway (ko04912). The pathways with the most enriched genes were the PI3K-Akt signaling pathway (ko04151), MAPK signaling pathway (ko04010) and mTOR signaling pathway (ko04150). The top 20 significantly enriched KEGG pathways were screened according to P ≤
≤ 0.05 (Fig. 4C, D).
0.05 (Fig. 4C, D).
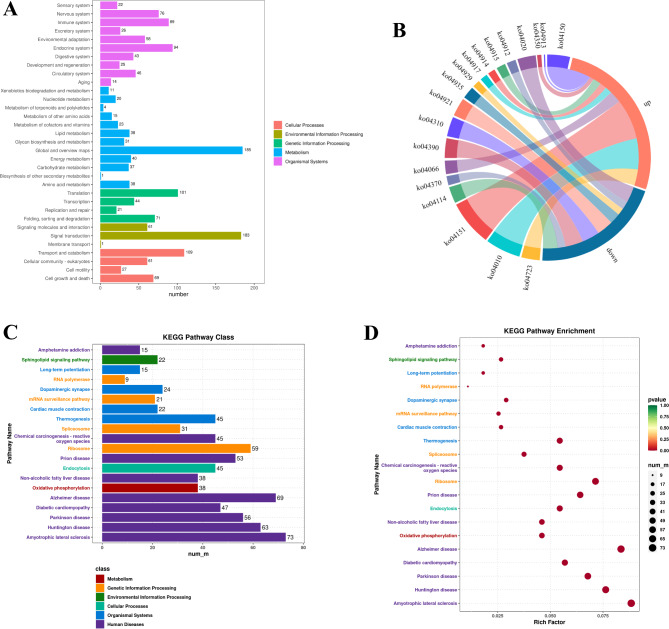
KEGG analysis of differentially expressed genes in the hypothalamus. (A) Distribution of differentially expressed gene KEGG terms in different KEGG Classes; (B) Chord diagram of sheep reproduction-related pathways in the hypothalamus; (C) Distribution of the top 20 KEGG terms in different KEGG Classes; (D) The top 20 significantly enriched KEGG terms for differentially expressed genes.
KEGG metabolic pathway analysis of DEGs in small-tailed Han sheep (XH group)
Further KEGG enrichment analysis was performed on the upregulated and downregulated DEGs of small-tailed Han sheep. The results showed that 467 upregulated DEGs participated in 314 KEGG pathways, 18 reproduction-related signaling pathways were screened, the pathways with the most enriched genes being the PI3K-Akt signaling pathway (ko04151), MAPK signaling pathway (ko04010), and mTOR signaling pathway (ko04150) (Table S8). The top 20 KEGG pathways that were significantly enriched were screened according to P ≤
≤ 0.05 (Fig. 5A, B), and many signaling pathways associated with tumorigenesis and cancer were also enriched, which may indicate that the hypothalamus regulates the cell activities. Overall, 359 downregulated DEGs participated in 304 KEGG pathways, and 18 reproduction-related signaling pathways, including the VEGF signaling pathway (ko04370), Hippo signaling pathway (ko04390), and GnRH signaling pathway (ko04912), were screened (Table S8). These downregulated genes were also enriched in apoptosis-related signaling pathways. The top 20 KEGG pathways that were significantly enriched were screened according to P
0.05 (Fig. 5A, B), and many signaling pathways associated with tumorigenesis and cancer were also enriched, which may indicate that the hypothalamus regulates the cell activities. Overall, 359 downregulated DEGs participated in 304 KEGG pathways, and 18 reproduction-related signaling pathways, including the VEGF signaling pathway (ko04370), Hippo signaling pathway (ko04390), and GnRH signaling pathway (ko04912), were screened (Table S8). These downregulated genes were also enriched in apoptosis-related signaling pathways. The top 20 KEGG pathways that were significantly enriched were screened according to P ≤
≤ 0.05 (Fig. 5C, D), in which the metabolism pathway was the class with the most enriched downregulated DEGs.
