Abstract
Free full text

Arbitrarily primed polymerase chain reaction as a rapid method to differentiate crossed from independent Pseudomonas cepacia infections in cystic fibrosis patients.
Abstract
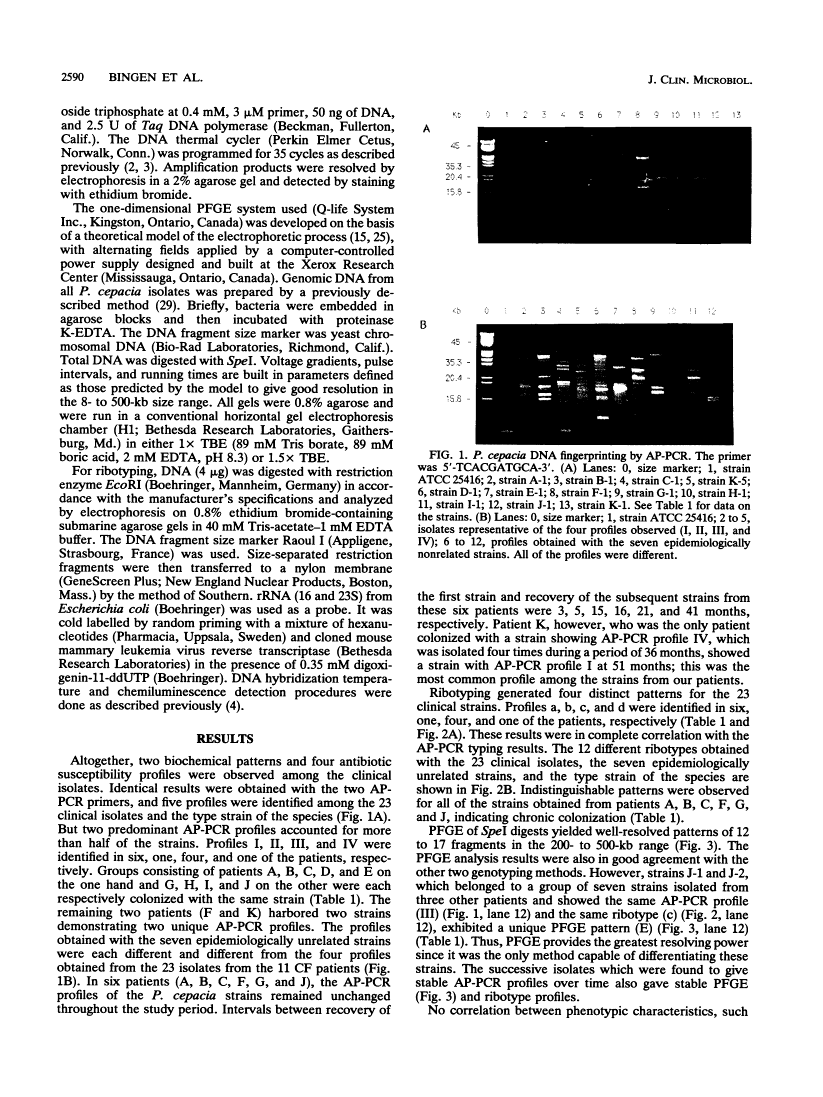
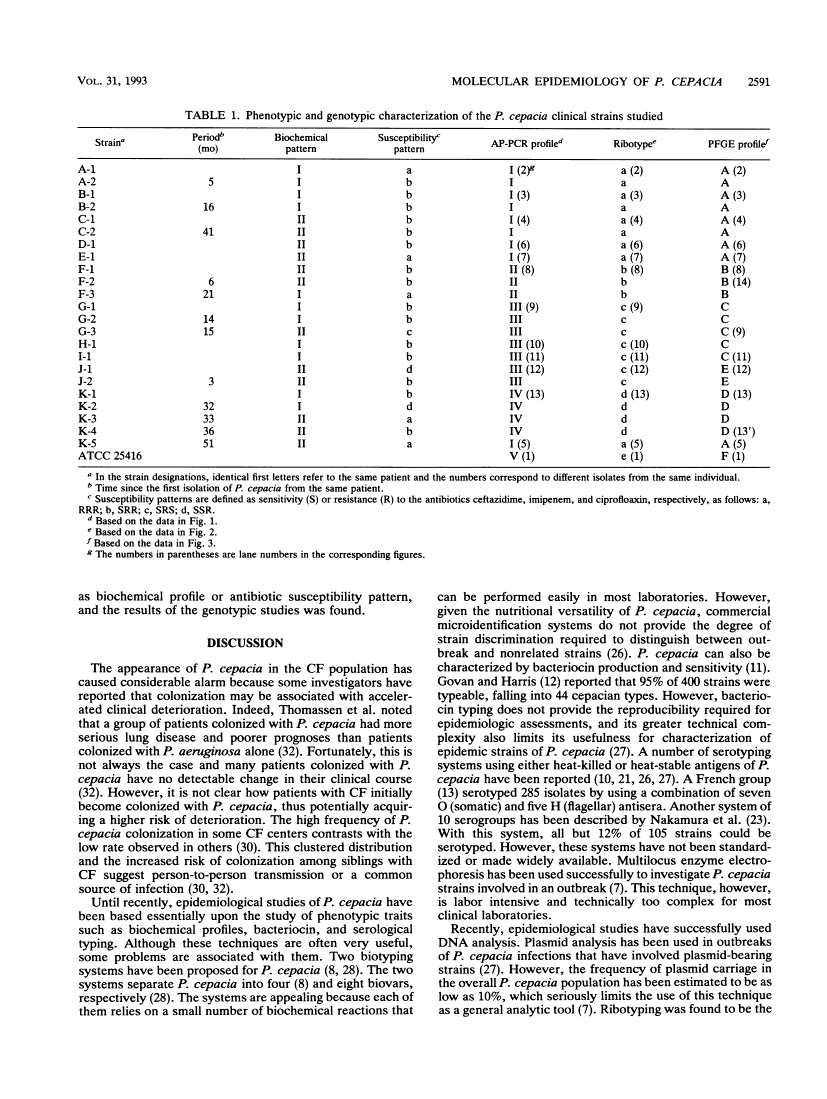
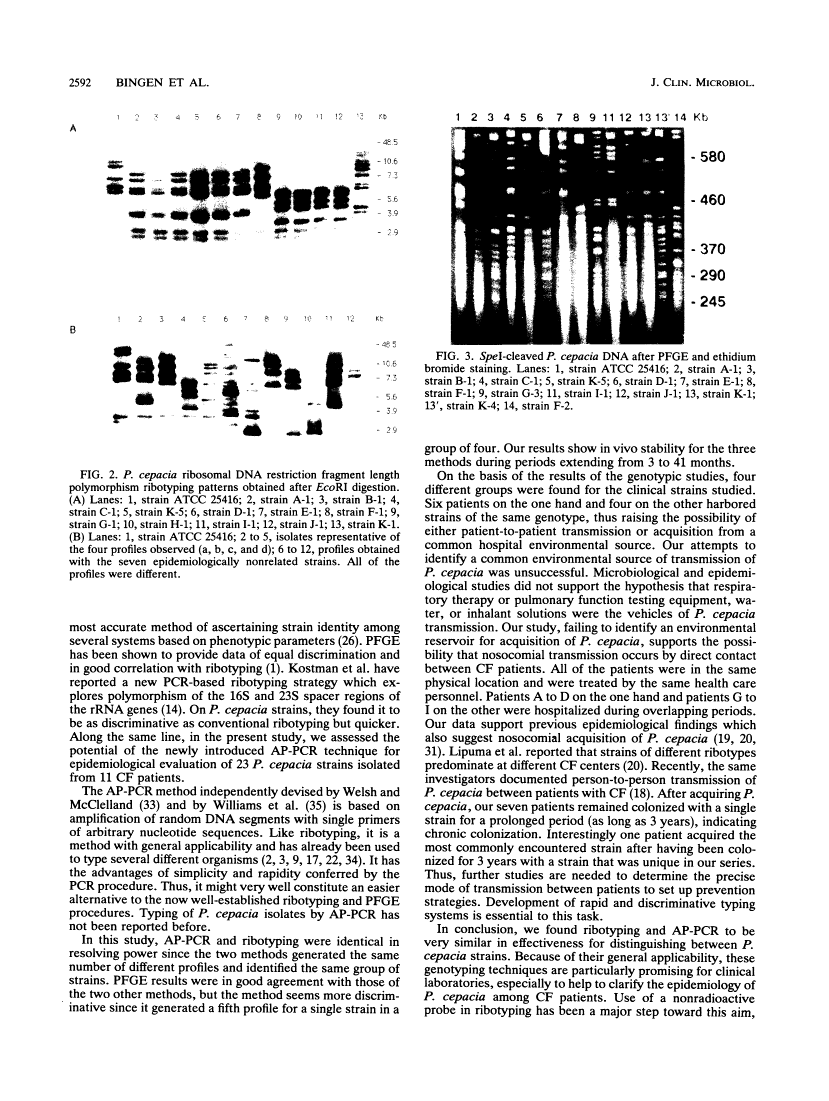
We used DNA fingerprinting by the arbitrarily primed polymerase chain reaction (AP-PCR) technique for an epidemiological investigation of 23 Pseudomonas cepacia isolates obtained from 11 cystic fibrosis (CF) patients attending our CF center. This approach was compared with ribotyping, pulsed-field gel electrophoresis (PFGE), and conventional phenotypic typing. AP-PCR and ribotyping were identical in resolving power, since the two methods generated four different profiles and identified the same group of strains. Six patients on the one hand and four on the other harbored strains of the same genotype, thus raising the possibility of either patient-to-patient transmission or acquisition from a common hospital environmental source. PFGE results were in good agreement with those of the other two methods, but PFGE seems more discriminative since it generated a fifth profile for a single strain in a group of four. Our results show in vivo stability for the three methods during a period extending from 3 to 41 months. These genotypic techniques are particularly promising for clinical laboratories to help to clarify the epidemiology of P. cepacia in CF patients. The AP-PCR method constitutes an easier alternative to the well-established ribotyping method. AP-PCR provides the quickest results with minimal technical complexity. However, our results suggest that it is less discriminative than the labor-intensive PFGE method.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson DJ, Kuhns JS, Vasil ML, Gerding DN, Janoff EN. DNA fingerprinting by pulsed field gel electrophoresis and ribotyping to distinguish Pseudomonas cepacia isolates from a nosocomial outbreak. J Clin Microbiol. 1991 Mar;29(3):648–649. [Europe PMC free article] [Abstract] [Google Scholar]
