Abstract
Free full text

Structure and Function of Plasmid pColD157 of Enterohemorrhagic Escherichia coli O157 and Its Distribution among Strains from Patients with Diarrhea and Hemolytic-Uremic Syndrome
Abstract
In this study, pColD157, a 6.7-kb colicinogenic plasmid of enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain CL40cu, was characterized by restriction mapping and determination of its complete nucleotide sequence. The sequence consists of 6,675 bp and shows a high degree of similarity to the nucleotide sequence of colicinogenic plasmids pColD-CA23 and pColK. Seven potential genes were located on pColD157, three of which were closely related (>97.9%) to the colicin D structural gene and the corresponding immunity and lysis genes of plasmid pColD-CA23, and these were therefore designated cda, cdi, and cdl, respectively, using the reference extension -CL40 for differentiation. The adjacent 3′ region is related to the origin of replication of pColD-CA23. In contrast, the remaining part of the plasmid harbors a cluster of genes, closely related to the mobilization genes of pColK, which is followed by a 0.3-kb stretch homologous to the pColK resolution function. These determinants were designated mbdA, mbdB, mbdC, and mbdD and cdr, respectively. Southern blot analysis was performed with a probe specific for the cda gene of pColD157 and two groups of EHEC O157:H7 isolates from patients with diarrhea or hemolytic-uremic syndrome resident in Germany. Whereas 16 of 46 E. coli O157 strains isolated between 1987 and 1991 harbored plasmid pColD157, only 1 of 50 strains isolated during 1996 carried this plasmid. In addition, all strains harboring plasmid pColD157 were shown to have colicinogenic activity.
Following the first documented outbreaks of enterohemorrhagic Escherichia coli (EHEC) O157:H7 infections in 1982 (11), these bacteria are increasingly being recognized as important human diarrheal pathogens and the predominant cause of hemolytic-uremic syndrome (HUS) worldwide (16, 17). The main pathogenic property of EHEC strains is the production of Shiga toxins (Stx), which are thought to be responsible for the systemic symptoms of bloody diarrhea and HUS (27, 43, 44). In addition, the LEE locus, a 35-kb pathogenicity island that is located in the EHEC chromosome, encodes proteins involved in the intimate adherence to and cytoskeletal damage of intestinal cells (24). Moreover, E. coli O157 strains were shown to almost invariably carry a 90-kb plasmid (pO157) (23) encoding possible additional virulence traits such as EHEC hemolysin (36), the bifunctional catalase-peroxidase KatP (9), a secreted serine protease (10), and a type II secretion pathway system (37).
A number of studies have reported that, in addition to pO157, several smaller plasmids occur sporadically in E. coli O157 strains (28, 29, 33). One of these, which is approximately 6.6 kb, was found in up to 62% of strains isolated in North America (33) and the United Kingdom (39). This plasmid appeared to be associated with a colicinogenic phenotype of E. coli O157:H7 strains (15, 39). Furthermore, one strain harboring this plasmid produced large amounts of an 87-kDa protein similar to but not identical to colicin D and was therefore named colicin D157 (7).
Although the colicin D encoded by plasmid pColD-CA23 has been analyzed extensively at the molecular level (14, 34), no further information on the nature of the colicin D-related plasmid occurring in EHEC O157 strains is available. Since the production of colicins by enteropathogenic bacteria is thought to be of importance for pathogenic activity in the colon (7), and thus to represent an accessory factor in virulence for some EHEC strains, we analyzed the structure of pColD157 and determined its distribution among E. coli O157 strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli O157:H7 strain CL40 was isolated from a patient with HUS (22) and was kindly provided by M. A. Karmali, Toronto, Ontario, Canada. This strain harbored two plasmids of 90 and 6.7 kb and was cured of the 90-kb plasmid by ethidium bromide treatment (21). This revealed strain CL40cu, possessing the 6.7-kb plasmid only. E. coli O157:H7 strain EDL933 was isolated from a patient with hemorrhagic colitis (26) and was used as a negative control for hybridization and functional colicin assays.
