Abstract
Free full text

Evaluation of Biolog system for identification of some gram-negative bacteria of clinical importance.
Abstract
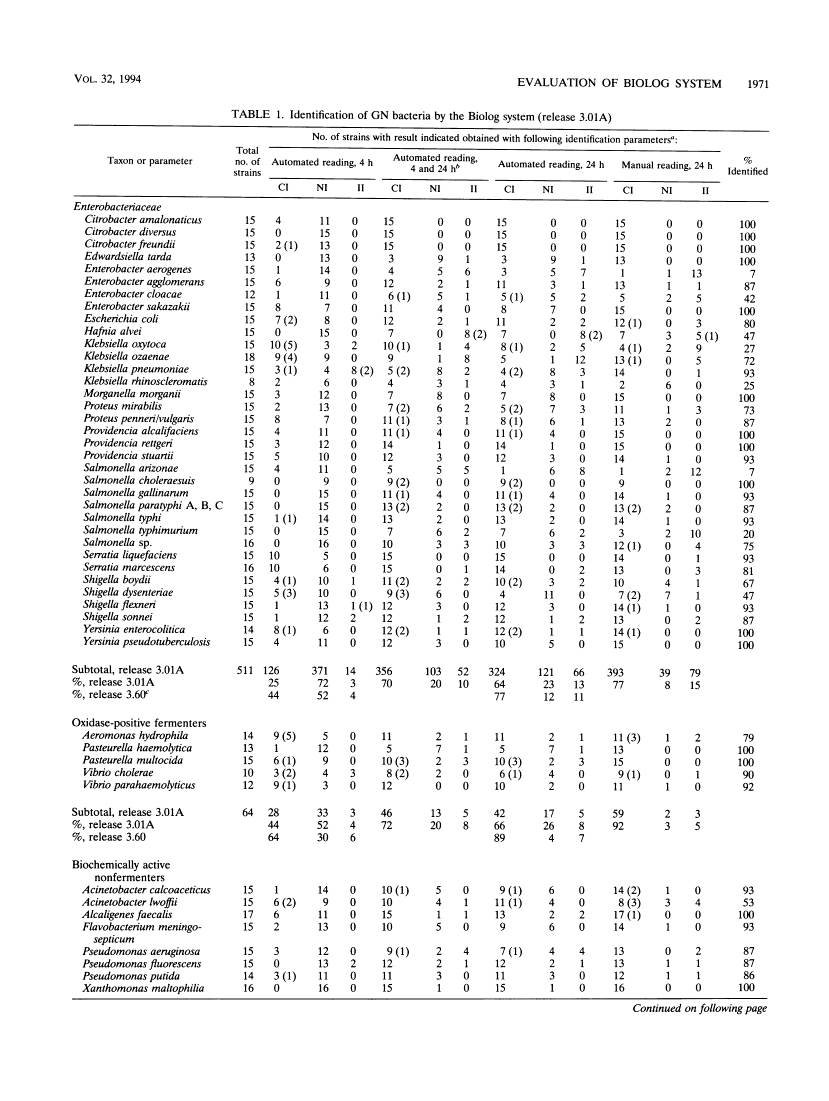
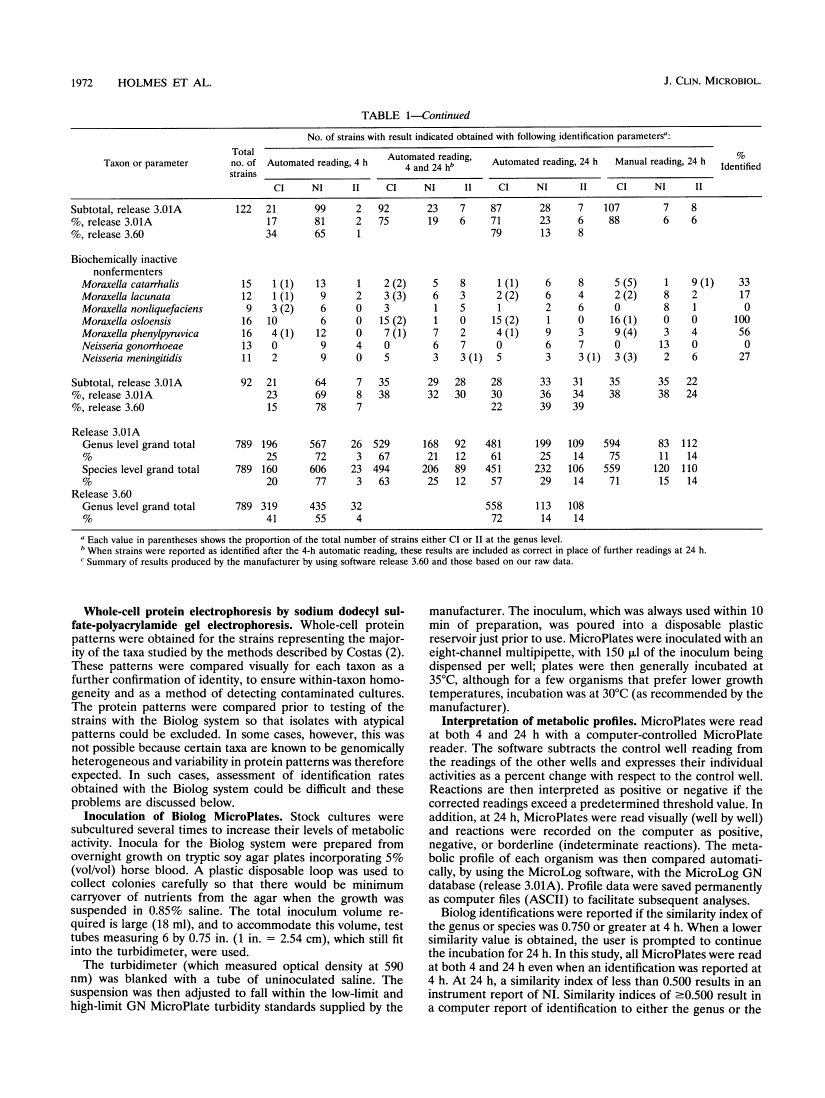
The Biolog system (Biolog, Inc., Hayward, Calif.) was evaluated for the identification of 55 gram-negative taxa (789 strains) likely to be encountered commonly in clinical laboratories. The Biolog system performed best with oxidase-positive fermenters and biochemically active nonfermenters and had the most problems with unreactive nonfermenters. It gave significantly better results when the MicroPlates were read manually rather than when they were read by the automated reader. Plates read manually gave the following performances: oxidase-positive fermenters, five taxa, 64 strains, 92% correct, 3% not identified, and 5% incorrect; biochemically active nonfermenters, eight taxa, 122 strains, 88% correct, 6% not identified, and 6% incorrect; members of the family Enterobacteriaceae, 35 taxa, 511 strains, 77% correct, 8% not identified, and 15% incorrect; unreactive nonfermenters, seven taxa, 92 strains, 38% correct, 24% not identified, and 38% incorrect. We found the system easy to use, but while for 39 of 55 of the taxa an identification rate of > 70% was achieved, problems were encountered, particularly with identification of capsulated strains of some Enterobacter and Klebsiella taxa, as well as the least biochemically active Moraxella and Neisseria strains.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carnahan AM, Joseph SW, Janda JM. Species identification of Aeromonas strains based on carbon substrate oxidation profiles. J Clin Microbiol. 1989 Sep;27(9):2128–2129. [Europe PMC free article] [Abstract] [Google Scholar]
- Holmes B, Dawson CA, Pinning CA. A revised probability matrix for the identification of gram-negative, aerobic, rod-shaped, fermentative bacteria. J Gen Microbiol. 1986 Nov;132(11):3113–3135. [Abstract] [Google Scholar]
- Holmes B, Pinning CA, Dawson CA. A probability matrix for the identification of gram-negative, aerobic, non-fermentative bacteria that grow on nutrient agar. J Gen Microbiol. 1986 Jul;132(7):1827–1842. [Abstract] [Google Scholar]
- Klingler JM, Stowe RP, Obenhuber DC, Groves TO, Mishra SK, Pierson DL. Evaluation of the Biolog automated microbial identification system. Appl Environ Microbiol. 1992 Jun;58(6):2089–2092. [Europe PMC free article] [Abstract] [Google Scholar]
