Abstract
Free full text

Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant.
Abstract
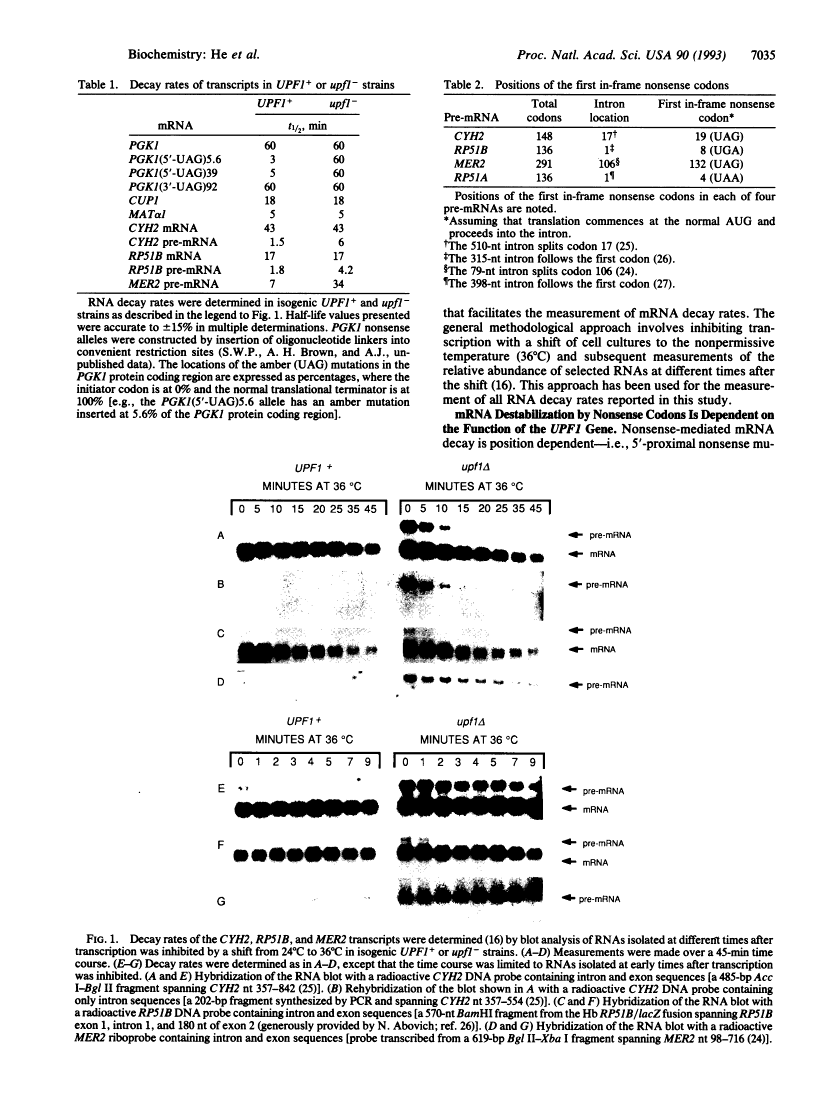
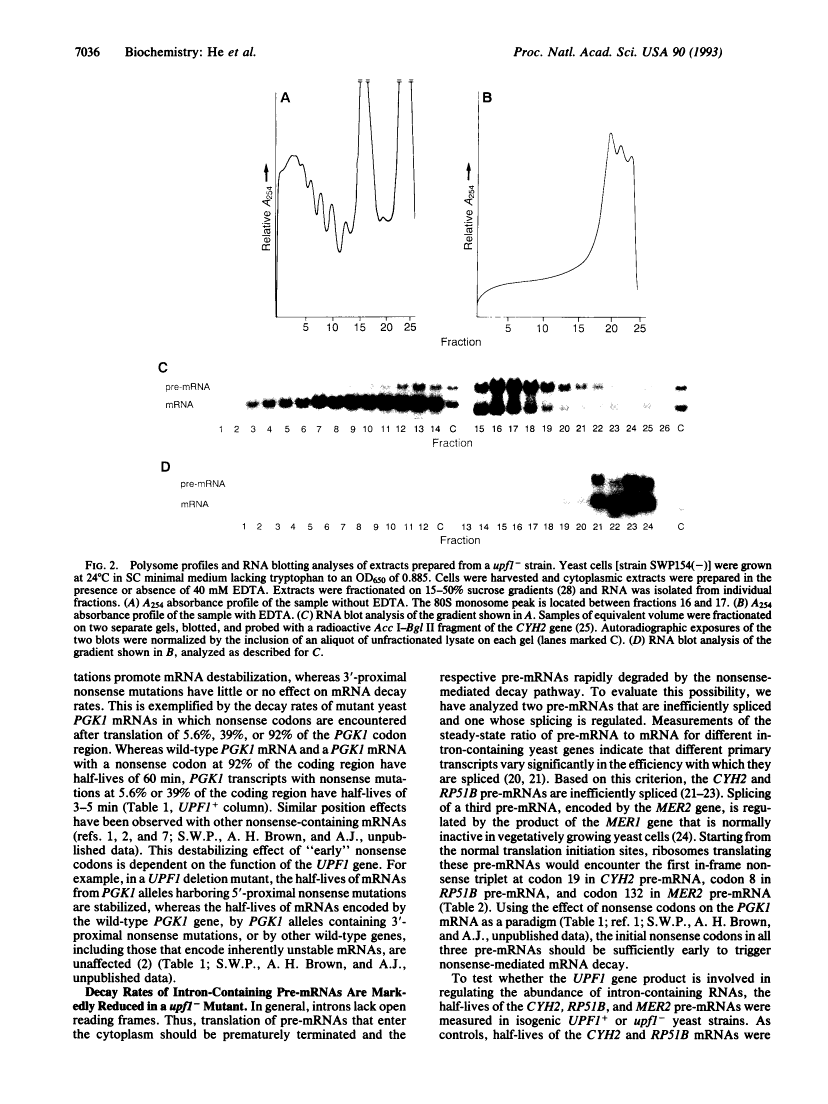
Nonsense-mediated mRNA decay, the accelerated turnover of mRNAs transcribed from genes containing early nonsense mutations, is dependent on the product of the UPF1 gene in yeast. Mutations that inactivate UPF1 lead to the selective stabilization of mRNAs containing early nonsense mutations but have no effect on the half-lives of almost all other mRNAs. Since the transcripts of nonsense alleles are not typical cellular constituents, we sought to identify those RNAs that comprise normal substrates of the nonsense-mediated mRNA decay pathway. Many yeast pre-mRNAs contain early in-frame nonsense codons and we consider it possible that a role of this pathway is to accelerate the degradation of pre-mRNAs present in the cytoplasm. Consistent with this hypothesis, we find that, in a strain lacking UPF1 function, the CYH2, RP51B, and MER2 pre-mRNAs are stabilized 2- to 5-fold and are associated with ribosomes. We conclude that a major source of early nonsense codon-containing cytoplasmic transcripts in yeast is pre-mRNAs and that the UPF1 protein may be part of a cellular system that ensures that potentially deleterious nonsense fragments of polypeptides do not accumulate.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. [Abstract] [Google Scholar]
- Barker GF, Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991 May;11(5):2760–2768. [Europe PMC free article] [Abstract] [Google Scholar]
- Baumann B, Potash MJ, Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. [Europe PMC free article] [Abstract] [Google Scholar]
- Gaspar ML, Meo T, Bourgarel P, Guenet JL, Tosi M. A single base deletion in the Tfm androgen receptor gene creates a short-lived messenger RNA that directs internal translation initiation. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8606–8610. [Europe PMC free article] [Abstract] [Google Scholar]
- Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990 Jan;17(1):77–79. [Abstract] [Google Scholar]
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. [Europe PMC free article] [Abstract] [Google Scholar]