In addition, the wild-type E. coli O157:H7 and O157:H− strains used in this study were isolated during two periods from patients with HUS and diarrhea throughout Germany. One group, 46 E. coli O157:H7 and O157:H− strains, were obtained between 1987 and 1991 and are described elsewhere (1, 5). Twenty-five of these were from patients with HUS, and 21 were from patients with diarrhea. The second group, 50 E. coli O157:H7 and O157:H− strains, were isolated during 1996. Twenty-five strains were from patients with HUS, and 25 were from patients with diarrhea. These strains were detected by PCR with stx1- and stx2-specific primers (38) and were serotyped by the method described by Bockemühl et al. (3). The strains were from different geographical regions in Germany and were epidemiologically unrelated.
E. coli K-12 C600 transformed with plasmid pColD-CA23 (34) was kindly provided by V. Braun, Tübingen, Germany, and served as a positive control for colicin tests. Laboratory strain E. coli DH5α (Gibco BRL, Eggenstein, Germany) was used as the host for recombinant plasmids. Plasmids pK18 (30) and pBluescript IIKS+ (Stratagene, Heidelberg, Germany) were used for cloning experiments.
Bacteria were routinely grown in Luria broth containing 1.0% (wt/vol) NaCl, 1.0% (wt/vol) tryptone, and 0.5% (wt/vol) yeast extract (pH 7.5). For maintenance of recombinant plasmids, ampicillin or kanamycin was added to a final concentration of 250 or 50 μg ml−1, respectively.
Colicin production assay.
Patch tests were performed as described by Pugsley (32). Briefly, fresh colonies of the test strains were transferred with a sterile toothpick onto a lawn seeded with 106 E. coli C600 indicator cells. Zones of inhibition were determined after 18 h of incubation at 37°C.
General recombinant DNA techniques.
Recombinant plasmids for DNA sequencing were purified with the Nucleobond AX100 Preparation Kit (Macherey-Nagel, Düren, Germany). For all other purposes, the method of Birnboim and Doly (2) was used. Restriction endonuclease digestion was performed according to the supplier’s instructions (Gibco BRL). Restriction fragments were separated on 0.8 to 2.0% (wt/vol) agarose gels and stained with 0.1% (wt/vol) ethidium bromide. A 1-kb ladder (Gibco BRL) was used as the molecular mass standard. The determination of restriction fragment sizes was carried out by comparing the migrations of restriction fragments with the migrations on a calibration graph of the molecular mass markers separated on the same gel. For cloning experiments, restriction fragments were isolated and purified from unstained gels by using a Prep-a-Gene kit (Bio-Rad, Munich, Germany) and ligated into vectors pK18 or pBluescript IIKS+ with T4 DNA ligase. Transformation experiments were carried out as described by Hanahan (18).
DNA sequencing.
Nucleotide sequences were determined by Taq cycle sequencing with universal and reverse primers for pUC and M13 vectors and with customized primers as described previously (36). Nucleotide sequence analysis and searches for homologous DNA sequences in the EMBL and GenBank database libraries as well as for homologous amino acid sequences in the Swiss-Prot and Piro databases were performed with the program package HUSAR (Heidelberg UNIX Sequence Analysis Resources; German Cancer Research Center, Heidelberg, Germany and the Lasergene program package (Dnastar, Madison, Wis.).
Southern blot hybridization.
An internal 587-bp fragment of the colicin D structural gene (cda-CL40) derived from pColD157 was used as the gene probe. This fragment was amplified with primers colD-1 (5′-GTA AAT CTG CCT GTT CGT GGA C-3′) and colD-2 (5′-CCT TTT TCT CTT CGG TAT GTT C-3′) and was then labeled randomly with digoxigenin-11-dUTP. The PCR conditions for amplification of the 587-bp fragment were as follows: denaturation at 94°C for 30 s, annealing at 57°C for 60 s, and extension at 72°C for 60 s for 30 cycles of amplification.
DNA was transferred from agarose gels to Zeta probe nylon membranes (Bio-Rad) by standard methods (35). For hybridization assays, the Non-radioactive DNA Labeling and Detection Kit (Boehringer GmbH, Mannheim, Germany) was used according to the manufacturer’s instructions. Hybridization was performed by incubating the membrane in hybridization solution for 18 h in a 60°C water bath. The specific washing step was carried out two times at 60°C for 15 min in a solution made of 0.03-fold SSC (4.5 mM sodium chloride plus 0.45 mM sodium citrate) and 0.1% (wt/vol) sodium dodecyl sulfate to effect a stringency of 95%.
Nucleotide sequence accession number.
