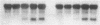Abstract
Free full text

The origin recognition complex in silencing, cell cycle progression, and DNA replication.
Abstract
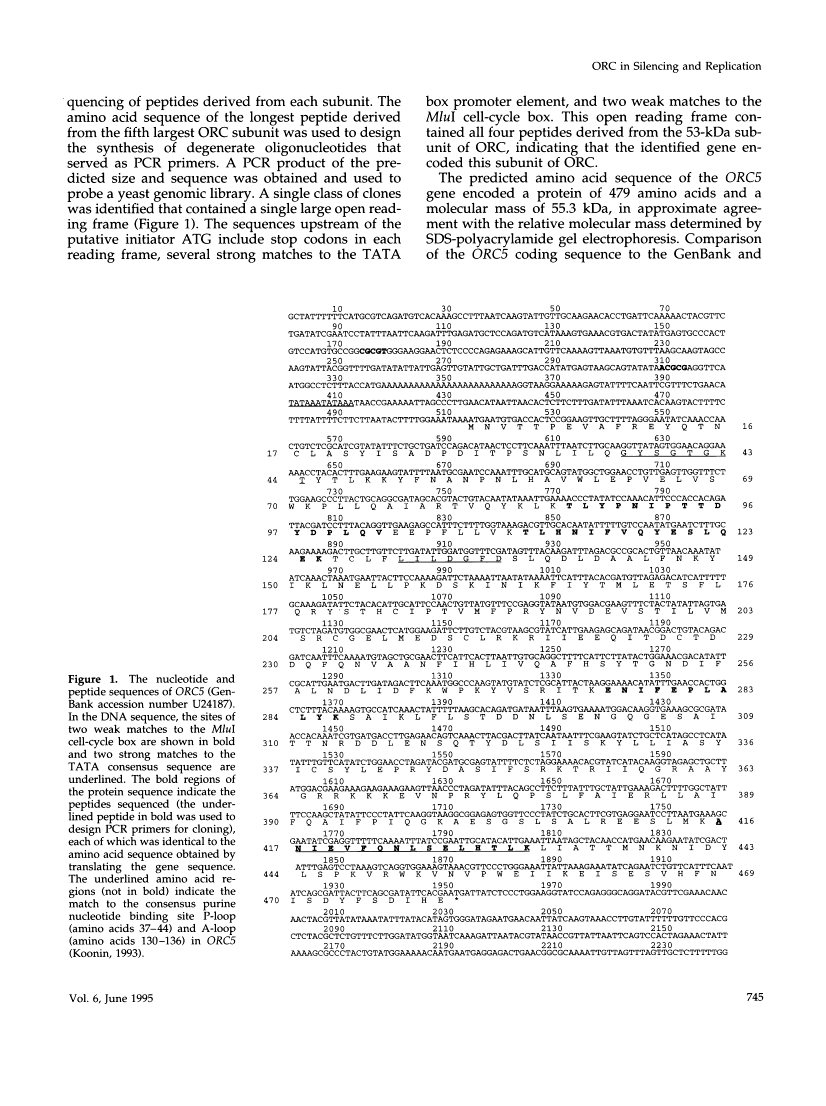
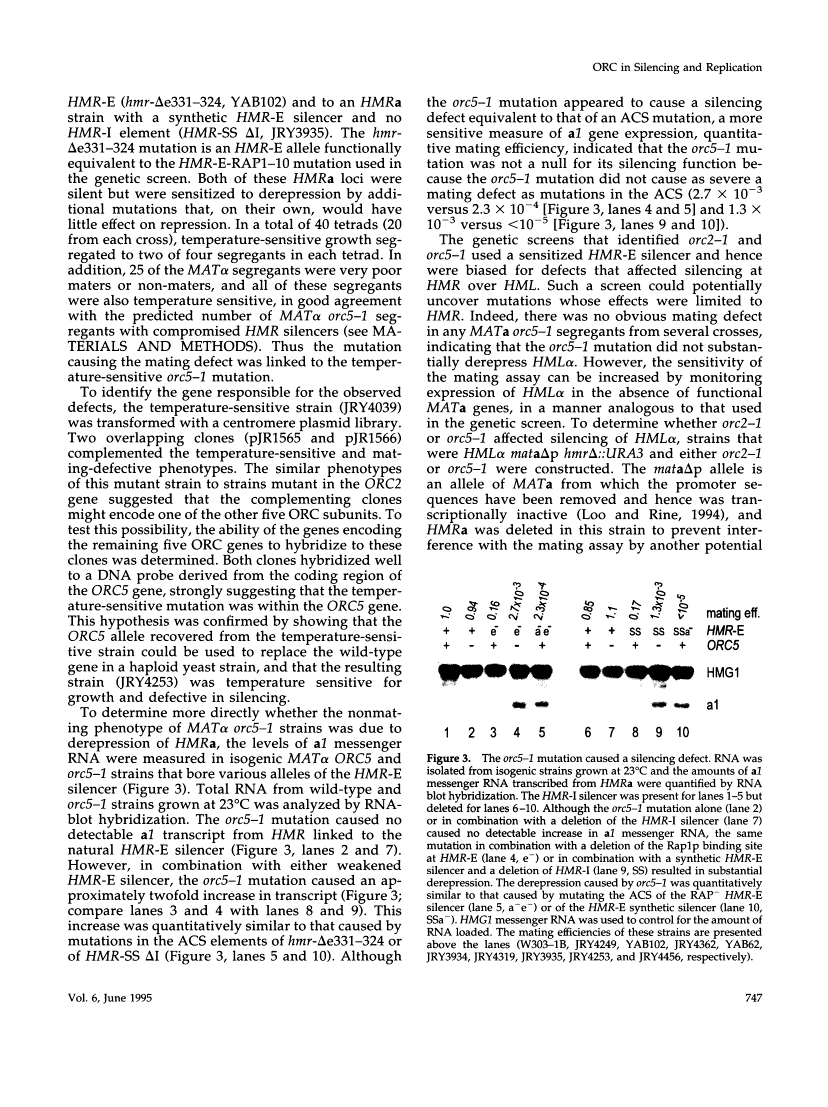
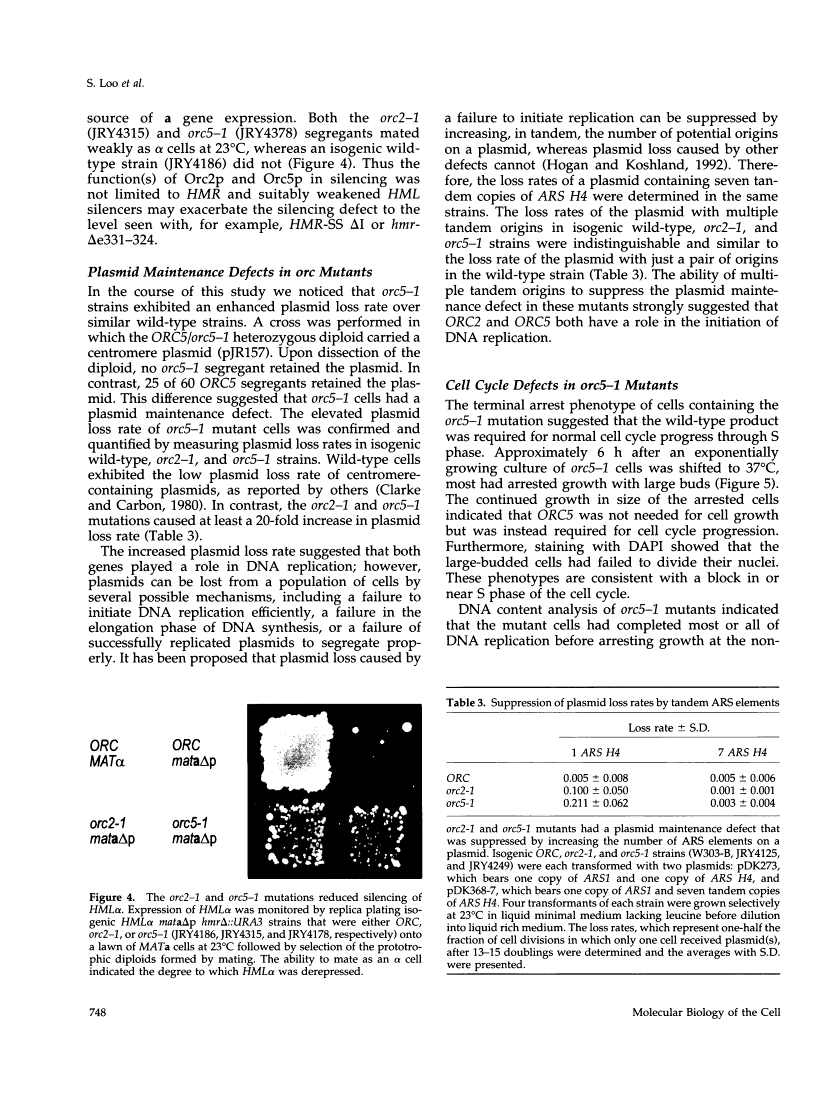
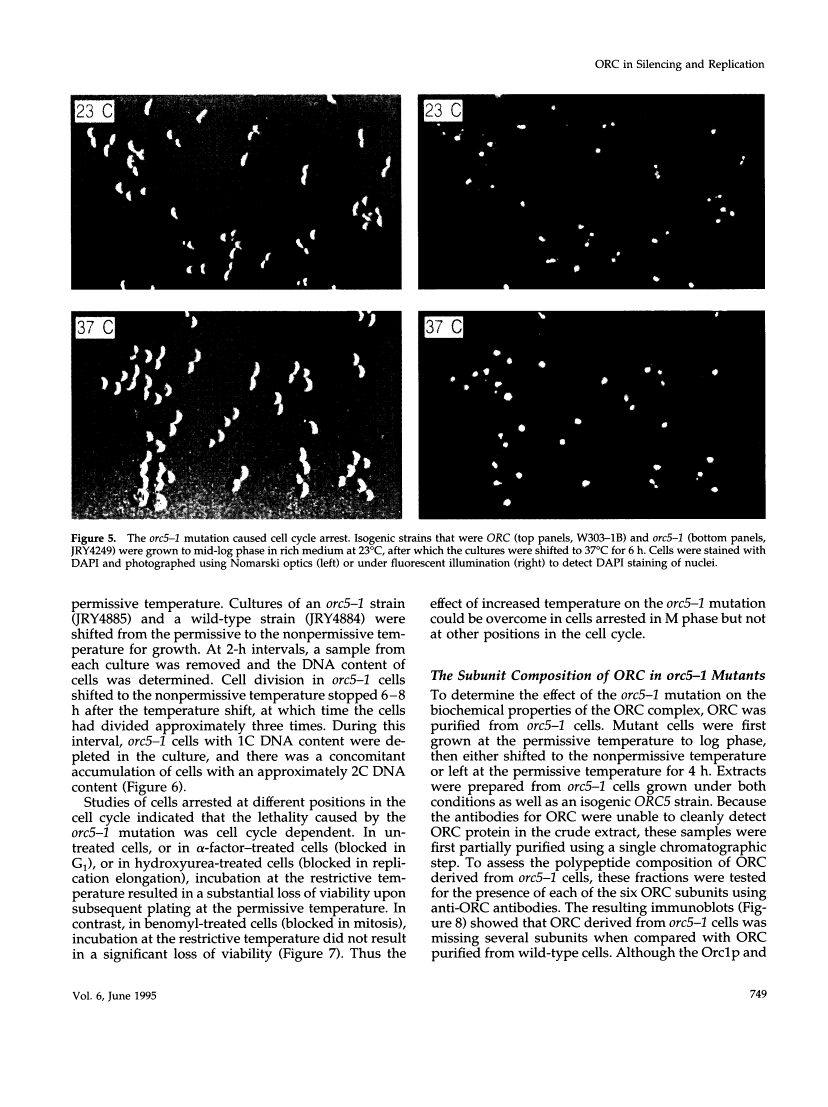
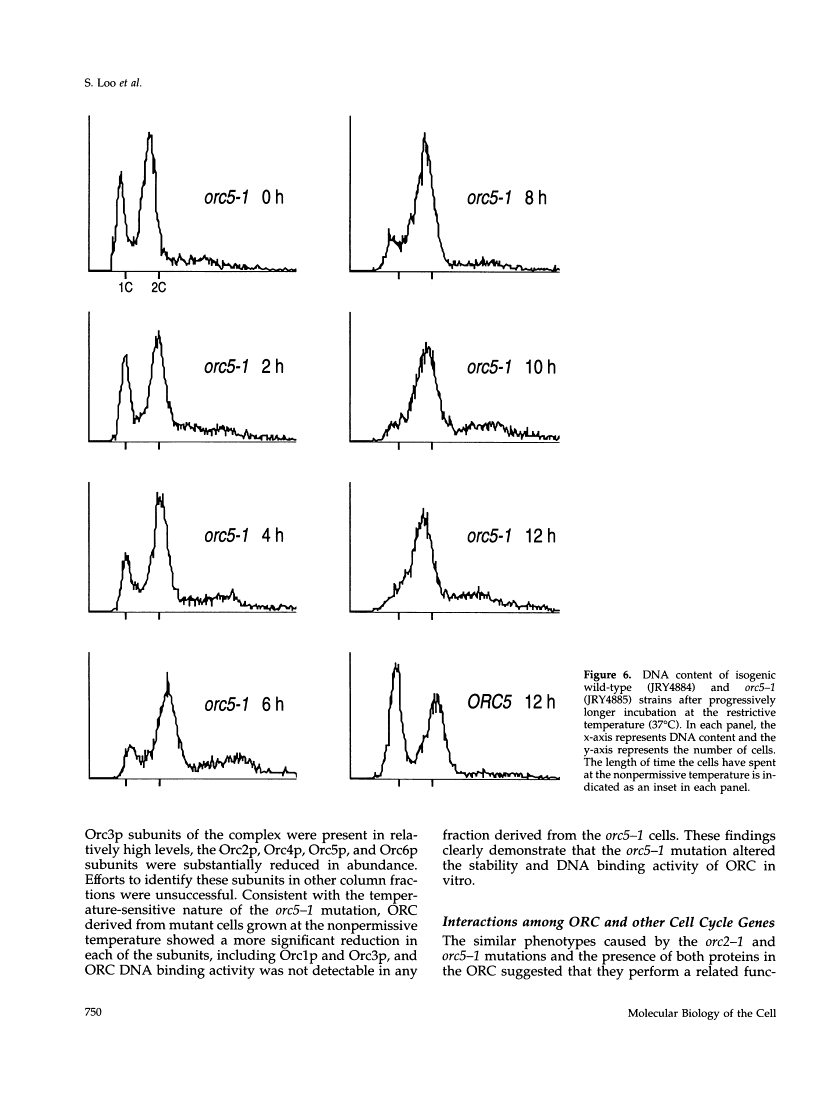
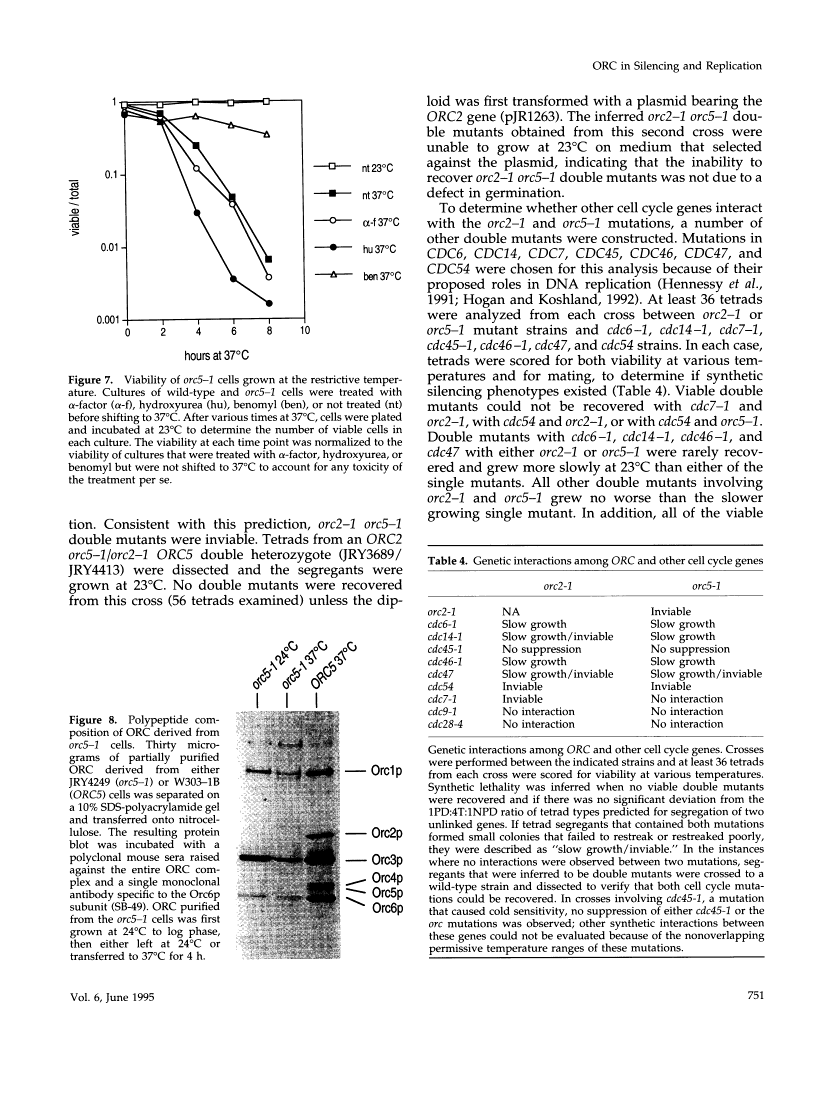
This report describes the isolation of ORC5, the gene encoding the fifth largest subunit of the origin recognition complex, and the properties of mutants with a defective allele of ORC5. The orc5-1 mutation caused temperature-sensitive growth and, at the restrictive temperature, caused cell cycle arrest. At the permissive temperature, the orc5-1 mutation caused an elevated plasmid loss rate that could be suppressed by additional tandem origins of DNA replication. The sequence of ORC5 revealed a potential ATP binding site, making Orc5p a candidate for a subunit that mediates the ATP-dependent binding of ORC to origins. Genetic interactions among orc2-1 and orc5-1 and other cell cycle genes provided further evidence for a role for the origin recognition complex (ORC) in DNA replication. The silencing defect caused by orc5-1 strengthened previous connections between ORC and silencing, and combined with the phenotypes caused by orc2 mutations, suggested that the complex itself functions in both processes.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (3.7M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984 Jul 5;176(3):307–331. [Abstract] [Google Scholar]
