Abstract
Free full text

Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives.
Abstract
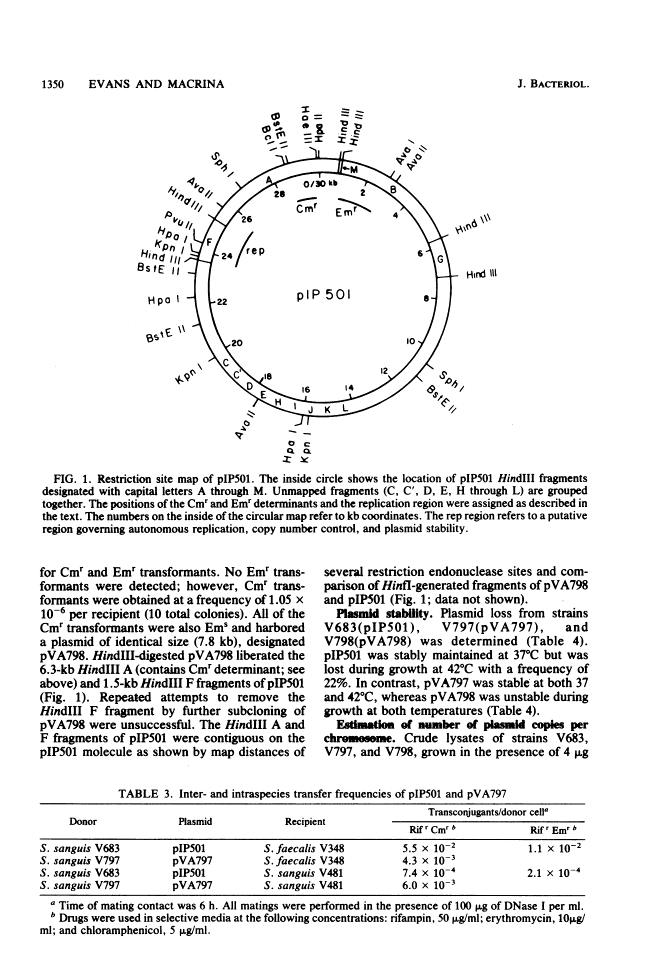
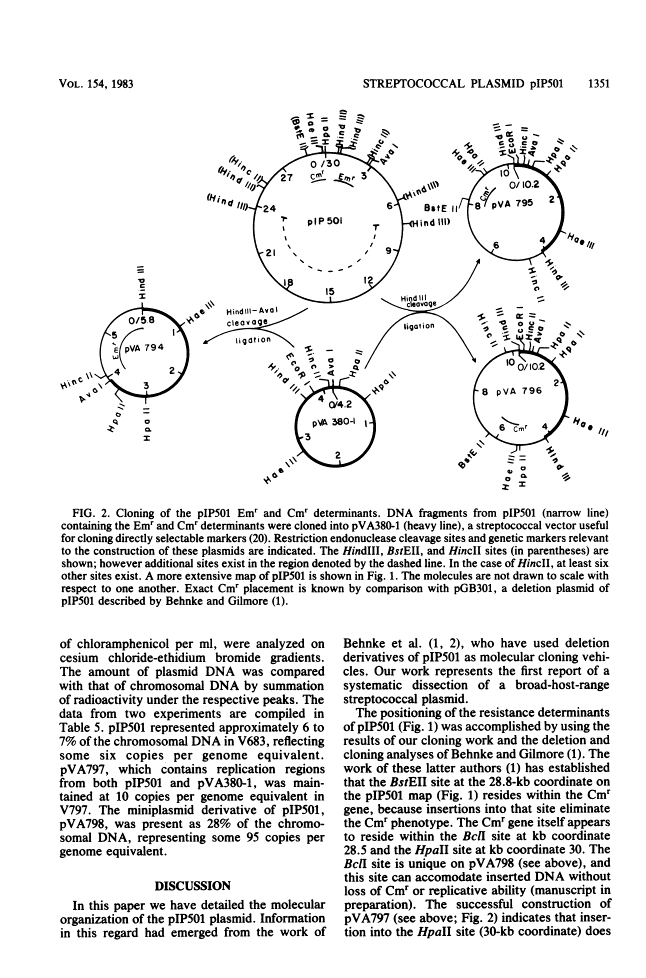
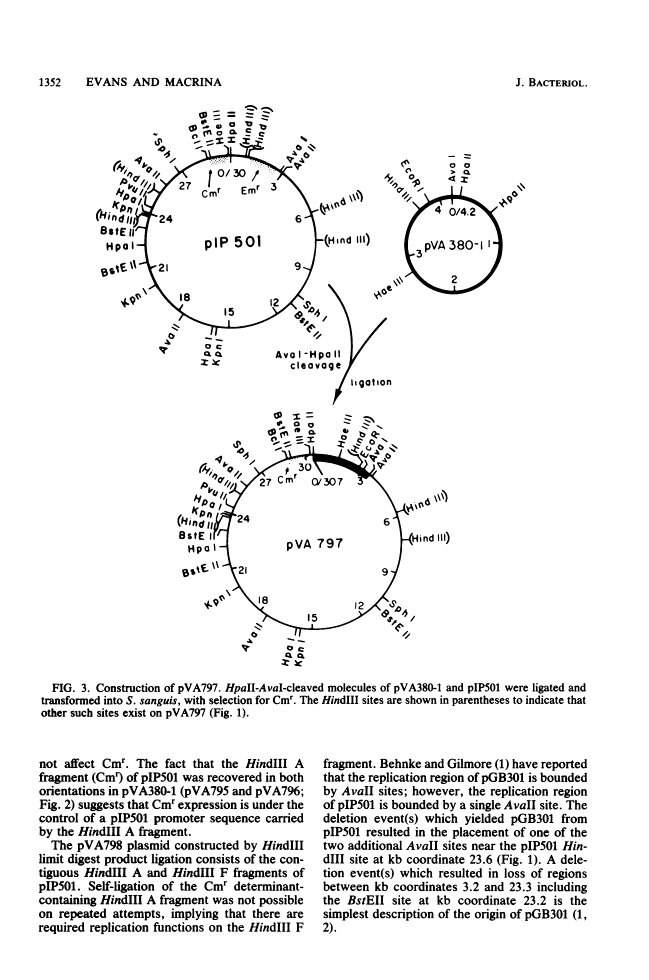
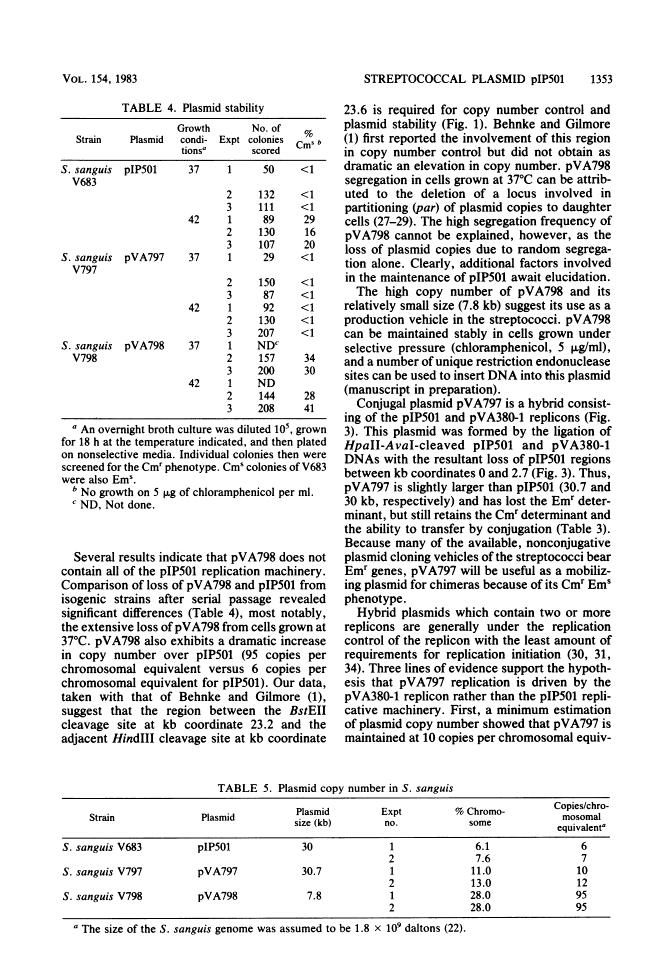
The streptococcal resistance plasmid pIP501 (30 kilobase pairs [kb]) encodes resistance to chloramphenicol (Cmr) and erythromycin (Emr) and is capable of conjugative transfer among numerous streptococcal species. By using a streptococcal host-vector recombinant DNA system, the Cmr and Emr determinants of pIP501 were localized to 6.3-kb HindIII and 2.1-kb HindIII-AvaI fragments, respectively. pIP501 was lost at a frequency of 22% in Streptococcus sanguis cells grown at 42 degrees C but was stable in cells grown at 37 degrees C (less than 1% frequency of loss). Sequences from a cryptic multicopy plasmid, pVA380-1, were substituted for the pIP501 Emr determinant in vitro, and the resulting recombinant plasmid, designated pVA797, was recovered in transformed S. sanguis cells. The replication of pVA797 was governed by the pVA380-1 sequences based on temperature-stable replication and incompatibility with pVA380-1-derived replicons. The self-ligation of partially cleaved HindIII pIP501 DNA fragments allowed the localization of a pIP501 region involved in autonomous plasmid replication. A small pIP501 derivative (pVA798) obtained from this experiment had a greatly increased copy number but was unstably inherited. Our data indicate that the sequences encoding the resistance determinants and some of the plasmid replication machinery are relatively clustered on the pIP501 molecule. The properties of pVA797 and pVA798 indicate that these molecules will enhance current streptococcal genetic systems from the standpoint of conjugative mobilization (pVA797) and gene amplification (pVA798).
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.1M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behnke D, Gilmore MS. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184(1):115–120. [Abstract] [Google Scholar]
- Behnke D, Gilmore MS, Ferretti JJ. Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol Gen Genet. 1981;182(3):414–421. [Abstract] [Google Scholar]
