Abstract
Free full text

DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain.
Abstract
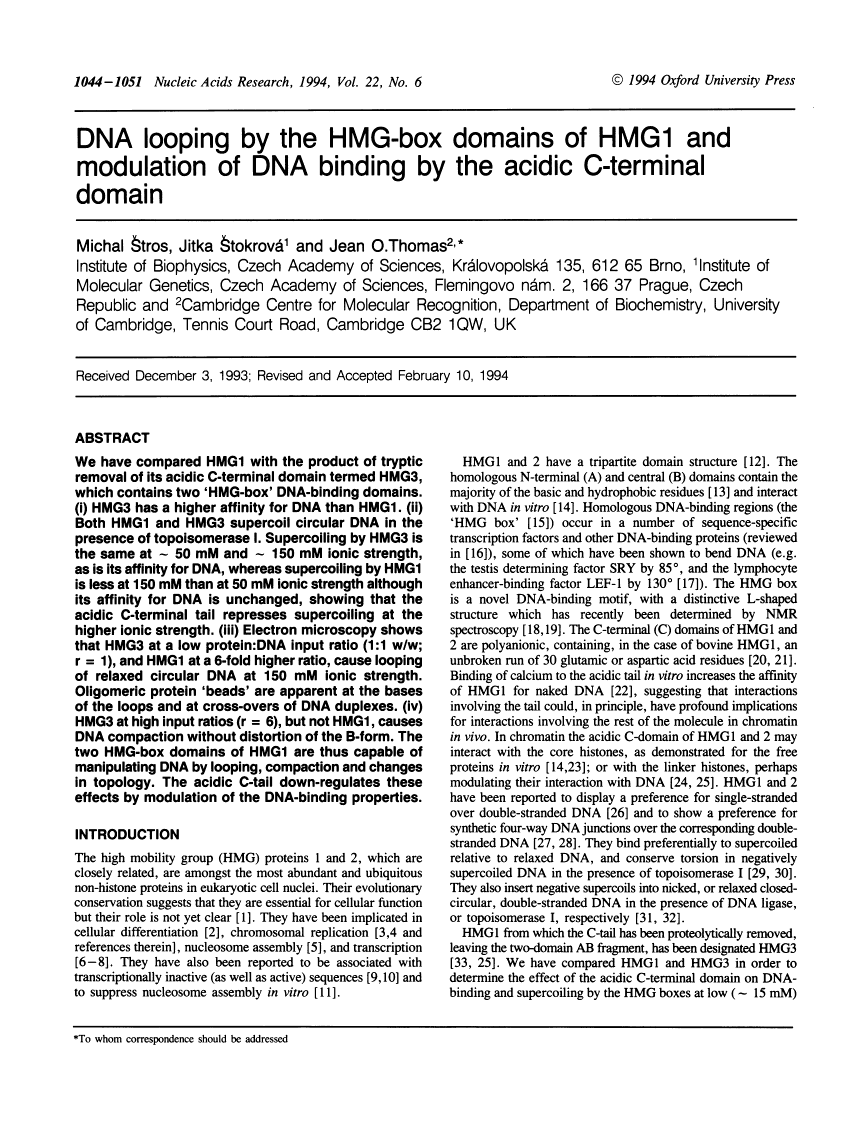
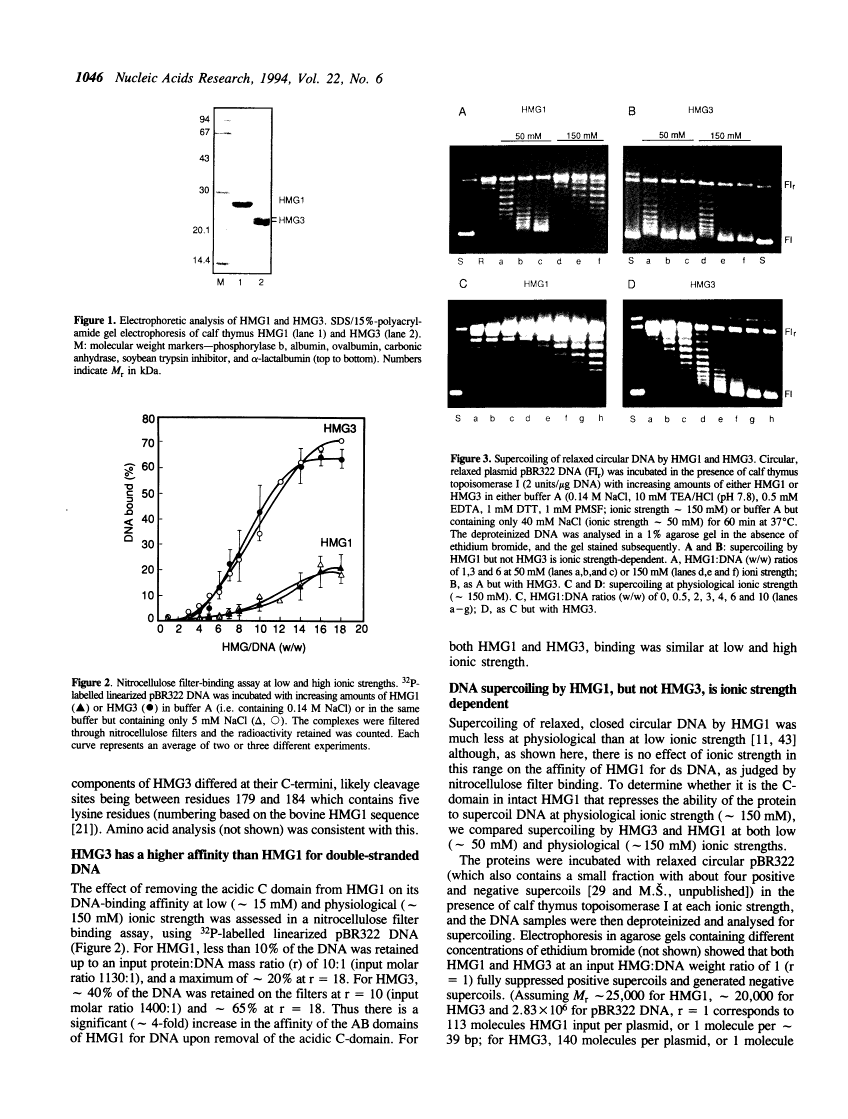
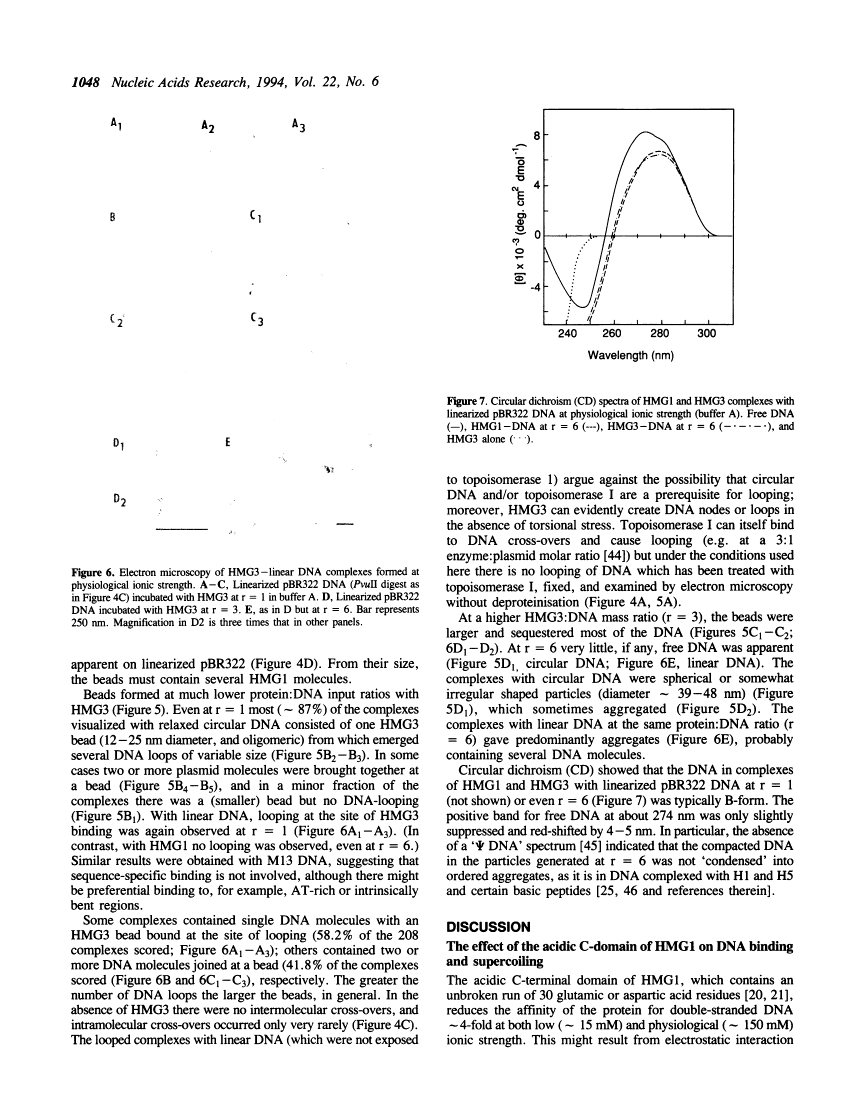
We have compared HMG1 with the product of tryptic removal of its acidic C-terminal domain termed HMG3, which contains two 'HMG-box' DNA-binding domains. (i) HMG3 has a higher affinity for DNA than HMG1. (ii) Both HMG1 and HMG3 supercoil circular DNA in the presence of topoisomerase I. Supercoiling by HMG3 is the same at approximately 50 mM and approximately 150 mM ionic strength, as is its affinity for DNA, whereas supercoiling by HMG1 is less at 150 mM than at 50 mM ionic strength although its affinity for DNA is unchanged, showing that the acidic C-terminal tail represses supercoiling at the higher ionic strength. (iii) Electron microscopy shows that HMG3 at a low protein:DNA input ratio (1:1 w/w; r = 1), and HMG1 at a 6-fold higher ratio, cause looping of relaxed circular DNA at 150 mM ionic strength. Oligomeric protein 'beads' are apparent at the bases of the loops and at cross-overs of DNA duplexes. (iv) HMG3 at high input ratios (r = 6), but not HMG1, causes DNA compaction without distortion of the B-form. The two HMG-box domains of HMG1 are thus capable of manipulating DNA by looping, compaction and changes in topology. The acidic C-tail down-regulates these effects by modulation of the DNA-binding properties.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. [Abstract] [Google Scholar]
- Seyedin SM, Pehrson JR, Cole RD. Loss of chromosomal high mobility group proteins HMG1 and HMG2 when mouse neuroblastoma and Friend erythroleukemia cells become committed to differentiation. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5988–5992. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonne C, Sautiere P, Duguet M, de Recondo AM. Identification of a single-stranded DNA binding protein from rat liver with high mobility group protein 1. J Biol Chem. 1982 Mar 25;257(6):2722–2725. [Abstract] [Google Scholar]
- Alexandrova EA, Beltchev BG. Acetylated HMG1 protein interacts specifically with homologous DNA polymerase alpha in vitro. Biochem Biophys Res Commun. 1988 Aug 15;154(3):918–927. [Abstract] [Google Scholar]
- Bonne-Andrea C, Harper F, Sobczak J, De Recondo AM. Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 1984 May;3(5):1193–1199. [Europe PMC free article] [Abstract] [Google Scholar]