0.05 (Fig. 5C, D), in which the metabolism pathway was the class with the most enriched downregulated DEGs.
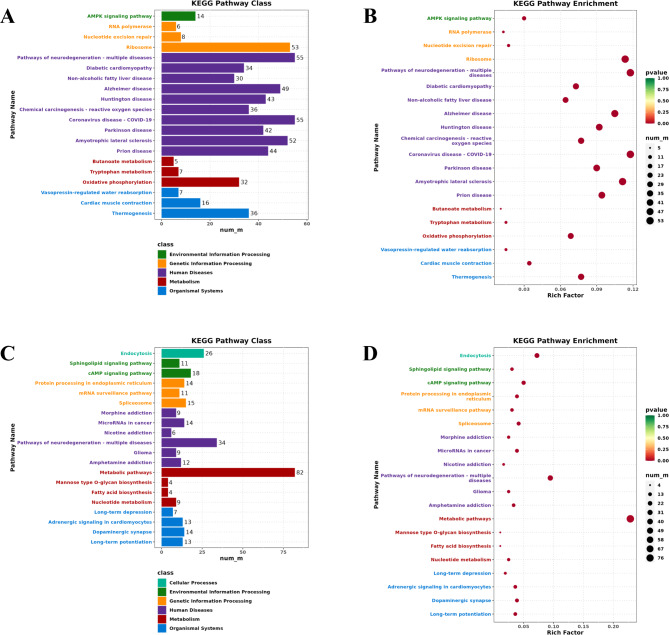
KEGG analysis of upregulated and downregulated genes in the hypothalamus of Small-tailed Han Sheep. (A) Distribution of the top 20 KEGG terms for upregulated genes in different KEGG Classes; (B) The top 20 significantly enriched KEGG terms for upregulated genes; (C) Distribution of the top 20 KEGG terms for downregulated genes in different KEGG Classes; (D) The top 20 significantly enriched KEGG terms for downregulated genes.
Regulatory network construction
Based on the GO and KEGG annotation results, 150 genes related to reproductive processes and body development were selected from the 2,114 DEGs, and a protein-protein interaction (PPI) regulatory network was constructed, including 124 nodes and 280 edges (Fig. 6A, Table S9). The Degree analysis method in the CytoHubba plugin was used to screen core genes from the top-ranking DEGs, and combined with GO annotation and KEGG pathway analysis results, a regulatory network of nine core genes (GSK3B, PPP2R1B, PPP2CB, YWHAQ, YWHAZ, YWHAH, MAP2K1, FOXO1, and IRS1) was constructed (Fig. 6B). These genes were involved in reproduction-related KEGG pathways, including the PI3K-Akt signaling pathway (ko04151), oocyte meiosis pathway (ko04114), Hippo signaling pathway (ko04390) and MAPK signaling pathway (ko04010).
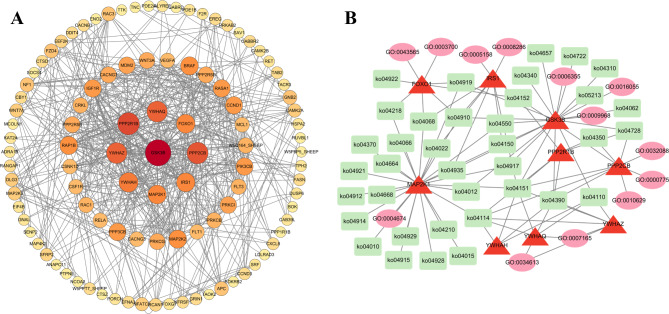
Analysis of regulatory network construction. (A) Genes interaction network of differentially expressed genes (DEGs); (B) Regulatory network constructed based on core genes.
qRT-PCR validation of the DEGs
To validate the RNA-seq results, eight DEGs were randomly selected for expression level detection. The expression patterns of these genes were tested using qRT-PCR and compared with the RNA-seq results. The results showed that the RNA-seq and qRT-PCR results exhibited similar expression patterns (Fig. 7), indicating that the RNA-seq data were reliable.