- Bingen E, Boissinot C, Desjardins P, Cave H, Brahimi N, Lambert-Zechovsky N, Denamur E, Blot P, Elion J. Arbitrarily primed polymerase chain reaction provides rapid differentiation of Proteus mirabilis isolates from a pediatric hospital. J Clin Microbiol. 1993 May;31(5):1055–1059. [Europe PMC free article] [Abstract] [Google Scholar]
- Bingen E, Cavé H, Aujard Y, Lambert-Zechovsky N, Desjardins P, Elion J, Denamur E. Molecular analysis of multiply recurrent meningitis due to Escherichia coli K1 in an infant. Clin Infect Dis. 1993 Jan;16(1):82–85. [Abstract] [Google Scholar]
- Bingen E, Denamur E, Lambert-Zechovsky N, Aujard Y, Brahimi N, Geslin P, Elion J. Analysis of DNA restriction fragment length polymorphism extends the evidence for breast milk transmission in Streptococcus agalactiae late-onset neonatal infection. J Infect Dis. 1992 Mar;165(3):569–573. [Abstract] [Google Scholar]
- Bingen E, Denamur E, Picard B, Goullet P, Lambert-Zechovsky N, Foucaud P, Navarro J, Elion J. Molecular epidemiological analysis of Pseudomonas aeruginosa strains causing failure of antibiotic therapy in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1992 May;11(5):432–437. [Abstract] [Google Scholar]
- Bingen EH, Denamur E, Lambert-Zechovsky NY, Bourdois A, Mariani-Kurkdjian P, Cezard JP, Navarro J, Elion J. DNA restriction fragment length polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltophilia bacteremia. J Clin Microbiol. 1991 Jul;29(7):1348–1350. [Europe PMC free article] [Abstract] [Google Scholar]
- Carson LA, Anderson RL, Panlilio AL, Beck-Sague CM, Miller JM. Isoenzyme analysis of Pseudomonas cepacia as an epidemiologic tool. Am J Med. 1991 Sep 16;91(3B):252S–255S. [Abstract] [Google Scholar]
- Esanu JG, Schubert RH. Zur Taxonomie und Nomenklatur von Pseudomonas cepacia. Zentralbl Bakteriol Orig A. 1973 Oct;224(4):478–483. [Abstract] [Google Scholar]
- Fekete A, Bantle JA, Halling SM, Stich RW. Amplification fragment length polymorphism in Brucella strains by use of polymerase chain reaction with arbitrary primers. J Bacteriol. 1992 Dec;174(23):7778–7783. [Europe PMC free article] [Abstract] [Google Scholar]
- Goldmann DA, Klinger JD. Pseudomonas cepacia: biology, mechanisms of virulence, epidemiology. J Pediatr. 1986 May;108(5 Pt 2):806–812. [Abstract] [Google Scholar]