- Axelrod A, Rine J. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):1080–1091. [Europe PMC free article] [Abstract] [Google Scholar]
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. [Abstract] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. [Abstract] [Google Scholar]
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. [Abstract] [Google Scholar]
- Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. [Abstract] [Google Scholar]
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991 Sep;5(9):1589–1600. [Abstract] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993 Nov 5;75(3):531–541. [Abstract] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. [Abstract] [Google Scholar]
- Culotti J, Hartwell LH. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp Cell Res. 1971 Aug;67(2):389–401. [Abstract] [Google Scholar]
- Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. [Abstract] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. [Abstract] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994 Aug 26;265(5176):1243–1246. [Abstract] [Google Scholar]
- Dubey DD, Davis LR, Greenfeder SA, Ong LY, Zhu JG, Broach JR, Newlon CS, Huberman JA. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991 Oct;11(10):5346–5355. [Europe PMC free article] [Abstract] [Google Scholar]
- Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992 Jan;12(1):155–163. [Europe PMC free article] [Abstract] [Google Scholar]
- Foss M, McNally FJ, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993 Dec 17;262(5141):1838–1844. [Abstract] [Google Scholar]
- Foss M, Rine J. Molecular definition of the PAS1-1 mutation which affects silencing in Saccharomyces cerevisiae. Genetics. 1993 Nov;135(3):931–935. [Europe PMC free article] [Abstract] [Google Scholar]
- Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995 Apr 15;9(8):911–924. [Abstract] [Google Scholar]
- Fox TD, Folley LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, Costanzo MC. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. [Abstract] [Google Scholar]
- George JW, Brosh RM, Jr, Matson SW. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994 Jan 14;235(2):424–435. [Abstract] [Google Scholar]
- Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4062–4065. [Europe PMC free article] [Abstract] [Google Scholar]
- Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991 Sep;10(9):2707–2715. [Europe PMC free article] [Abstract] [Google Scholar]
- Hennessy KM, Clark CD, Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. [Abstract] [Google Scholar]
- Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. [Abstract] [Google Scholar]
- Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990 Jul;9(7):2321–2329. [Europe PMC free article] [Abstract] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. [Abstract] [Google Scholar]
- Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3098–3102. [Europe PMC free article] [Abstract] [Google Scholar]
- Hollingsworth RE, Jr, Sclafani RA. DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6272–6276. [Europe PMC free article] [Abstract] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. [Europe PMC free article] [Abstract] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993 May;13(5):2899–2908. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6286–6290. [Europe PMC free article] [Abstract] [Google Scholar]