The complete nucleotide sequence of plasmid pColD157 was submitted to the EMBL database library and was given the accession no. Y10412.
RESULTS
Mapping, cloning, and sequencing of the 6.7-kb plasmid pColD157.
A total of 18 restriction enzymes were tested during construction of a restriction map of the 6.7-kb plasmid, and 9 of these (BamHI, BglII, EcoRI, HindIII, PstI, PvuI, SalI, SphI, and XbaI) were unable to cleave the plasmid. In contrast, HaeIII and Sau3AI showed more than 10 restriction sites each and were rejected. The endonucleases ClaI, DraI, EcoRV, HincII, KpnI, PvuII, and SmaI displayed an appropriate number of restriction sites (<10) and were used for mapping. The restriction map of pColD157 was established by comparing and combining the restriction fragment sizes resulting from single and double digests (Fig. (Fig.1).1). Comparison with the restriction map of pColD-CA23 (34) suggested that there was a close relationship between the two plasmids, and this prompted us to define the DraI site, which is flanked by two HincII sites, as the starting point of the map (Fig. (Fig.1).1).
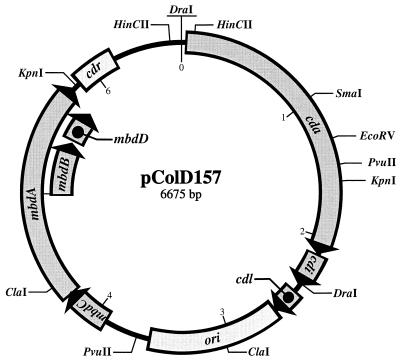
Restriction and gene map of the 6,675-bp plasmid pColD157 derived from E. coli O157 strain CL40cu. The restriction sites for ClaI, DraI, EcoRV, HincII, KpnI, PvuI, PvuII, and SmaI were determined experimentally and are indicated in the figure. Seven open reading frames depicted here are considered to be genes. cda, cdi, and cdl are homologuous to the colicin D structural, lysis, and immunity genes of pColD-CA23, respectively, and the mbdC, mbdA, mbdB, and mbdD genes are strongly related to the mobilization genes (mbk) of pColK. The characteristics of the genes and proteins involved are described in the text and in Table Table2.2. The numbering inside the circle is the molecular size (in kilobase pairs).
KpnI and ClaI restriction fragments were chosen for subcloning into pK18 and pBluescript IIKS+, respectively. The resulting plasmids were transformed into E. coli DH5α and were used for sequencing. Nucleotide sequence analysis revealed that the original plasmid had a total of 6,675 bp. All restriction sites determined by restriction analysis occurred in the sequence within less than 0.05 kb of the estimated point. In addition, one site for PvuI and a further site for ClaI were detected, but these were not revealed in restriction experiments with E. coli O157:H7 strain CL40cu, probably because of methylation. The positions of the restriction sites (on the basis of the nucleotide sequence) and the respective restriction fragment sizes are presented in Table Table1.1.
TABLE 1
Positions of restriction sites and sizes of restriction fragments in the map of pColD157
pColD157
| Restriction enzyme | No. of restriction sites | Position (bp) of restriction site | Length of restriction fragments (bp) |
|---|---|---|---|
| ClaI | 3 | 2,133,a 3,040, 4,342 | 1,301, 5,374 (4,467, 907)a |
| DraI | 2 | 1, 2,367 | 2,367, 4,308 |
| EcoRV | 1 | 1,303 | 6,675 |
| HincII | 2 | 93, 6,589 | 179, 6,496 |
| KpnI | 2 | 1,593, 5,841 | 2,427, 4,248 |
| PvuI | 1 | 6,074a | 6,675a |
| PvuII | 2 | 1,484, 3,678 | 2,194, 4,481 |
| SmaI | 1 | 974 | 6,675 |
Sequence analysis.