- Miller JM, Rhoden DL. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J Clin Microbiol. 1991 Jun;29(6):1143–1147. [Europe PMC free article] [Abstract] [Google Scholar]
- Stager CE, Davis JR. Automated systems for identification of microorganisms. Clin Microbiol Rev. 1992 Jul;5(3):302–327. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.32.8.1970-1975.1994
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/32/8/1970.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/32/8/1970
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/32/8/1970.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Basic concepts, recent advances, and future perspectives in the diagnosis of bovine mastitis.
J Vet Sci, 25(1):e18, 01 Jan 2024
Cited by: 0 articles | PMID: 38311330 | PMCID: PMC10839174
Review Free full text in Europe PMC
Statistical prediction of microbial metabolic traits from genomes.
PLoS Comput Biol, 19(12):e1011705, 19 Dec 2023
Cited by: 3 articles | PMID: 38113208 | PMCID: PMC10729968
Strong alkaline electrolyzed water efficiently inactivates SARS-CoV-2, other viruses, and Gram-negative bacteria.
Biochem Biophys Res Commun, 575:36-41, 23 Aug 2021
Cited by: 7 articles | PMID: 34455219 | PMCID: PMC8381626
Comparative Genome Analysis of the Lignocellulose Degrading Bacteria Citrobacter freundii so4 and Sphingobacterium multivorum w15.
Front Microbiol, 11:248, 03 Mar 2020
Cited by: 9 articles | PMID: 32194522 | PMCID: PMC7065263
Monomorium ant is a carrier for pathogenic and potentially pathogenic bacteria.
BMC Res Notes, 12(1):230, 16 Apr 2019
Cited by: 3 articles | PMID: 30992046 | PMCID: PMC6469133
Go to all (30) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evaluation of two BBL Crystal systems for identification of some clinically important gram-negative bacteria.
J Clin Microbiol, 32(9):2221-2224, 01 Sep 1994
Cited by: 26 articles | PMID: 7814550 | PMCID: PMC263971
Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification.
J Clin Microbiol, 29(6):1143-1147, 01 Jun 1991
Cited by: 44 articles | PMID: 1864931 | PMCID: PMC269959
Evaluation of RapID onE system for identification of 379 strains in the family Enterobacteriaceae and oxidase-negative, gram-negative nonfermenters.
J Clin Microbiol, 32(4):931-934, 01 Apr 1994
Cited by: 10 articles | PMID: 8027345 | PMCID: PMC263165
Evaluation of the MicroScan rapid neg ID3 panel for identification of Enterobacteriaceae and some common gram-negative nonfermenters.
J Clin Microbiol, 38(10):3577-3580, 01 Oct 2000
Cited by: 7 articles | PMID: 11015366 | PMCID: PMC87439