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. [Abstract] [Google Scholar]
- Morse DE, Yanofsky C. Polarity and the degradation of mRNA. Nature. 1969 Oct 25;224(5217):329–331. [Abstract] [Google Scholar]
- Nilsson G, Belasco JG, Cohen SN, von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. [Europe PMC free article] [Abstract] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. [Europe PMC free article] [Abstract] [Google Scholar]
- Peltz SW, Brewer G, Bernstein P, Hart PA, Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1(2):99–126. [Abstract] [Google Scholar]
- Culbertson MR, Underbrink KM, Fink GR. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980 Aug;95(4):833–853. [Europe PMC free article] [Abstract] [Google Scholar]
- Fink GR. Pseudogenes in yeast? Cell. 1987 Apr 10;49(1):5–6. [Abstract] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. [Europe PMC free article] [Abstract] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. [Europe PMC free article] [Abstract] [Google Scholar]
- Baim SB, Pietras DF, Eustice DC, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. [Europe PMC free article] [Abstract] [Google Scholar]
- Steel LF, Jacobson A. Translational control of ribosomal protein synthesis during early Dictyostelium discoideum development. Mol Cell Biol. 1987 Mar;7(3):965–972. [Europe PMC free article] [Abstract] [Google Scholar]
- Pikielny CW, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. [Abstract] [Google Scholar]
- Swida U, Thüroff E, Käufer NF. Intron mutations that affect the splicing efficiency of the CYH2 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 May;203(2):300–304. [Abstract] [Google Scholar]
- Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993 Mar 26;72(6):893–901. [Abstract] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991 Sep 20;66(6):1257–1268. [Abstract] [Google Scholar]
- Käufer NF, Fried HM, Schwindinger WF, Jasin M, Warner JR. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983 May 25;11(10):3123–3135. [Europe PMC free article] [Abstract] [Google Scholar]
- Abovich N, Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. [Europe PMC free article] [Abstract] [Google Scholar]
- Teem JL, Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. [Europe PMC free article] [Abstract] [Google Scholar]