In order to detect regions which are able to encode proteins, open reading frames on both strands were determined. A total of more than 25 frames were found by using ATG as the translational start codon and TAG, TAA, or TGA as the stop codon. Because of this, only those which were preceded by a Shine-Dalgarno sequence and which were longer than 120 bp were considered to be genes, and by using these criteria, seven open reading frames were found (Table (Table2).2). Database library searches were then performed to assess the functions of these putative genes. As a first step, the nucleotide sequence of each frame was subjected to a database homology search with the sequences in the GenBank and EMBL libraries. For two of the frames, we found striking sequence similarities to the genes cda (colicin D activity) and cdi (colicin D immunity) of the colicinogenic plasmid pColD-CA23 (34). Because these genes are organized in an operon structure together with cdl (colicin D lysis) (14), we manually performed a similarity search using the cdl sequence (25), which is not recorded in any database library. In fact, we found a third gene to be very similar to cdl of pColD-CA23. Since the colicin D determinants of pColD157 and pColD-CA23 were nearly identical, we used the same gene designations and introduced reference extensions for differentiation (e.g., cda-CA23 versus cda-CL40).
TABLE 2
Characterization of open reading frames and other features of pColD157 and comparison with other colicinogenic factors
factors
| Designation of ORFa or region | Description | Length of ORF or region (bp) | Map position (bp) | Homologous gene | % Matching base pairs | % Matching amino acids | Reference |
|---|---|---|---|---|---|---|---|
| cda-CL40 | Colicin D activity | 2,091 | 25–2,116 | cda-CA23 | 99.8 | 99.4 | 34 |
| cdi-CL40 | Colicin D immunity | 261 | 2,115–2,376 | cdi-CA23 | 99.2 | 98.9 | 34 |
| cdl-CL40 | Colicin D lysis | 144 | 2,448–2592 | cdl-CA23 | 97.9 | 100 | 25 |
| mbdA | pColD157 mobility | 1,596 | 4,213–5,809 | mbkA-pColK |  SNAb SNAb | 92.5 | 4 |
| mbdB | pColD157 mobility | 558 | 4,901–5,459 | mbkB-pColK | SNA | 83.3 | 4 |
| mbdC | pColD157 mobility | 345 | 3,876–4,221 | mbkC-pColK | SNA | 90.7 | 4 |
| mbdD | pColD157 mobility | 231 | 5,468–5,699 | mbkD-pColK | SNA | 97.4 | 4 |
| ori | Origin of replication | 998 | 2,595–3,592 | ori-pColD-CA23 | 90.2 | 31 | |
| cdr | Multimer resolution | 316 | 5,865–6,180 | ckr-pColK | 98.1 | 42 |
In the next step, we translated each open reading frame and compared the deduced amino acid sequences with those in protein database libraries (Swiss-Prot and Piro). For a cluster of genes located downstream from the colicin D operon, there was a high degree of similarity to the mobilization proteins (Mbk) of another colicinogenic plasmid, pColK (4). However, some of the putative genes encoding the mobilization genes of pColD157 were longer than the respective genes described in pColK. Shortening of the pColD157 open reading frames by using another ATG start codon and translation of the shortened open reading frames increased the similarity between these proteins. The putative mobilization genes of pColD157 were designated mbdA, mbdB, mbdC, and mbdD, according to the standard nomenclature for mobilization genes of colicinogenic factors.
In a search for other functions, the entire nucleotide sequence of the plasmid was finally entered into the EMBL database library. One region resembling the pColD-CA23 origin of replication (46) was found, and another region was closely related to the pColK resolution function (40). Less striking homologies to pColE1, pColA, and pColB were also found, reflecting the general relationship between colicinogenic plasmids. The genes and functions of pColD157 are summarized in Table Table2.2. To illustrate their relative positions, we extended the restriction map which is reproduced in Fig. Fig.11.
Plasmid profile and Southern blot hybridization.
In order to assess the distribution of pColD157, we investigated two groups of E. coli O157:H7 and O157:H− strains isolated from the stools of patients with diarrhea or HUS. Forty-six strains were isolated between 1987 and 1991, and another 50 E. coli O157 strains were isolated during 1996. The strains were from patients distributed throughout Germany.
Plasmid analysis demonstrated that all E. coli O157 strains except strain CL40cu harbored a large plasmid of approximately 90 kb. Subsequent hybridization with the cda-CL40-specific probe revealed that 16 of 46 strains (35%) isolated between 1987 and 1991 but only 1 of 50 strains (2%) isolated in 1996 were positive with this probe. Results for representative examples of the investigated strains are presented in Fig. Fig.2.2. The multiple hybridization signals which occurred in Fig. Fig.2B2B (lanes 1, 2, 3, 4, 5, and 9) depended on the presence of various plasmid conformations (data not shown). By analysis of the distribution of plasmid pColD157 among strains from patients with diarrhea and HUS, we found no correlation between the presence of plasmid pColD157 in E. coli O157 isolates and the symptoms of the patients.