- Kassir Y, Simchen G. Regulation of mating and meiosis in yeast by the mating-type region. Genetics. 1976 Feb;82(2):187–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. [Abstract] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993 Jun 11;21(11):2541–2547. [Europe PMC free article] [Abstract] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992 Dec;56(4):543–560. [Europe PMC free article] [Abstract] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994 Jun 17;264(5166):1768–1771. [Abstract] [Google Scholar]
- McNally FJ, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Nov;11(11):5648–5659. [Europe PMC free article] [Abstract] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990 Feb 16;247(4944):841–845. [Abstract] [Google Scholar]
- Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JF. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993 Nov 4;366(6450):87–89. [Abstract] [Google Scholar]
- Miller AM, Nasmyth KA. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984 Nov 15;312(5991):247–251. [Abstract] [Google Scholar]
- Mortimer RK, Contopoulou CR, King JS. Genetic and physical maps of Saccharomyces cerevisiae, Edition 11. Yeast. 1992 Oct;8(10):817–902. [Abstract] [Google Scholar]
- Nasmyth KA. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. [Abstract] [Google Scholar]
- Park EC, Szostak JW. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990 Sep;10(9):4932–4934. [Europe PMC free article] [Abstract] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989 Nov 17;59(4):637–647. [Abstract] [Google Scholar]
- Rivier DH, Rine J. An origin of DNA replication and a transcription silencer require a common element. Science. 1992 May 1;256(5057):659–663. [Abstract] [Google Scholar]
- Rowley A, Dowell SJ, Diffley JF. Recent developments in the initiation of chromosomal DNA replication: a complex picture emerges. Biochim Biophys Acta. 1994 Apr 6;1217(3):239–256. [Abstract] [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheng Z, Charbonneau H. The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J Biol Chem. 1993 Mar 5;268(7):4728–4733. [Abstract] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):237–249. [Europe PMC free article] [Abstract] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994 May 19;369(6477):245–247. [Abstract] [Google Scholar]
- Wan J, Xu H, Grunstein M. CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem. 1992 Jun 5;267(16):11274–11280. [Abstract] [Google Scholar]
- Yan H, Merchant AM, Tye BK. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993 Nov;7(11):2149–2160. [Abstract] [Google Scholar]
Associated Data
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.6.6.741
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc301233?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1091/mbc.6.6.741
Article citations
An essential Noc3p dimerization cycle mediates ORC double-hexamer formation in replication licensing.
Life Sci Alliance, 6(3):e202201594, 04 Jan 2023