- Gonzalez CF, Vidaver AK. Bacteriocin, plasmid and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. J Gen Microbiol. 1979 Jan;110(1):161–170. [Abstract] [Google Scholar]
- Govan JR, Harris G. Typing of Pseudomonas cepacia by bacteriocin susceptibility and production. J Clin Microbiol. 1985 Oct;22(4):490–494. [Europe PMC free article] [Abstract] [Google Scholar]
- Heidt A, Monteil H, Richard C. O and H serotyping of Pseudomonas cepacia. J Clin Microbiol. 1983 Sep;18(3):738–740. [Europe PMC free article] [Abstract] [Google Scholar]
- Kostman JR, Edlind TD, LiPuma JJ, Stull TL. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992 Aug;30(8):2084–2087. [Europe PMC free article] [Abstract] [Google Scholar]
- Lalande M, Noolandi J, Turmel C, Rousseau J, Slater GW. Pulsed-field electrophoresis: application of a computer model to the separation of large DNA molecules. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8011–8015. [Europe PMC free article] [Abstract] [Google Scholar]
- Larsen GY, Stull TL, Burns JL. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J Clin Microbiol. 1993 Apr;31(4):788–792. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehmann PF, Lin D, Lasker BA. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992 Dec;30(12):3249–3254. [Europe PMC free article] [Abstract] [Google Scholar]
- LiPuma JJ, Dasen SE, Nielson DW, Stern RC, Stull TL. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet. 1990 Nov 3;336(8723):1094–1096. [Abstract] [Google Scholar]
- LiPuma JJ, Fisher MC, Dasen SE, Mortensen JE, Stull TL. Ribotype stability of serial pulmonary isolates of Pseudomonas cepacia. J Infect Dis. 1991 Jul;164(1):133–136. [Abstract] [Google Scholar]
- LiPuma JJ, Mortensen JE, Dasen SE, Edlind TD, Schidlow DV, Burns JL, Stull TL. Ribotype analysis of Pseudomonas cepacia from cystic fibrosis treatment centers. J Pediatr. 1988 Nov;113(5):859–862. [Abstract] [Google Scholar]
- Mazurier SI, Wernars K. Typing of Listeria strains by random amplification of polymorphic DNA. Res Microbiol. 1992 Jun;143(5):499–505. [Abstract] [Google Scholar]