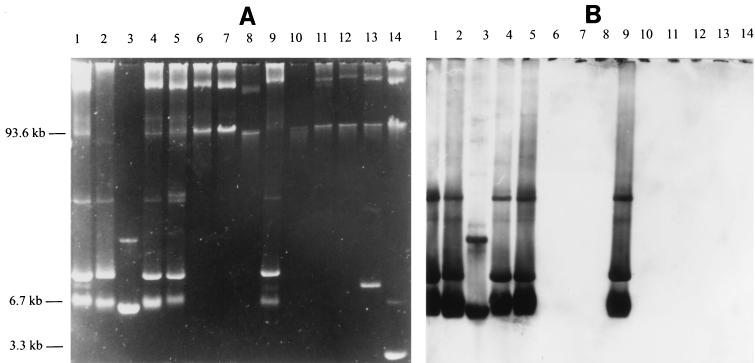
Agarose gel electrophoresis (A) and Southern blot hybridization (B) with a colicin D (cda-CL40)-specific probe of whole plasmid preparations of representative E. coli strains used in this study. Lanes 1, 2, and 4 to 7, representative isolates from 1987 to 1991; lanes 8 to 13 isolates from 1996. Lanes: 1, CL40; 2, CL40cu; 3, non-O157, colicin D-producing reference strain C600/pColD-CA23; 4, 5769/87; 5, 4739/91; 6, 703/88; 7, 702/88; 8, 4049/96; 9, 3645/96; 10, 4232/96; 11, 4140/96; 12, 5028/96; 13, 5268/96; 14, plasmids (93.6 and 3.3 kb) of the E. coli O157:H7 reference strain EDL933. The multiple hybridization signals in lanes 1 to 5 and 9 in panel B depend on the different conformations of colicin D plasmids.
Colicinogenic activity.
All strains used in this study were assayed for colicinogenic activity by using laboratory strain E. coli C600 as a sensitive indicator strain. All E. coli O157 strains harboring the 6.7-kb plasmid pColD157 and E. coli K-12 C600/pColDCA-23 showed colicinogenic activity in a patch test, examples of the results of which are presented in Fig. Fig.3.3. In contrast, none of the other strains inhibited the growth of the indicator strain E. coli C600.

Patch test for colicinogenic activities of E. coli strains. Fresh colonies of the strains were transferred with a sterile toothpick onto a lawn of 106 E. coli C600 indicator cells and were incubated overnight at 37°C. The strains tested and the numbering of these strains are the same as those described in the legend to Fig. Fig.22.
DISCUSSION
The 6.7-kb plasmid pColD157 of enterohemorrhagic E. coli O157:H7 appears to consist of two structurally different parts. A 2.5-kb DNA stretch is almost identical to a part of plasmid pColD-CA23, which harbors genes for colicin activity (cda), immunity to the produced colicin (cdi), and a lysis function involved in colicin release (cdl). Thus, pColD157 should enable its host to produce and release a colicin similar to colicin D and to resist its action. This conclusion is supported by the results of other investigators showing colicin D-like colicin production and immunity to colicin D-CA23 in cultures of O157 strains harboring a 6.6-kb plasmid and by our results demonstrating colicinogenic activity. Since the amino acid sequences deduced for colicin D and the O157 colicin were nearly identical, and this was also true for the immunity proteins, we propose that the genes be differentiated by use of reference extensions (e.g., cda-CA23 versus cda-CL40) but not the establishment of a new colicin type. Despite this, the associated plasmids, as a whole, differ in size and structure, and this justifies keeping the name pColD157 for colicinogenic O157 plasmids, as proposed by Bradley et al. (7).
Four open reading frames, the deduced amino acid sequences of which showed strong homology to the hypothetical mobilization proteins (MbkA to MbkD) of plasmid pColK, were found within another region of the plasmid (3.8 to 5.8 kb). The mbk genes are, in turn, closely related to the pColA and pColE1 mobilization genes and may promote plasmid mobilization by a broad range of helper plasmids, as found for pColE1 (4).