- Stoute JA, Marzluff WF. HMG-proteins 1 and 2 are required for transcription of chromatin by endogenous RNA polymerase. Biochem Biophys Res Commun. 1982 Aug 31;107(4):1279–1284. [Abstract] [Google Scholar]
- Tremethick DJ, Molloy PL. Effects of high mobility group proteins 1 and 2 on initiation and elongation of specific transcription by RNA polymerase II in vitro. Nucleic Acids Res. 1988 Dec 9;16(23):11107–11123. [Europe PMC free article] [Abstract] [Google Scholar]
- Singh J, Dixon GH. High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry. 1990 Jul 3;29(26):6295–6302. [Abstract] [Google Scholar]
- Blanco J, States JC, Dixon GH. General method for isolation of DNA sequences that interact with specific nuclear proteins in chromosomes: binding of the high mobility group protein HMG-T to a subset of the protamine gene family. Biochemistry. 1985 Dec 31;24(27):8021–8028. [Abstract] [Google Scholar]
- Postnikov YV, Shick VV, Belyavsky AV, Khrapko KR, Brodolin KL, Nikolskaya TA, Mirzabekov AD. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991 Feb 25;19(4):717–725. [Europe PMC free article] [Abstract] [Google Scholar]
- Waga S, Mizuno S, Yoshida M. Nonhistone proteins HMG1 and HMG2 suppress the nucleosome assembly at physiological ionic strength. Biochim Biophys Acta. 1989 Mar 1;1007(2):209–214. [Abstract] [Google Scholar]
- Reeck GR, Isackson PJ, Teller DC. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982 Nov 4;300(5887):76–78. [Abstract] [Google Scholar]
- Cary PD, Turner CH, Mayes E, Crane-Robinson C. Conformation and domain structure of the non-histone chromosomal proteins, HMG 1 and 2. Isolation of two folded fragments from HMG 1 and 2. Eur J Biochem. 1983 Mar 15;131(2):367–374. [Abstract] [Google Scholar]
- Carballo M, Puigdomènech P, Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2(10):1759–1764. [Europe PMC free article] [Abstract] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. [Abstract] [Google Scholar]
- Ner SS. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. [Abstract] [Google Scholar]
- Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. [Abstract] [Google Scholar]
- Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. [Europe PMC free article] [Abstract] [Google Scholar]
- Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993 Jul 25;21(15):3427–3436. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaplan DJ, Duncan CH. Full length cDNA sequence for bovine high mobility group 1 (HMG1) protein. Nucleic Acids Res. 1988 Nov 11;16(21):10375–10375. [Europe PMC free article] [Abstract] [Google Scholar]
- Walker JM, Hastings JR, Johns EW. A novel continuous sequence of 41 aspartic and glutamic residues in a non-histone chromosomal protein. Nature. 1978 Jan 19;271(5642):281–282. [Abstract] [Google Scholar]
- Stros M, Bernués J, Querol E. Calcium modulates the binding of high-mobility-group protein 1 to DNA. Biochem Int. 1990 Aug;21(5):891–899. [Abstract] [Google Scholar]
- Bernués J, Espel E, Querol E. Identification of the core-histone-binding domains of HMG1 and HMG2. Biochim Biophys Acta. 1986 May 5;866(4):242–251. [Abstract] [Google Scholar]
- Kohlstaedt LA, Sung EC, Fujishige A, Cole RD. Non-histone chromosomal protein HMG1 modulates the histone H1-induced condensation of DNA. J Biol Chem. 1987 Jan 15;262(2):524–526. [Abstract] [Google Scholar]
- Stros M, Vorlícková M. Non-histone chromosomal protein HMG1 reduces the histone H5-induced changes in c.d. spectra of DNA: the acidic C-terminus of HMG1 is necessary for binding to H5. Int J Biol Macromol. 1990 Oct;12(5):282–288. [Abstract] [Google Scholar]
- Isackson PJ, Fishback JL, Bidney DL, Reeck GR. Preferential affinity of high molecular weight high mobility group non-histone chromatin proteins for single-stranded DNA. J Biol Chem. 1979 Jul 10;254(13):5569–5572. [Abstract] [Google Scholar]
- Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. [Abstract] [Google Scholar]
- Bianchi ME, Falciola L, Ferrari S, Lilley DM. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheflin LG, Spaulding SW. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry. 1989 Jun 27;28(13):5658–5664. [Abstract] [Google Scholar]
- Sheflin LG, Fucile NW, Spaulding SW. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993 Apr 6;32(13):3238–3248. [Abstract] [Google Scholar]
- Javaherian K, Liu JF, Wang JC. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science. 1978 Mar 24;199(4335):1345–1346. [Abstract] [Google Scholar]
- Mathis DJ, Kindelis A, Spadafora C. HMG proteins (1 + 2) form beaded structures when complexed with closed circular DNA. Nucleic Acids Res. 1980 Jun 25;8(12):2577–2590. [Europe PMC free article] [Abstract] [Google Scholar]
- Goodwin GH, Walker JM, Johns EW. Studies on the degradation of high mobility group non-histone chromosomal proteins. Biochim Biophys Acta. 1978 Jun 22;519(1):233–242. [Abstract] [Google Scholar]
- Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. [Abstract] [Google Scholar]
- Pil PM, Chow CS, Lippard SJ. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9465–9469. [Europe PMC free article] [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Clark DJ, Thomas JO. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. [Abstract] [Google Scholar]
- Hill CS, Thomas JO. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. [Abstract] [Google Scholar]
- Espejo RT, Lebowitz J. A simple electrophoretic method for the determination of superhelix density of closed circular DNAs and for observation of their superhelix density heterogeneity. Anal Biochem. 1976 May 7;72:95–103. [Abstract] [Google Scholar]
- Portmann R, Schaffner W, Birnstiel M. Partial denaturation mapping of cloned histone DNA from the sea urchin Psammechinus miliaris. Nature. 1976 Nov 4;264(5581):31–34. [Abstract] [Google Scholar]
- Stokrová J, Vojtisková M, Palecek E. Electron microscopy of supercoiled pEJ4 DNA containing homopurine.homopyrimidine sequences. J Biomol Struct Dyn. 1989 Apr;6(5):891–898. [Abstract] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. [Europe PMC free article] [Abstract] [Google Scholar]
- Duguet M, Bonne C, de Recondo AM. Single-strand deoxyribonucleic acid binding protein from rat liver changes the helical structure of deoxyribonucleic acid. Biochemistry. 1981 Jun 9;20(12):3598–3603. [Abstract] [Google Scholar]
- Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990 Dec;9(13):4555–4562. [Europe PMC free article] [Abstract] [Google Scholar]
- Fasman GD, Schaffhausen B, Goldsmith L, Adler A. Conformational changes associated with f-1 histone-deoxyribonucleic acid complexes. Circular dichroism studies. Biochemistry. 1970 Jul 7;9(14):2814–2822. [Abstract] [Google Scholar]
- Hill CS, Martin SR, Thomas JO. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. [Europe PMC free article] [Abstract] [Google Scholar]
- Carballo M, Puigdomènech P, Tancredi T, Palau J. Interaction between domains in chromosomal protein HMG-1. EMBO J. 1984 Jun;3(6):1255–1261. [Europe PMC free article] [Abstract] [Google Scholar]
- Cary PD, Turner CH, Leung I, Mayes E, Crane-Robinson C. Conformation and domain structure of the non-histone chromosomal proteins HMG 1 and 2. Domain interactions. Eur J Biochem. 1984 Sep 3;143(2):323–330. [Abstract] [Google Scholar]