- Nakamura Y, Hyodo S, Chonan E, Shigeta S, Yabuuchi E. Serological classification of Pseudomonas cepacia by somatic antigen. J Clin Microbiol. 1986 Jul;24(1):152–154. [Europe PMC free article] [Abstract] [Google Scholar]
- Noolandi J, Slater GW, Lim HA, Viovy JL. Generalized tube model of biased reptation for gel electrophoresis of DNA. Science. 1989 Mar 17;243(4897):1456–1458. [Abstract] [Google Scholar]
- Rabkin CS, Jarvis WR, Anderson RL, Govan J, Klinger J, LiPuma J, Martone WJ, Monteil H, Richard C, Shigeta S, et al. Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev Infect Dis. 1989 Jul-Aug;11(4):600–607. [Abstract] [Google Scholar]
- Rabkin CS, Jarvis WR, Martone WJ. Current status of Pseudomonas cepacia typing systems. Eur J Epidemiol. 1987 Dec;3(4):343–346. [Abstract] [Google Scholar]
- Richard C, Monteil H, Mégraud F, Chatelain R, Laurent B. Caractères phénotypiques de 100 souches de Pseudomonas cepacia. Proposition d'un schéma de biovars. Ann Biol Clin (Paris) 1981;39(1):9–15. [Abstract] [Google Scholar]
- Tablan OC, Chorba TL, Schidlow DV, White JW, Hardy KA, Gilligan PH, Morgan WM, Carson LA, Martone WJ, Jason JM, et al. Pseudomonas cepacia colonization in patients with cystic fibrosis: risk factors and clinical outcome. J Pediatr. 1985 Sep;107(3):382–387. [Abstract] [Google Scholar]
- Taylor RF, Dalla Costa L, Kaufmann ME, Pitt TL, Hodson ME. Pseudomonas cepacia pulmonary infection in adults with cystic fibrosis: is nosocomial acquisition occurring? J Hosp Infect. 1992 Jul;21(3):199–204. [Abstract] [Google Scholar]
- Thomassen MJ, Demko CA, Klinger JD, Stern RC. Pseudomonas cepacia colonization among patients with cystic fibrosis. A new opportunist. Am Rev Respir Dis. 1985 May;131(5):791–796. [Abstract] [Google Scholar]
- Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. [Europe PMC free article] [Abstract] [Google Scholar]
- Welsh J, Pretzman C, Postic D, Saint Girons I, Baranton G, McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992 Jul;42(3):370–377. [Abstract] [Google Scholar]
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.31.10.2589-2593.1993
