Abstract
Free full text

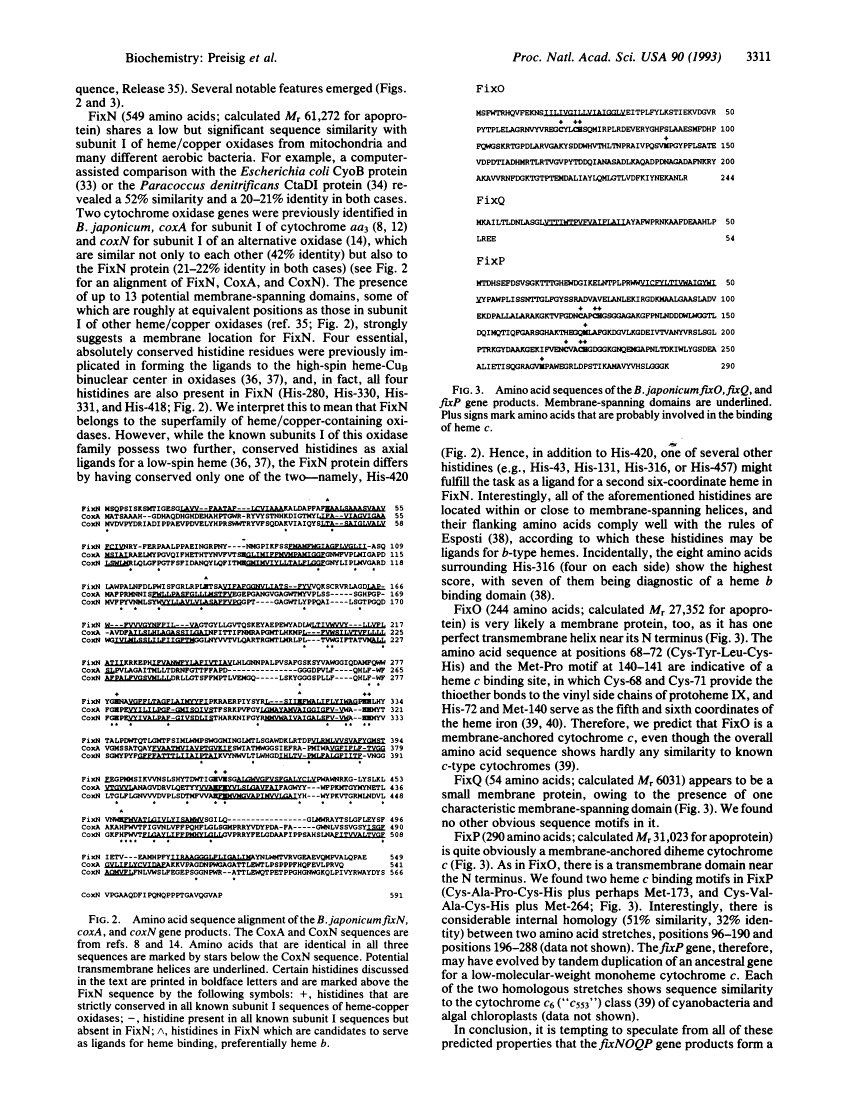
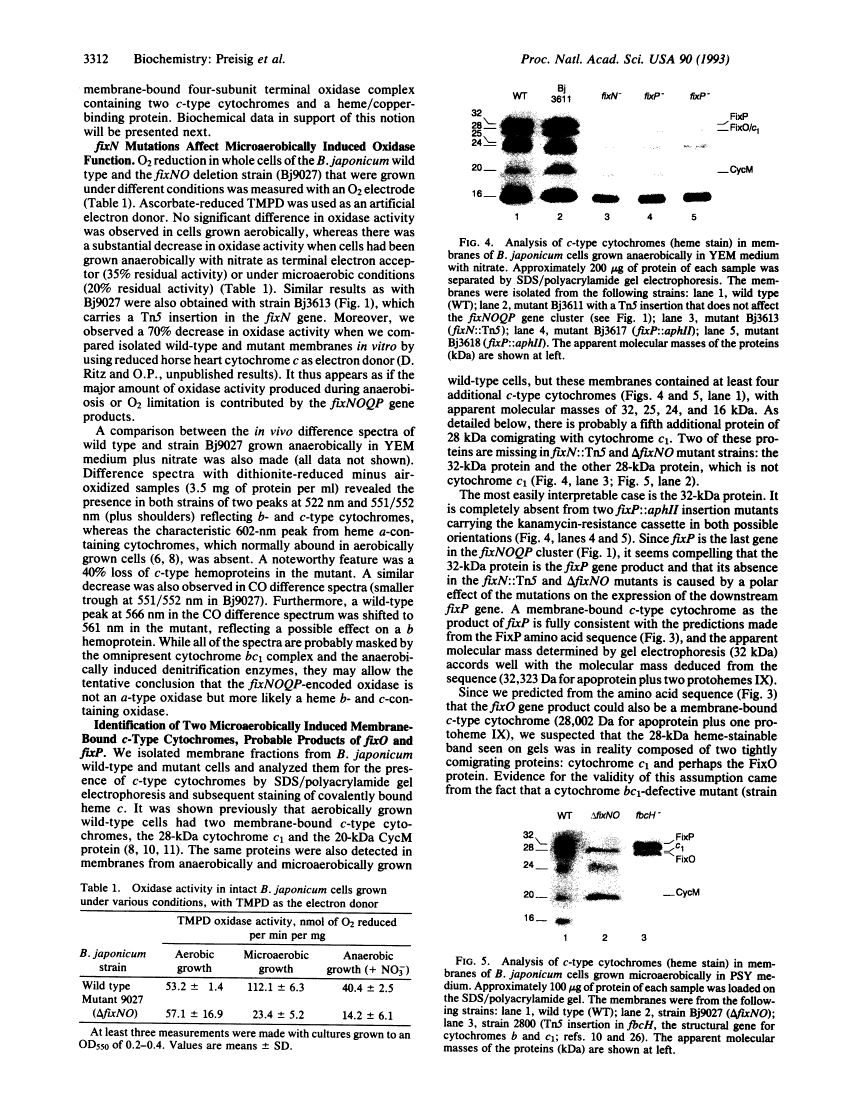
Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis.