- Marekov LN, Beltchev BG, Pivec L. High mobility group proteins HMG1 and HMG2 do not decrease the melting temperature of DNA. Biochem Biophys Res Commun. 1984 May 16;120(3):782–788. [Abstract] [Google Scholar]
- Butler AP, Mardian JK, Olins DE. Nonhistone chromosomal protein HMG 1 interactions with DNA. Fluorescence and thermal denaturation studies. J Biol Chem. 1985 Sep 5;260(19):10613–10620. [Abstract] [Google Scholar]
- Stros M, Kleinwächter V. Thermal denaturation and fluorescence study of nucleosomes containing non-histone chromosomal protein HMG2. Biochim Biophys Acta. 1987 Nov 20;910(2):163–170. [Abstract] [Google Scholar]
- Makiguchi K, Chida Y, Yoshida M, Shimura K. Mg2+-dependent unwinding of DNA by nonhistone chromosomal protein HMG(1 + 2) from pig thymus as determined by DNA melting temperature analysis. J Biochem. 1984 Feb;95(2):423–429. [Abstract] [Google Scholar]
- Yoshida M. High glutamic and aspartic region in nonhistone protein HMG(1+2) unwinds DNA double helical structure. J Biochem. 1987 Jan;101(1):175–180. [Abstract] [Google Scholar]
- Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonne C, Duguet M, de Recondo AM. Single-strand DNA binding protein from rat liver: interactions with supercoiled DNA. Nucleic Acids Res. 1980 Nov 11;8(21):4955–4968. [Europe PMC free article] [Abstract] [Google Scholar]
- Haykinson MJ, Johnson RC. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993 Jun;12(6):2503–2512. [Europe PMC free article] [Abstract] [Google Scholar]
- Su W, Jackson S, Tjian R, Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991 May;5(5):820–826. [Abstract] [Google Scholar]
- Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5670–5674. [Europe PMC free article] [Abstract] [Google Scholar]
- Topal MD, Thresher RJ, Conrad M, Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991 Feb 19;30(7):2006–2010. [Abstract] [Google Scholar]
- Ghrir R, Mignotte B, Guéride M. Amino terminal sequence of the mitochondrial protein mtDBP-C: similarity with nonhistone chromosomal proteins HMG 1 and 2. Biochimie. 1991 May;73(5):615–616. [Abstract] [Google Scholar]
- Brown JW, Anderson JA. The binding of the chromosomal protein HMG-2a to DNA regions of reduced stabilities. J Biol Chem. 1986 Jan 25;261(3):1349–1354. [Abstract] [Google Scholar]
- Shooter KV, Goodwin GH, Johns EW. Interactions of a purified non-histone chromosomal protein with DNA and histone. Eur J Biochem. 1974 Sep 1;47(2):263–270. [Abstract] [Google Scholar]