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/31/10/2589.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/31/10/2589
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/31/10/2589.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.31.10.2589-2593.1993
Article citations
Utility of random amplification of polymorphic DNA assay and BOX-A PCR in molecular characterization of Streptococcus pneumoniae isolates recovered from various ophthalmic infections.
Ophthalmic Res, 38(1):36-43, 13 Oct 2005
Cited by: 2 articles | PMID: 16224199
Species distribution and ribotype diversity of Burkholderia cepacia complex isolates from French patients with cystic fibrosis.
J Clin Microbiol, 42(10):4824-4827, 01 Oct 2004
Cited by: 27 articles | PMID: 15472352 | PMCID: PMC522310
[Epidemiology of nosocomial infections due to Pseudomonas aeruginosa, Burkholderia cepacia and Stenotrophomonas maltophilia].
Pathol Biol (Paris), 53(6):341-348, 01 Jul 2005
Cited by: 15 articles | PMID: 16004946
Review
Epidemiologic investigation of Burkholderia cepacia acquisition in two pediatric intensive care units.
Infect Control Hosp Epidemiol, 24(9):707-710, 01 Sep 2003
Cited by: 18 articles | PMID: 14510255
Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex.
J Clin Microbiol, 39(3):1073-1078, 01 Mar 2001
Cited by: 84 articles | PMID: 11230429 | PMCID: PMC87875
Go to all (31) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Investigation of nosocomial respiratory infection due to Pseudomonas cepacia by arbitrarily primed polymerase chain reaction.
Diagn Microbiol Infect Dis, 23(3):77-83, 01 Nov 1995
Cited by: 2 articles | PMID: 8849650
Burkholderia (Pseudomonas) cepacia and cystic fibrosis: the epidemiology in Belgium.
Acta Clin Belg, 51(4):222-230, 01 Jan 1996
Cited by: 16 articles | PMID: 8858887
Comparison of different PCR approaches for characterization of Burkholderia (Pseudomonas) cepacia isolates.
J Clin Microbiol, 33(12):3304-3307, 01 Dec 1995
Cited by: 21 articles | PMID: 8586722 | PMCID: PMC228693
Transmissibility of Pseudomonas cepacia infection in clinic patients and lung-transplant recipients with cystic fibrosis.
N Engl J Med, 331(15):981-987, 01 Oct 1994
Cited by: 65 articles | PMID: 7521938