- Bougueleret L, Bieth G, Horodniceanu T. Conjugative R plasmids in group C and G streptococci. J Bacteriol. 1981 Feb;145(2):1102–1105. [Europe PMC free article] [Abstract] [Google Scholar]
- Clewell DB, Yagi Y, Dunny GM, Schultz SK. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. [Europe PMC free article] [Abstract] [Google Scholar]
- Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3479–3483. [Europe PMC free article] [Abstract] [Google Scholar]
- Engel HW, Soedirman N, Rost JA, van Leeuwen WJ, van Embden JD. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J Bacteriol. 1980 May;142(2):407–413. [Europe PMC free article] [Abstract] [Google Scholar]
- Gasson MJ, Davies FL. High-frequency conjugation associated with Streptococcus lactis donor cell aggregation. J Bacteriol. 1980 Sep;143(3):1260–1264. [Europe PMC free article] [Abstract] [Google Scholar]
- Gibson EM, Chace NM, London SB, London J. Transfer of plasmid-mediated antibiotic resistance from streptococci to lactobacilli. J Bacteriol. 1979 Jan;137(1):614–619. [Europe PMC free article] [Abstract] [Google Scholar]
- Haas MJ, Davies J. Characterization of the plasmids comprising the "R factor" R5 and their relationships to other R plasmids. Plasmid. 1980 May;3(3):260–277. [Abstract] [Google Scholar]
- Hansen JB, Olsen RH. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. [Europe PMC free article] [Abstract] [Google Scholar]
- Hershfield V. Plasmids mediating multiple drug resistance in group B streptococcus: transferability and molecular properties. Plasmid. 1979 Jan;2(1):137–149. [Abstract] [Google Scholar]
- Horodniceanu T, Bouanchaud DH, Bieth G, Chabbert YA. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1976 Nov;10(5):795–801. [Europe PMC free article] [Abstract] [Google Scholar]
- Jacob AE, Hobbs SJ. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974 Feb;117(2):360–372. [Europe PMC free article] [Abstract] [Google Scholar]
- Lawson JW, Gooder H. Growth and development of competence in the group H streptococci. J Bacteriol. 1970 Jun;102(3):820–825. [Europe PMC free article] [Abstract] [Google Scholar]
