Abstract
Free full text

Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3.
Abstract
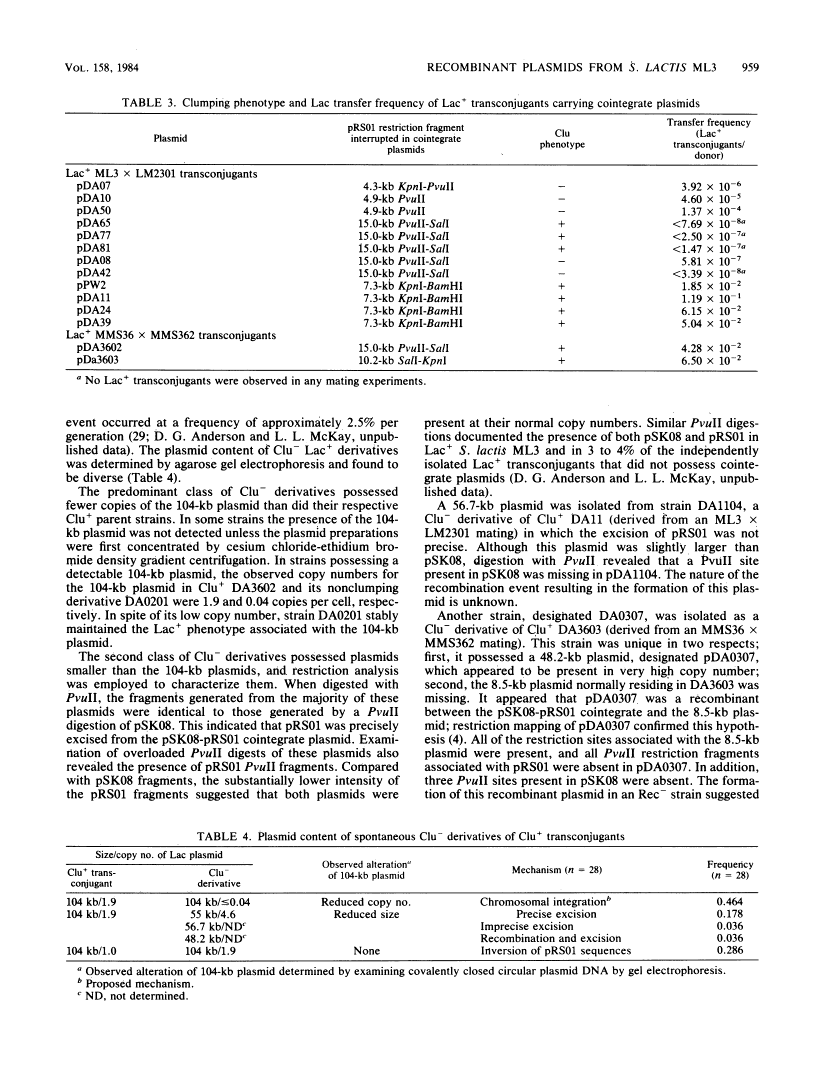
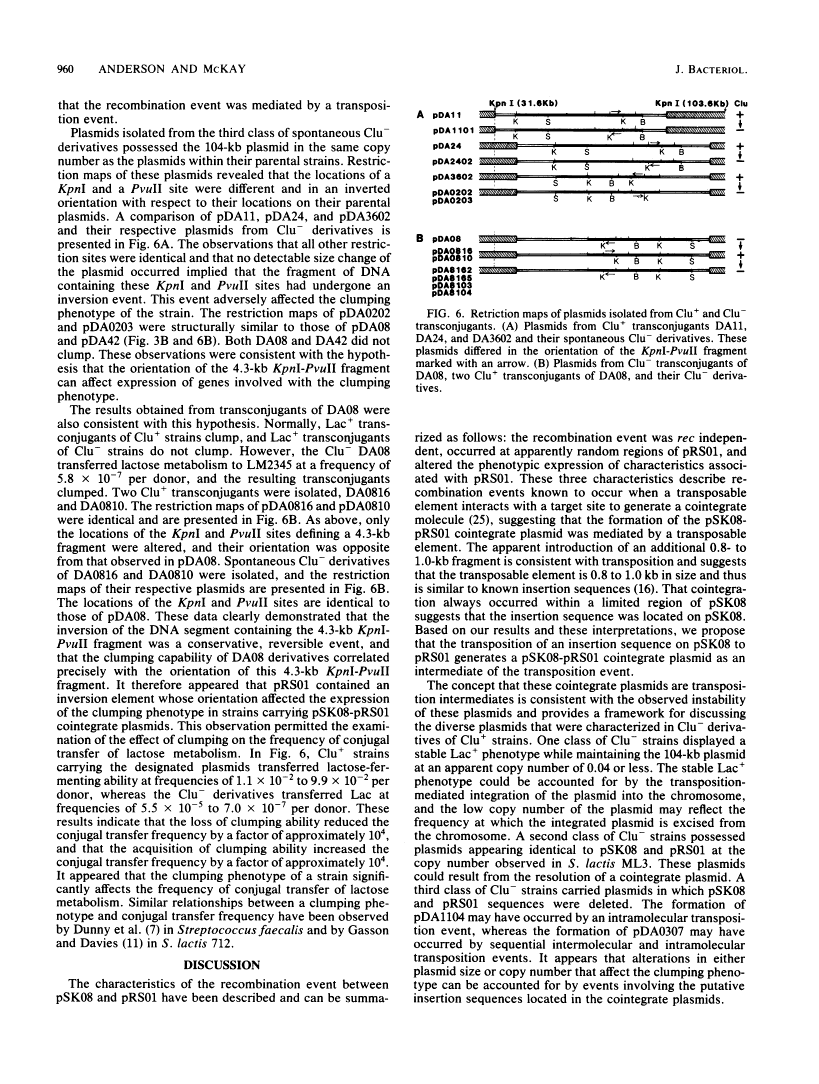
Restriction mapping was employed to characterize the 104-kilobase (kb) cointegrate lactose plasmids from 15 independent transconjugants derived from Streptococcus lactis ML3 as well as the 55-kb lactose plasmid ( pSK08 ) and a previously uncharacterized 48.4-kb plasmid ( pRS01 ) from S. lactis ML3. The data revealed that the 104-kb plasmids were cointegrates of pSK08 and pRS01 and were structurally distinct. The replicon fusion event occurred within adjacent 13.8- or 7.3-kb PvuII fragments of pSK08 and interrupted apparently random regions of pRS01 . Correlation of the transconjugants' clumping and conjugal transfer capabilities with the interrupted region of pRS01 identified pRS01 regions coding for these properties. In the 104-kb plasmids, the pRS01 region was present in both orientations with respect to the pSK08 region. The replicon fusion occurred in recombination-deficient (Rec-) strains and appeared to introduce a 0.8 to 1.0-kb segment of DNA within the junction fragments. The degeneration of the cointegrate plasmids was monitored by examining the lactose plasmids from nonclumping derivatives of clumping transconjugants. These plasmids displayed either precise or imprecise excision of pRS01 sequences or had dramatically reduced copy numbers. Both alterations occurred by rec-independent mechanisms. Alterations of a transconjugant 's clumping phenotype also occurred by rec-independent inversion of a 4.3-kb KpnI-PvuII fragment within the pRS01 sequences of the cointegrate plasmid.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.9M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson DG, McKay LL. Plasmids, loss of lactose metabolism, and appearance of partial and full lactose-fermenting revertants in Streptococcus cremoris B1. J Bacteriol. 1977 Jan;129(1):367–377. [Europe PMC free article] [Abstract] [Google Scholar]
- Anderson DG, McKay LL. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. [Europe PMC free article] [Abstract] [Google Scholar]
- Anderson DG, McKay LL. In Vivo Cloning of lac Genes in Streptococcus lactis ML3. Appl Environ Microbiol. 1984 Feb;47(2):245–249. [Europe PMC free article] [Abstract] [Google Scholar]
- Clements MB, Syvanen M. Isolation of polA mutation that affects transposition of insertion sequences and transposons. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):201–204. [Abstract] [Google Scholar]
- Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3479–3483. [Europe PMC free article] [Abstract] [Google Scholar]
