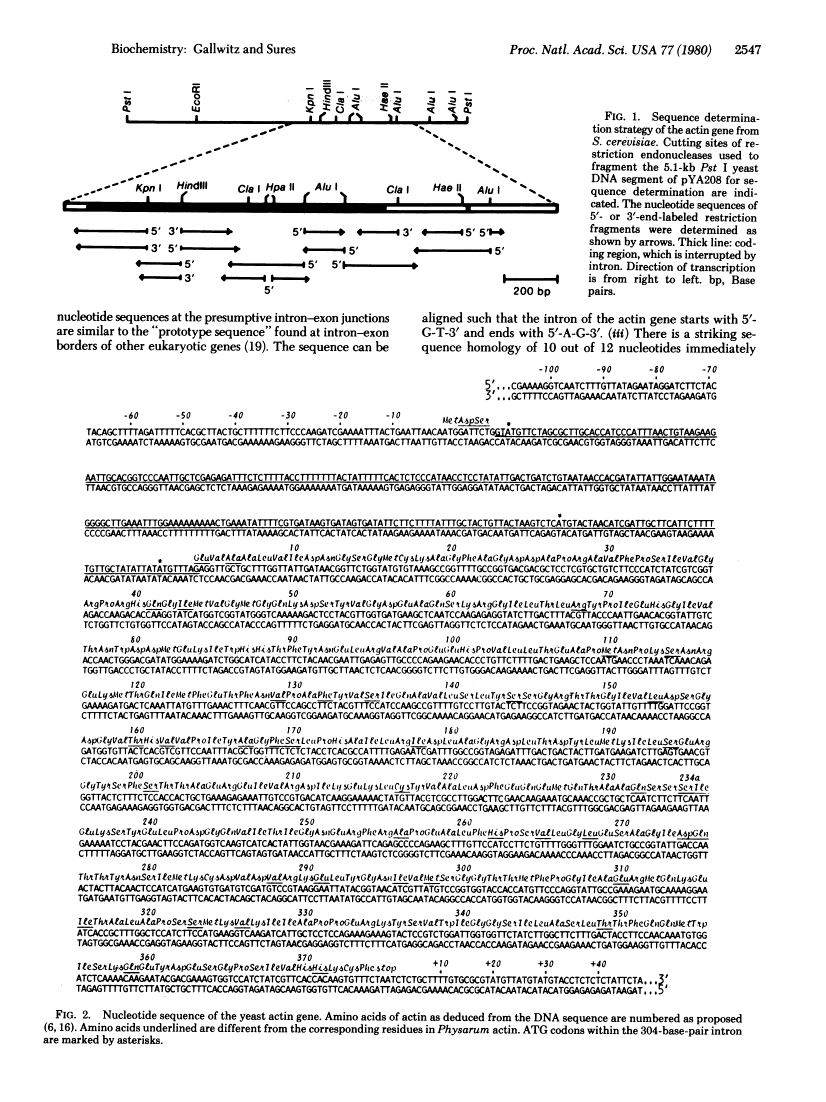
Abstract
Free full text

Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae.
Abstract
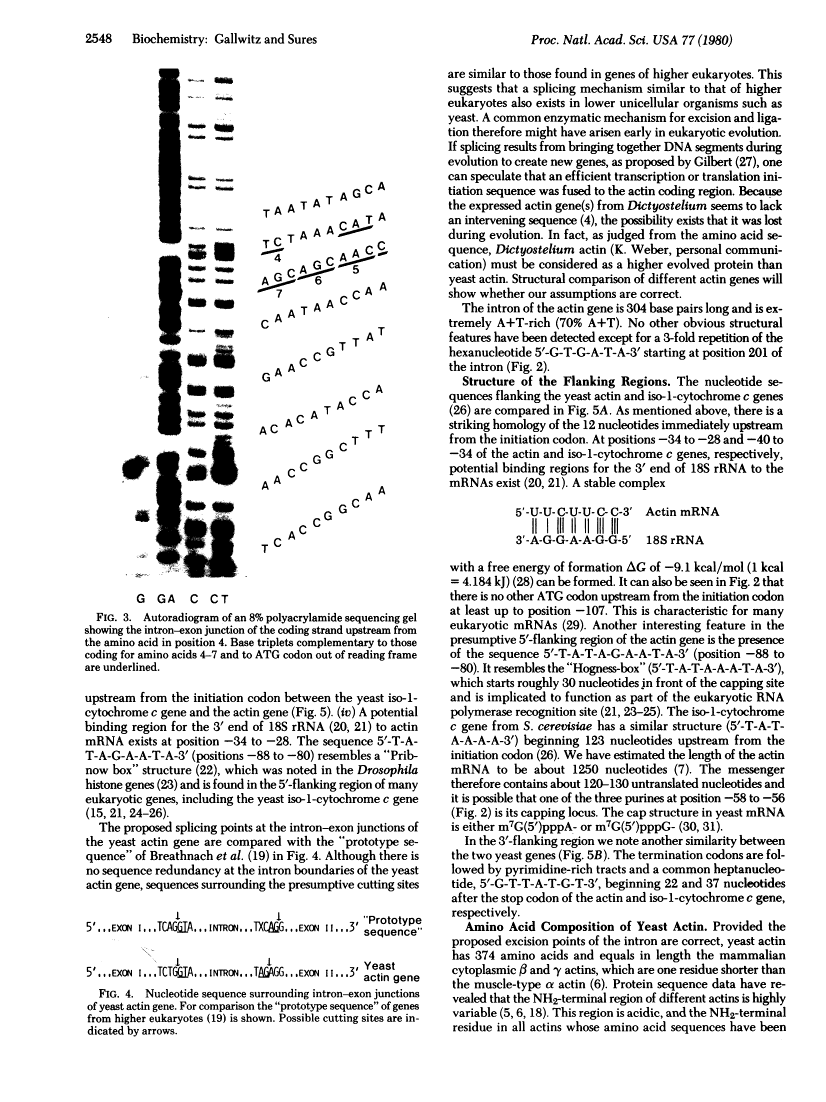
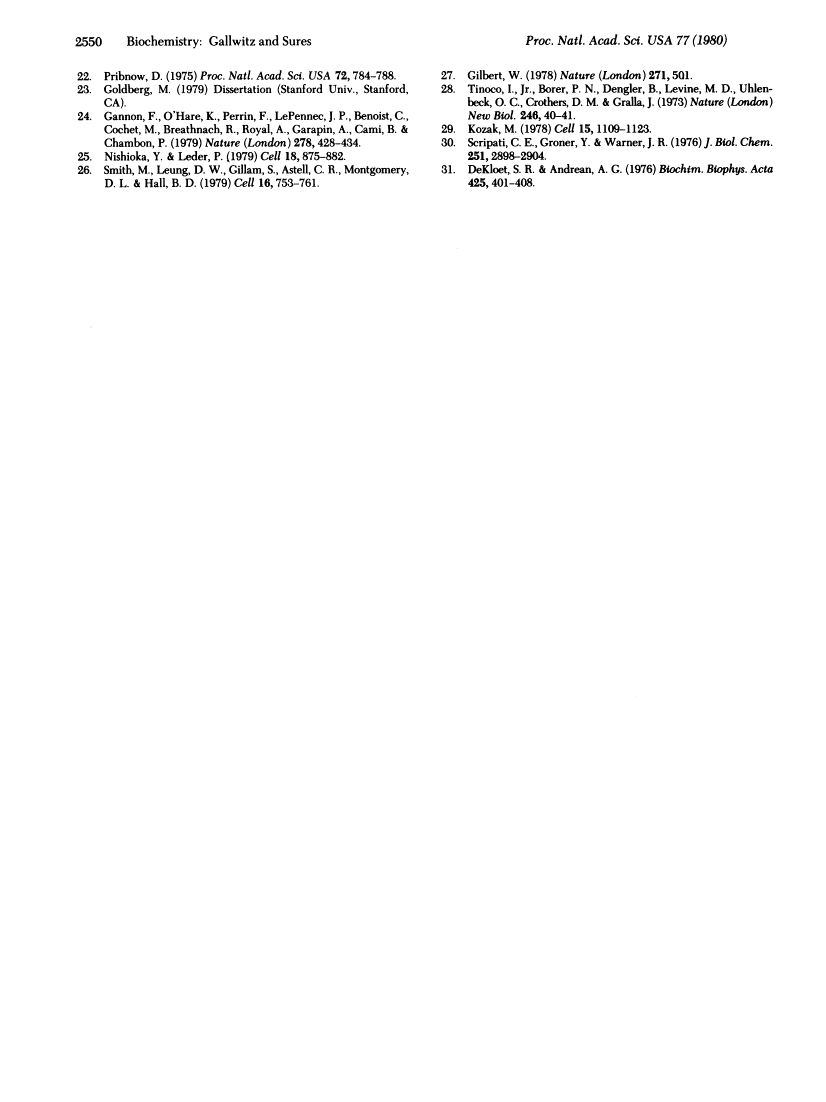
The complete nucleotide sequence of the actin gene from Saccharomyces cerevisiae has been determined. The coding region is interrupted by a 304-base-pair intervening sequence that is located within the triplet coding for amino acid 4. DNA sequences of the intron-exon junctions are similar to those found in higher eukaryotes and can be aligned such that the intron starts with the dinucleotide 5'-G-T-3' and ends with 5'-A-G-3'. Regions fo homology within the sequences upstream from the initiation codon and those following the termination codon have been detected between the yeast iso-1-cytochrome c gene and the actin gene. As deduced from the nucleotide sequence, yeast actin has 374 amino acid residues. Its primary structure, especially the NH2-terminal third of the protein, is highly conserved during evolution.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1006K), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Korn ED. Biochemistry of actomyosin-dependent cell motility (a review). Proc Natl Acad Sci U S A. 1978 Feb;75(2):588–599. [Europe PMC free article] [Abstract] [Google Scholar]
- Storti RV, Rich A. Chick cytoplasmic actin and muscle actin have different structural genes. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2346–2350. [Europe PMC free article] [Abstract] [Google Scholar]
- Kindle KL, Firtel RA. Identification and analysis of Dictyostelium actin genes, a family of moderately repeated genes. Cell. 1978 Nov;15(3):763–778. [Abstract] [Google Scholar]
- Vandekerckhove J, Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J Mol Biol. 1978 Dec 25;126(4):783–802. [Abstract] [Google Scholar]
- Vandekerckhove J, Weber K. The amino acid sequence of Physarum actin. Nature. 1978 Dec 14;276(5689):720–721. [Abstract] [Google Scholar]
- Gallwitz D, Seidel R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Mar 11;8(5):1043–1059. [Europe PMC free article] [Abstract] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith HO, Birnstiel ML. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. [Europe PMC free article] [Abstract] [Google Scholar]