Abstract
We report the discovery of a Bradyrhizobium japonicum gene cluster (fixNOQP) in which mutations resulted in defective soybean root-nodule bacteroid development and symbiotic nitrogen fixation. The predicted, DNA-derived protein sequences suggested that FixN is a heme b and copper-binding oxidase subunit, FixO a monoheme cytochrome c, FixQ a polypeptide of 54 amino acids, and FixP a diheme cytochrome c and that they are all membrane-bound. The isolation and analysis of membrane proteins from B. japonicum wild-type and mutant cells revealed two c-type cytochromes of 28 and 32 kDa as the likely products of the fixO and fixP genes and showed that both were synthesized only under oxygen-limited growth conditions. Furthermore, fixN insertion and fixNO deletion mutants grown microaerobically or anaerobically (with nitrate) exhibited a strong decrease in whole-cell oxidase activity as compared with the wild type. The data suggest that the fixNOQP gene products are induced at low oxygen concentrations and constitute a member of the bacterial heme/copper cytochrome oxidase superfamily. The described features are compatible with the postulate that this oxidase complex is specifically required to support bacterial respiration in endosymbiosis.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.3M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- O'Brian MR, Maier RJ. Molecular aspects of the energetics of nitrogen fixation in Rhizobium-legume symbioses. Biochim Biophys Acta. 1989 May 30;974(3):229–246. [Abstract] [Google Scholar]
- Bott M, Bolliger M, Hennecke H. Genetic analysis of the cytochrome c-aa3 branch of the Bradyrhizobium japonicum respiratory chain. Mol Microbiol. 1990 Dec;4(12):2147–2157. [Abstract] [Google Scholar]
- Thöny-Meyer L, Stax D, Hennecke H. An unusual gene cluster for the cytochrome bc1 complex in Bradyrhizobium japonicum and its requirement for effective root nodule symbiosis. Cell. 1989 May 19;57(4):683–697. [Abstract] [Google Scholar]
- Bott M, Ritz D, Hennecke H. The Bradyrhizobium japonicum cycM gene encodes a membrane-anchored homolog of mitochondrial cytochrome c. J Bacteriol. 1991 Nov;173(21):6766–6772. [Europe PMC free article] [Abstract] [Google Scholar]
- Gabel C, Maier RJ. Nucleotide sequence of the coxA gene encoding subunit I of cytochrome aa3 of Bradyrhizobium japonicum. Nucleic Acids Res. 1990 Oct 25;18(20):6143–6143. [Europe PMC free article] [Abstract] [Google Scholar]
- Keister DL, Marsh SS. Hemoproteins of Bradyrhizobium japonicum Cultured Cells and Bacteroids. Appl Environ Microbiol. 1990 Sep;56(9):2736–2741. [Europe PMC free article] [Abstract] [Google Scholar]
- Bott M, Preisig O, Hennecke H. Genes for a second terminal oxidase in Bradyrhizobium japonicum. Arch Microbiol. 1992;158(5):335–343. [Abstract] [Google Scholar]
- Appleby CA, James P, Hennecke H. Characterization of three soluble c-type cytochromes isolated from soybean root nodule bacteroids of Bradyrhizobium japonicum strain CC705. FEMS Microbiol Lett. 1991 Oct 1;67(2):137–144. [Abstract] [Google Scholar]
- Rossbach S, Loferer H, Acuña G, Appleby CA, Hennecke H. Cloning, sequencing and mutational analysis of the cytochrome c552 gene (cycB) from Bradyrhizobium japonicum strain 110. FEMS Microbiol Lett. 1991 Oct 1;67(2):145–152. [Abstract] [Google Scholar]
- Tully RE, Sadowsky MJ, Keister DL. Characterization of cytochromes c550 and c555 from Bradyrhizobium japonicum: cloning, mutagenesis, and sequencing of the c555 gene (cycC). J Bacteriol. 1991 Dec;173(24):7887–7895. [Europe PMC free article] [Abstract] [Google Scholar]
- Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983 Aug;135(2):103–109. [Abstract] [Google Scholar]
- Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991 Jan;225(1):38–48. [Abstract] [Google Scholar]
- Daniel RM, Appleby CA. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P 450 , other haemoproteins, nitrate and nitrite reductases. Biochim Biophys Acta. 1972 Sep 20;275(3):347–354. [Abstract] [Google Scholar]