- Goodwin GH, Shooter KV, Johns EW. Interaction of a non-histone chromatin protein (high-mobility group protein 2) with DNA. Eur J Biochem. 1975 Jun;54(2):427–433. [Abstract] [Google Scholar]
- Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992 Apr 10;256(5054):234–237. [Abstract] [Google Scholar]
- Pontiggia A, Negri A, Beltrame M, Bianchi ME. Protein HU binds specifically to kinked DNA. Mol Microbiol. 1993 Feb;7(3):343–350. [Abstract] [Google Scholar]
- Lilley DM. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/22.6.1044
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc307928?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/22.6.1044
Article citations
O-GlcNAcylation of High Mobility Group Box 1 (HMGB1) Alters Its DNA Binding and DNA Damage Processing Activities.
J Am Chem Soc, 143(39):16030-16040, 21 Sep 2021
Cited by: 7 articles | PMID: 34546745 | PMCID: PMC8636066
Interactions of HMGB Proteins with the Genome and the Impact on Disease.
Biomolecules, 11(10):1451, 02 Oct 2021
Cited by: 21 articles | PMID: 34680084 | PMCID: PMC8533419
Review Free full text in Europe PMC
Immunological Significance of HMGB1 Post-Translational Modification and Redox Biology.
Front Immunol, 11:1189, 10 Jun 2020
Cited by: 64 articles | PMID: 32587593 | PMCID: PMC7297982
Review Free full text in Europe PMC
Chromosomal Organization and Regulation of Genetic Function in Escherichia coli Integrates the DNA Analog and Digital Information.
EcoSal Plus, 9(1), 01 Feb 2020
Cited by: 7 articles | PMID: 32056535
Ligation of free HMGB1 to TLR2 in the absence of ligand is negatively regulated by the C-terminal tail domain.
Mol Med, 24(1):19, 04 May 2018
Cited by: 20 articles | PMID: 30134807 | PMCID: PMC6016865
Go to all (80) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Calcium binding to HMG1 protein induces DNA looping by the HMG-box domains.
FEBS Lett, 344(2-3):201-206, 01 May 1994
Cited by: 22 articles | PMID: 8187884
The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal.
J Mol Biol, 304(2):135-149, 01 Nov 2000
Cited by: 50 articles | PMID: 11080451
Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA.
Eur J Biochem, 251(1-2):427-434, 01 Jan 1998
Cited by: 26 articles | PMID: 9492314
HMG1 and 2: architectural DNA-binding proteins.
Biochem Soc Trans, 29(pt 4):395-401, 01 Aug 2001
Cited by: 160 articles | PMID: 11497996
Review
Funding
Funders who supported this work.