- Manrow RE, Jacobson A. Identification and characterization of developmentally regulated mRNP proteins of Dictyostelium discoideum. Dev Biol. 1986 Jul;116(1):213–227. [Abstract] [Google Scholar]
- Larkin JC, Thompson JR, Woolford JL., Jr Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol Cell Biol. 1987 May;7(5):1764–1775. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.90.15.7034
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc47070?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.90.15.7034
Article citations
Nonsense-mediated mRNA decay of metal-binding activator MAC1 is dependent on copper levels and 3'-UTR length in Saccharomyces cerevisiae.
Curr Genet, 70(1):5, 06 May 2024
Cited by: 0 articles | PMID: 38709348
Genetic screens in Saccharomyces cerevisiae identify a role for 40S ribosome recycling factors Tma20 and Tma22 in nonsense-mediated decay.
G3 (Bethesda), 14(3):jkad295, 01 Mar 2024
Cited by: 5 articles | PMID: 38198768 | PMCID: PMC10917514
Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p.
Curr Issues Mol Biol, 46(1):244-261, 29 Dec 2023
Cited by: 1 article | PMID: 38248319 | PMCID: PMC10814901
Post-transcriptional Regulation of Gene Expression via Unproductive Splicing.
Acta Naturae, 16(1):4-13, 01 Jan 2024
Cited by: 0 articles | PMID: 38698955 | PMCID: PMC11062102
Mechanistic toxicology in light of genetic compensation.
Toxicol Sci, kfad113, 06 Nov 2023
Cited by: 0 articles | PMID: 37941503 | PMCID: PMC10823772
Review Free full text in Europe PMC
Go to all (185) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae.
RNA, 3(3):234-244, 01 Mar 1997
Cited by: 54 articles | PMID: 9056761 | PMCID: PMC1369476
The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon.
Genes Dev, 5(12a):2303-2314, 01 Dec 1991
Cited by: 328 articles | PMID: 1748286
Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution.
Mol Cell Biol, 23(3):842-851, 01 Feb 2003
Cited by: 54 articles | PMID: 12529390 | PMCID: PMC140708
NMD monitors translational fidelity 24/7.
Curr Genet, 63(6):1007-1010, 23 May 2017
Cited by: 34 articles | PMID: 28536849 | PMCID: PMC5668330
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: GM23549
Grant ID: GM27757
Grant ID: R01 GM027757