- Ramseier TM, Göttfert M. Codon usage and G + C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156(4):270–276. [Abstract] [Google Scholar]
- Thöny-Meyer L, James P, Hennecke H. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5001–5005. [Europe PMC free article] [Abstract] [Google Scholar]
- Kahn D, David M, Domergue O, Daveran ML, Ghai J, Hirsch PR, Batut J. Rhizobium meliloti fixGHI sequence predicts involvement of a specific cation pump in symbiotic nitrogen fixation. J Bacteriol. 1989 Feb;171(2):929–939. [Europe PMC free article] [Abstract] [Google Scholar]
- Batut J, Daveran-Mingot ML, David M, Jacobs J, Garnerone AM, Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989 Apr;8(4):1279–1286. [Europe PMC free article] [Abstract] [Google Scholar]
- Anthamatten D, Scherb B, Hennecke H. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992 Apr;174(7):2111–2120. [Europe PMC free article] [Abstract] [Google Scholar]
- Tinoco I, Jr, Borer PN, Dengler B, Levin MD, Uhlenbeck OC, Crothers DM, Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. [Abstract] [Google Scholar]
- Chepuri V, Lemieux L, Au DC, Gennis RB. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990 Jul 5;265(19):11185–11192. [Abstract] [Google Scholar]
- Raitio M, Jalli T, Saraste M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987 Sep;6(9):2825–2833. [Europe PMC free article] [Abstract] [Google Scholar]
- Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990 Nov;23(4):331–366. [Abstract] [Google Scholar]
- Minagawa J, Mogi T, Gennis RB, Anraku Y. Identification of heme and copper ligands in subunit I of the cytochrome bo complex in Escherichia coli. J Biol Chem. 1992 Jan 25;267(3):2096–2104. [Abstract] [Google Scholar]
- Gennis RB. Site-directed mutagenesis studies on subunit I of the aa3-type cytochrome c oxidase of Rhodobacter sphaeroides: a brief review of progress to date. Biochim Biophys Acta. 1992 Jul 17;1101(2):184–187. [Abstract] [Google Scholar]
- Esposti MD. Prediction and comparison of the haem-binding sites in membrane haemoproteins. Biochim Biophys Acta. 1989 Dec 7;977(3):249–265. [Abstract] [Google Scholar]
- Matsushita K, Shinagawa E, Adachi O, Ameyama M. o-Type cytochrome oxidase in the membrane of aerobically grown Pseudomonas aeruginosa. FEBS Lett. 1982 Mar 22;139(2):255–258. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.90.8.3309
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc46289?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.90.8.3309
Article citations
Pangenome Evolution Reconciles Robustness and Instability of Rhizobial Symbiosis.
mBio, 13(3):e0007422, 13 Apr 2022
Cited by: 10 articles | PMID: 35416699 | PMCID: PMC9239051
How Rhizobia Adapt to the Nodule Environment.
J Bacteriol, 203(12):e0053920, 01 Feb 2021
Cited by: 26 articles | PMID: 33526611 | PMCID: PMC8315917
Review Free full text in Europe PMC
Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation.
Mol Syst Biol, 16(6):e9419, 01 Jun 2020
Cited by: 21 articles | PMID: 32490601 | PMCID: PMC7268258
Metabolic Analyses of Nitrogen Fixation in the Soybean Microsymbiont Sinorhizobium fredii Using Constraint-Based Modeling.
mSystems, 5(1):e00516-19, 18 Feb 2020
Cited by: 9 articles | PMID: 32071157 | PMCID: PMC7029217
The effect of plant compartments on the Broussonetia papyrifera-associated fungal and bacterial communities.
Appl Microbiol Biotechnol, 104(8):3627-3641, 20 Feb 2020
Cited by: 4 articles | PMID: 32078018
Go to all (155) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum.
J Bacteriol, 178(6):1532-1538, 01 Mar 1996
Cited by: 192 articles | PMID: 8626278 | PMCID: PMC177835
Functional analysis of the fixNOQP region of Azorhizobium caulinodans.
J Bacteriol, 176(9):2560-2568, 01 May 1994
Cited by: 41 articles | PMID: 8169204 | PMCID: PMC205393
Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum.
J Biol Chem, 271(15):9114-9119, 01 Apr 1996
Cited by: 54 articles | PMID: 8621562
Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation.
Adv Microb Physiol, 40:191-231, 01 Jan 1998
Cited by: 32 articles | PMID: 9889979
Review