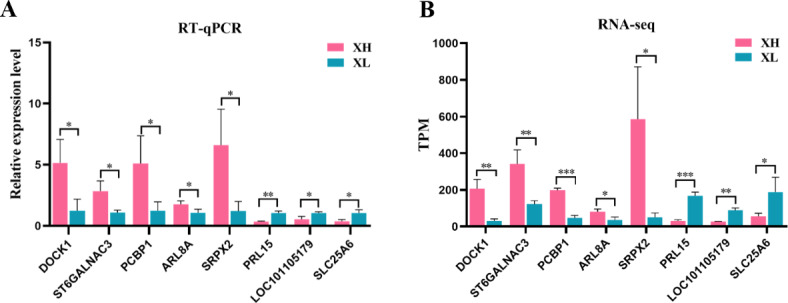
Validation of RNA-seq data by RT-qPCR. (A) RT-qPCR validation of selected differentially expressed genes (DEGs); (B) RNA-seq results of selected DEGs. RT-qPCR data are represented as relative gene expression levels, RNA-seq data are represented as TPM. ***p <
< 0.001, **p
0.001, **p <
< 0.01, *p
0.01, *p <
< 0.05.
0.05.
Discussion
Early studies have shown that sheep fertility traits can be regulated by a single gene or polygenes, especially for high-fecundity breeds31. For example, the BMP15 gene is one of the main genes affecting sheep fecundity and is critical for the normal development of follicles, and it can reduce the mortality of cumulus cells, increase the maturation of oocytes, and thus promote early follicle development32. As a neuroendocrine center, the hypothalamus regulates the secretion of pituitary hormones and the activities of their target glands, including reproduction and growth33, but there are few relevant studies on the mechanism underlying the influence of the hypothalamus on the reproductive performance of sheep. In this study, six full-length transcriptome sequencing of the hypothalamus from small-tailed Han sheep (XH) and Wadi sheep (XL) were performed to screen the DEGs, generate a regulatory network, and screen candidate genes that may be closely associated with sheep reproductive performance. Based on full-length transcriptome sequencing data, 2,114 DEGs were identified, and some signaling pathways closely associated with sheep reproduction, such as the oocyte meiosis, progesterone-mediated oocyte maturation, estrogen signaling pathway, ovarian steroidogenesis pathway, and VEGF signaling pathway, were obtained (Fig. 4B, Table S7).
Insulin-like growth factor 1 receptor (IGF-1R) is the only gene enriched in the ovarian steroidogenesis pathway, and its expression in the hypothalamus of the XH group was upregulated (Table S4). The ovarian steroidogenesis pathway is closely related to ovarian function and follicle development. IGF-1R is a receptor gene of insulin-like growth factor I (IGF-I) and plays an important role in IGF signal transduction. IGF synergizes with gonadotropins to promote ovarian cell growth and steroid production, thereby stimulating ovarian function34. In Bos taurus, IGF-I was found to promote embryonic development35. After the IGF-1R gene is knocked out in mice, mice suffer from severe intrauterine growth retardation, which is approximately 45% of the normal value36. In summary, we speculated that IGF-1R may play certain roles in the dynamic development of sheep follicles and the normal growth of the uterus through the ovarian steroidogenesis pathway.
Angiogenesis and vasodilation are two key factors that control the increase in placental blood flow. Increased placental blood flow can enhance the function of the placenta, resulting in healthier offspring37. The vascular endothelial growth factor (VEGF) family comprises VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGF-E and placenta growth factor (PGF). VEGF usually refers to VEGF-A, which promotes neovasculature formation and increases vascular permeability. Previous studies have shown that the expression of fibroblast growth factor-2 (FGF-2) and VEGF is positively correlated with the growth of placental blood vessels during pregnancy38. This study showed that the expression level of VEGFA in the XH group was significantly higher than that in the XL group (Table S4). VEGF-A may increase the growth of placental blood vessels and blood flow to the placenta during gestation in sheep, thus enhancing placental function and allowing the embryo to receive more adequate nutrition, which may be partly responsible for the slightly higher fertility of small-tailed Han sheep compared to Wadi sheep.