- Maniatis T, Jeffrey A, van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. [Abstract] [Google Scholar]
- Richardson CC. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. [Europe PMC free article] [Abstract] [Google Scholar]
- Klenow H, Overgaard-Hansen K, Patkar SA. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Sures I, Levy S, Kedes LH. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu RC, Elzinga M. Partial amino acid sequence of brain actin and its homology with muscle actin. Biochemistry. 1977 Dec 27;16(26):5801–5806. [Abstract] [Google Scholar]
- Koteliansky VE, Glukhova MA, Bejanian MV, Surguchov AP, Smirnov VN. Isolation and characterization of actin-like protein from yeast Saccharomyces cerevisiae. FEBS Lett. 1979 Jun 1;102(1):55–58. [Abstract] [Google Scholar]
- Vandekerckhove J, Weber K. The complete amino acid sequence of actins from bovine aorta, bovine heart, bovine fast skeletal muscle, and rabbit slow skeletal muscle. A protein-chemical analysis of muscle actin differentiation. Differentiation. 1979;14(3):123–133. [Abstract] [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. [Europe PMC free article] [Abstract] [Google Scholar]
- Hagenbüchle O, Santer M, Steitz JA, Mans RJ. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. [Abstract] [Google Scholar]
- Lomedico P, Rosenthal N, Efstratidadis A, Gilbert W, Kolodner R, Tizard R. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell. 1979 Oct;18(2):545–558. [Abstract] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. [Europe PMC free article] [Abstract] [Google Scholar]
- Gannon F, O'Hare K, Perrin F, LePennec JP, Benoist C, Cochet M, Breathnach R, Royal A, Garapin A, Cami B, et al. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. [Abstract] [Google Scholar]
- Nishioka Y, Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. [Abstract] [Google Scholar]
- Smith M, Leung DW, Gillam S, Astell CR, Montgomery DL, Hall BD. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. [Abstract] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. [Abstract] [Google Scholar]