Cited by: 0 articles | PMID: 36599624 | PMCID: PMC9813392
Circulating Placental Alkaline Phosphatase Expressing Exosomes in Maternal Blood Showed Temporal Regulation of Placental Genes.
Front Med (Lausanne), 8:758971, 24 Dec 2021
Cited by: 1 article | PMID: 35004728 | PMCID: PMC8739800
A human cancer cell line initiates DNA replication normally in the absence of ORC5 and ORC2 proteins.
J Biol Chem, 295(50):16949-16959, 28 Sep 2020
Cited by: 14 articles | PMID: 32989049 | PMCID: PMC7863895
Biochemical and functional characterization of a meiosis-specific Pch2/ORC AAA+ assembly.
Life Sci Alliance, 3(11):e201900630, 21 Aug 2020
Cited by: 4 articles | PMID: 32826290 | PMCID: PMC7442955
The interaction networks of the budding yeast and human DNA replication-initiation proteins.
Cell Cycle, 18(6-7):723-741, 19 Mar 2019
Cited by: 1 article | PMID: 30890025 | PMCID: PMC6464591
Go to all (123) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae.
Science, 262(5141):1838-1844, 01 Dec 1993
Cited by: 203 articles | PMID: 8266071
Roles for ORC in M phase and S phase.
Science, 279(5357):1733-1737, 01 Mar 1998
Cited by: 33 articles | PMID: 9497294
Analysis on origin recognition complex containing Orc5p with defective Walker A motif.
J Biol Chem, 279(9):8469-8477, 18 Nov 2003
Cited by: 11 articles | PMID: 14625297
The origin replication complex (ORC): the stone that kills two birds.
Bioessays, 16(4):233-235, 01 Apr 1994
Cited by: 0 articles | PMID: 8031299
Review
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: AI-20460
NIEHS NIH HHS (1)
Grant ID: P30ESO1896-12
NIGMS NIH HHS (1)
Grant ID: GM-31105