- Efstathiou JD, McKay LL. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977 Apr;130(1):257–265. [Europe PMC free article] [Abstract] [Google Scholar]
- Gasson MJ. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Gasson MJ, Davies FL. High-frequency conjugation associated with Streptococcus lactis donor cell aggregation. J Bacteriol. 1980 Sep;143(3):1260–1264. [Europe PMC free article] [Abstract] [Google Scholar]
- Jacob AE, Hobbs SJ. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974 Feb;117(2):360–372. [Europe PMC free article] [Abstract] [Google Scholar]
- Kempler GM, McKay LL. Genetic Evidence for Plasmid-Linked Lactose Metabolism in Streptococcus lactis subsp. diacetylactis. Appl Environ Microbiol. 1979 May;37(5):1041–1043. [Europe PMC free article] [Abstract] [Google Scholar]
- Klaenhammer TR, McKay LL, Baldwin KA. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. [Europe PMC free article] [Abstract] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. [Abstract] [Google Scholar]
- Kuhl SA, Larsen LD, McKay LL. Plasmid Profiles of Lactose-Negative and Proteinase-Deficient Mutants of Streptococcus lactis C10, ML(3), and M18. Appl Environ Microbiol. 1979 Jun;37(6):1193–1195. [Europe PMC free article] [Abstract] [Google Scholar]
- McKay LL, Baldwin KA. Stabilization of Lactose Metabolism in Streptococcus lactis C2. Appl Environ Microbiol. 1978 Aug;36(2):360–367. [Europe PMC free article] [Abstract] [Google Scholar]
- McKay LL, Baldwin KA, Efstathiou JD. Transductional evidence for plasmid linkage of lactose metabolism in streptococcus lactis C2. Appl Environ Microbiol. 1976 Jul;32(1):45–52. [Europe PMC free article] [Abstract] [Google Scholar]
- McKay LL, Baldwin KA, Walsh PM. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. [Europe PMC free article] [Abstract] [Google Scholar]
- McKay L, Miller A, 3rd, Sandine WE, Elliker PR. Mechanisms of lactose utilization by lactic acid streptococci: enzymatic and genetic analyses. J Bacteriol. 1970 Jun;102(3):804–809. [Europe PMC free article] [Abstract] [Google Scholar]
- Murphy E, Novick RP. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. [Abstract] [Google Scholar]
- Ohtsubo E, Zenilman M, Ohtsubo H, McCormick M, Machida C, Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. [Abstract] [Google Scholar]
- Snook RJ, McKay LL. Conjugal Transfer of Lactose-Fermenting Ability Among Streptococcus cremoris and Streptococcus lactis Strains. Appl Environ Microbiol. 1981 Nov;42(5):904–911. [Europe PMC free article] [Abstract] [Google Scholar]
- Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Site-specific recombination and its role in the life cycle of bacteriophage P1. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):297–309. [Abstract] [Google Scholar]
- Terzaghi BE, Sandine WE. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. [Europe PMC free article] [Abstract] [Google Scholar]
- Walsh PM, McKay LL. Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J Bacteriol. 1981 Jun;146(3):937–944. [Europe PMC free article] [Abstract] [Google Scholar]
- Walsh PM, McKay LL. Restriction endonuclease analysis of the lactose plasmid in Streptococcus lactis ML3 and two recombinant lactose plasmids. Appl Environ Microbiol. 1982 May;43(5):1006–1010. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.158.3.954-962.1984
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/158/3/954.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/158/3/954
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/158/3/954
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/130927101
Article citations
Evolved distal tail protein of skunaviruses facilitates adsorption to exopolysaccharide-encoding lactococci.
Microbiome Res Rep, 2(4):26, 18 Jul 2023
Cited by: 1 article | PMID: 38045920 | PMCID: PMC10688798
Host-encoded, cell surface-associated exopolysaccharide required for adsorption and infection by lactococcal P335 phage subtypes.
Front Microbiol, 13:971166, 04 Oct 2022
Cited by: 3 articles | PMID: 36267184 | PMCID: PMC9576995
Plasmid Complement of Lactococcus lactis NCDO712 Reveals a Novel Pilus Gene Cluster.
PLoS One, 11(12):e0167970, 12 Dec 2016
Cited by: 18 articles | PMID: 27941999 | PMCID: PMC5152845
An amazing journey.
Annu Rev Food Sci Technol, 6:1-17, 04 Feb 2015
Cited by: 2 articles | PMID: 25665173
Review
Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis.
PLoS One, 7(12):e51663, 11 Dec 2012
Cited by: 49 articles | PMID: 23240053 | PMCID: PMC3519859
Go to all (53) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Restriction endonuclease analysis of the lactose plasmid in Streptococcus lactis ML3 and two recombinant lactose plasmids.
Appl Environ Microbiol, 43(5):1006-1010, 01 May 1982
Cited by: 11 articles | PMID: 6285820 | PMCID: PMC244177
Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3.
J Bacteriol, 169(12):5481-5488, 01 Dec 1987
Cited by: 72 articles | PMID: 2824436 | PMCID: PMC213975
Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer.
Appl Environ Microbiol, 60(12):4413-4420, 01 Dec 1994
Cited by: 29 articles | PMID: 7811081 | PMCID: PMC202000
In vivo gene transfer systems and transposons.
Biochimie, 70(4):489-502, 01 Apr 1988
Cited by: 25 articles | PMID: 2844299
Review