- LeBlanc DJ, Hawley RJ, Lee LN, St Martin EJ. "Conjugal" transfer of plasmid DNA among oral streptococci. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3484–3487. [Europe PMC free article] [Abstract] [Google Scholar]
- Macrina FL, Tobian JA, Jones KR, Evans RP. Molecular cloning in the Streptococci. Basic Life Sci. 1982;19:195–210. [Abstract] [Google Scholar]
- Macrina FL, Jones KR, Wood PH. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J Bacteriol. 1980 Sep;143(3):1425–1435. [Europe PMC free article] [Abstract] [Google Scholar]
- Macrina FL, Keeler CL, Jr, Jones KR, Wood PH. Molecular characterization of unique deletion mutants of the streptococcal plasmid, pAM beta 1. Plasmid. 1980 Jul;4(1):8–16. [Abstract] [Google Scholar]
- Macrina FL, Reider JL, Virgili SS, Kopecko DJ. Survey of the extrachromosomal gene pool of Streptococcus mutans. Infect Immun. 1977 Jul;17(1):215–226. [Europe PMC free article] [Abstract] [Google Scholar]
- Macrina FL, Tobian JA, Jones KR, Evans RP, Clewell DB. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982 Oct;19(3):345–353. [Abstract] [Google Scholar]
- Macrina FL, Wood PH, Jones KR. Simple method for demonstrating small plasmid deoxyribonucleic acid molecules in oral streptococci. Appl Environ Microbiol. 1980 May;39(5):1070–1073. [Europe PMC free article] [Abstract] [Google Scholar]
- Meacock PA, Cohen SN. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. [Abstract] [Google Scholar]
- Nordström K, Molin S, Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid. 1980 Sep;4(2):215–227. [Abstract] [Google Scholar]
- Nordström K, Molin S, Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. II. Incompatibility properties of the partitioning system. Plasmid. 1980 Nov;4(3):332–339. [Abstract] [Google Scholar]
- Smith MD, Shoemaker NB, Burdett V, Guild WR. Transfer of plasmids by conjugation in Streptococcus pneumonias. Plasmid. 1980 Jan;3(1):70–79. [Abstract] [Google Scholar]
- Tobian JA, Macrina FL. Helper plasmid cloning in Streptococcus sanguis: cloning of a tetracycline resistance determinant from the Streptococcus mutans chromosome. J Bacteriol. 1982 Oct;152(1):215–222. [Europe PMC free article] [Abstract] [Google Scholar]