- Tinoco I, Jr, Borer PN, Dengler B, Levin MD, Uhlenbeck OC, Crothers DM, Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. [Abstract] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. [Abstract] [Google Scholar]
- Sripati CE, Groner Y, Warner JR. Methylated, blocked 5' termini of yeast mRNA. J Biol Chem. 1976 May 25;251(10):2898–2904. [Abstract] [Google Scholar]
- De Kloet SR, Andrean BA. Methylated nucleosides in polyadenylate-containing yeast messenger ribonucleic acid. Biochim Biophys Acta. 1976 Apr 2;425(4):401–408. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.77.5.2546
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc349438?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.77.5.2546
Article citations
Regulated pre-mRNA splicing: the ghostwriter of the eukaryotic genome.
Biochim Biophys Acta, 1819(6):538-545, 09 Jan 2012
Cited by: 15 articles | PMID: 22248620 | PMCID: PMC3345063
Review Free full text in Europe PMC
RNA splicing and debranching viewed through analysis of RNA lariats.
Mol Genet Genomics, 286(5-6):395-410, 08 Nov 2011
Cited by: 5 articles | PMID: 22065066
Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus).
J Clin Microbiol, 48(6):2154-2170, 31 Mar 2010
Cited by: 69 articles | PMID: 20357218 | PMCID: PMC2884488
RNA degradation in fission yeast mitochondria is stimulated by a member of a new family of proteins that are conserved in lower eukaryotes.
J Mol Biol, 367(3):681-691, 10 Jan 2007
Cited by: 5 articles | PMID: 17292401
The actin gene in Paracoccidioides brasiliensis: organization, expression and phylogenetic analyses.
Mycol Res, 111(pt 3):363-369, 04 Jan 2007
Cited by: 7 articles | PMID: 17363236
Go to all (212) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Isolation and sequence of the gene for actin in Saccharomyces cerevisiae.
Proc Natl Acad Sci U S A, 77(7):3912-3916, 01 Jul 1980
Cited by: 297 articles | PMID: 7001447 | PMCID: PMC349737
Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae.
Nucleic Acids Res, 13(11):3791-3804, 01 Jun 1985
Cited by: 46 articles | PMID: 3892483 | PMCID: PMC341278
The actin gene in yeast Saccharomyces cerevisiae: 5' and 3' end mapping, flanking and putative regulatory sequences.
Nucleic Acids Res, 9(23):6339-6350, 01 Dec 1981
Cited by: 41 articles | PMID: 6275358 | PMCID: PMC327607