In this study, dopaminergic synaptic signaling pathways, as well as the valine, leucine and isoleucine degradation signaling pathways were enriched. Some studies have shown that differences in GnRH levels are one of the reasons for differences in the numbers of lambs born39, while dopamine can affect the secretion of GnRH to accelerate the metabolism of valine, leucine and isoleucine in high-production ewes, providing essential energy for ewes with multiple pregnancies40. One study revealed that injecting a mixture of the branched-chain amino acids, isoleucine and valine during the late luteal phase of the estrus cycle could increase the ovulation rate of ewes41, and amino acid biosynthesis signaling pathway was also enriched in the hypothalamus. These identified pathways and the DEGs in the pathways can directly or indirectly affect the reproductive process of sheep, but the specific mechanism involved remains to be further studied.
This study identified nine hub genes through the PPI network, and the first was GSK3B, which was enriched in many reproduction-related pathways, such as the mTOR signaling pathway, WNT signaling pathway, and prolactin signaling pathway. GSK3B can affect the activation of proteins responsible for the regulation of the cell cycle, thus indirectly participating in the regulation of breast cell proliferation42; moreover, GSK3B can regulate protein synthesis in skeletal muscle and accelerate the regeneration of atrophic skeletal muscle43. In addition, the GSK3B gene is a signaling molecule of the WNT signaling pathway, and the WNT signaling pathway regulates the development of sheep embryos during the implantation period44. Therefore, we speculated that the GSK3B gene may indirectly affect the development of sheep embryos by regulating cell growth and proliferation. Additionally, many studies have reported the functions of other hub genes, for example MAP2K1 is involved in the regulation of steroidogenesis in mammalian ovaries, is ubiquitously expressed in the embryo and placenta, and plays a leading role in placental development. Loss of function of MAP2K1 in mice results in embryonic death45. The FOXO1 gene was significantly upregulated in the XH group (Table S4), which indirectly regulates follicle development by regulating the synthesis of lipids and sterols and FOXO1 plays an important role in follicle development and corpus luteum formation in mice46. In conclusion, these DEGs between the XH and XL groups may have an important impact on the fecundity of the ewes. The screened core genes can be used as key candidate genes to regulate sheep fecundity, but the specific mechanism involved in the regulation of sheep reproductive physiology needs further in-depth study. This study can provide reference data for further explorations of the mechanism of high reproductive performance in sheep and cultivating new sheep breeds with high fecundity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors express their gratitude to their colleagues for the support in sample collection. We are also grateful to small-tailed Han Sheep Breeding Farm in Jiaxiang County of Shandong Province and Shandong Binzhou Animal Science & Veterinary Medicine Academy for providing raw materials.
Author contributions
Tong Wang: contributed to conception, design, acquisition, analysis, interpretation, critically drafted manuscript and the images.Zhibin Ji: contributed to conception, design, acquisition, analysis, critically drafted manuscript and revised manuscript.Xue Xiao: contributed to analysis, interpretation and drafted the manuscript. Dejie Zhu: contributed to analysis, interpretation and critically revised manuscript. Hengyi Li: contributed to critically revised manuscript. Xinyu Li: contributed to conception, design, interpretation and critically revised manuscript.All authors approved the submitted version and agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved and the resolution documented in the literature.
Funding
This study was financially supported by grants from the National Key Research and Development Program of China (2021YFD1200900).
Data availability
The data that support the findings of this study are available from the tables and supplementary materials. The raw data of sequencing were deposited in GEO database (GSE275445).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
 [Europe PMC free article] [Abstract] [Google Scholar]
[Europe PMC free article] [Abstract] [Google Scholar]Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Funding
Funders who supported this work.
National Key Research and Develop-ment Program of China (1)
Grant ID: 2021YFD1200901