- Uhlin BE, Nordström K. Plasmid incompatibility and control of replication: copy mutants of the R-factor R1 in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):641–649. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.154.3.1347-1355.1983
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/154/3/1347.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/154/3/1347
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/154/3/1347
Citations & impact
Impact metrics
Citations of article over time
Article citations
Conjugation across Bacillus cereus and kin: A review.
Front Microbiol, 13:1034440, 04 Nov 2022
Cited by: 1 article | PMID: 36406448 | PMCID: PMC9673590
Review Free full text in Europe PMC
Listeria monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review.
Pathogens, 9(7):E528, 30 Jun 2020
Cited by: 53 articles | PMID: 32629911 | PMCID: PMC7400505
Review Free full text in Europe PMC
TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Nucleic Acids Res, 46(17):9201-9219, 01 Sep 2018
Cited by: 9 articles | PMID: 30060171 | PMCID: PMC6158623
Targeting Type IV Secretion System Proteins to Combat Multidrug-Resistant Gram-positive Pathogens.
J Infect Dis, 215(12):1836-1845, 01 Jun 2017
Cited by: 6 articles | PMID: 28863473
Go to all (65) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Molecular cloning of a pIP501 derivative yields a model replicon for the study of streptococcal conjugation.
J Gen Microbiol, 131(1):145-153, 01 Jan 1985
Cited by: 6 articles | PMID: 2985739
Intergeneric and intrageneric conjugal transfer of plasmid-encoded antibiotic resistance determinants in Leuconostoc spp.
Appl Environ Microbiol, 54(2):281-287, 01 Feb 1988
Cited by: 17 articles | PMID: 2833158 | PMCID: PMC202444
Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region.
J Bacteriol, 157(2):445-453, 01 Feb 1984
Cited by: 63 articles | PMID: 6319361 | PMCID: PMC215268
Molecular structure and evolution of the conjugative multiresistance plasmid pRE25 of Enterococcus faecalis isolated from a raw-fermented sausage.
Int J Food Microbiol, 88(2-3):325-329, 01 Dec 2003
Cited by: 29 articles | PMID: 14597005
Review
Funding
Funders who supported this work.
NIDCR NIH HHS (2)
Grant ID: DE00081
Grant ID: DE04224