All mobilization genes described above had the same structural organization, with two frames entirely overlapped by a third one. Moreover, our results provide evidence that mobD genes (mbkD, mbdD, etc.) are highly conserved between colicinogenic plasmids and are therefore in agreement with the findings of Boyd et al. (4). This is of particular interest since mbeD was recently shown to control the entry exclusion of pColE1 (45). In turn, the carboxy-terminal region of mobA genes is highly variable, and on comparing mbdA and mbkA, we discovered a frameshift mutation involving insertion of a single base at position 5743. In view of the given homologies it would appear that pColD157 can be efficiently mobilized by conjugative helper plasmids, but detailed investigations of O157 conjugation are required in order to answer this question.
Other features of pColD157 included a region highly similar to the origin of replication of pColD-CA23 and a region similar to the ColK multimer resolution function (ckr), which is known to direct monomerization by recombination at a specific crossover site and which may inhibit cell division in the presence of plasmid multimers. It is of interest that homology ceases at the crossover site.
In view of the high degree of similarity to pColD-CA23 in one part of the sequence and to pColK in the other part, pColD157 might have resulted from a recombination event between these two colicinogenic factors. This would support the modular conception for the evolution of colicin-determining plasmids.
The fact that pColD157 was absent from most EHEC O157 isolates obtained in Germany in 1996 brings its clinical importance into question. However, its prevalence in isolates obtained in other countries (33, 39) as well as in E. coli O157 isolates obtained in Germany in the years 1987 to 1991 was relatively high. In this study, no correlation between the presence of pColD157 in E. coli O157 isolates from stool specimens and the symptoms of the respective patients could be found. This is in accordance with the observations of Bradley et al. (7), who presented similar results concerning the colicinogeny of E. coli O157:H7 isolates. However, in their study, colicinogenic activity was found only in strains from patients and not in strains from asymptomatic carriers.
Colicins have been shown to be produced by a range of pathogenic E. coli isolates. Bradley (6) has demonstrated that 24% of 124 pathogenic E. coli strains including strains of serogroups O6, O26, O44, O55, O111, 114, O125, O126, O119, and O157 synthesized colicins. Among these strains, serogroups O55 and O157 had larger percentages of colicinogenic strains (33 and 50%, respectively) than the remainder of the serogroups. In addition, colicinogeny could be shown in Shigella spp. (20), uropathogenic E. coli, and E. coli isolated from human blood (13). Although the production of bacteriocins by a wide range of gram-negative and gram-positive bacteria has been reported, the role of this large group of bacterial toxins remains speculative.
Early studies of Branche et al. (8) and Emslie-Smith (12), who investigated the length of survival of colicinogenic bacteria in the intestine, revealed a significant correlation between the period of residence and the colicinogenicities of strains. Both groups found that resident serotypes were more likely than transient types to be colicinogenic. Moreover, a volunteer study performed by Smith and Huggins (41) revealed that after ingestion of a mixture of 10% colicin V-producing E. coli and 90% colicin V-negative E. coli, only the colicin V-producing E. coli strains were recovered at day 7. Although these experiments revealed a possible role of colicins in the succession of strains in the intestine, the results could not be confirmed by others. A more promising approach to determining the significance of colicinogeny would be to follow up the monitoring of strain succession with further studies on those strains which become residents or transients because of the presence or absence of colicin factors. Regarding enteric E. coli pathogens, it would be of interest to know whether long-term shedding of EHEC strains is related to the presence of colicinogenic factors. A direct role of colicins in human disease is not likely (19). Although changes in the intestinal flora after infection with colicin-producing pathogens have been observed, the direct action of colicins on eucaryotic cells could not be proven.
The most likely function of the colicins is to confer a selective advantage in the human intestine. However, since it is not likely that colicins in the gut would escape degradation by intestinal proteases, a possible role of colicins outside the normal habitat of E. coli was suspected. The production of colicins could be of importance among competing bacteria in the fecal-oral transmission route, e.g., to become numerically dominant in feces. However, the concentrations of major proteases such as trypsin, chymotrypsin, and elastase in feces are extremely low, so that colicins could be functional in the colon.
The assumed evolutionary advantage attributed to colicin production might play a role in the early stages of infection, and colicinogeny might raise the virulence of enterohemorrhagic E. coli O157:H7 and O157:H− by inhibiting the normal gut flora or by giving them an advantage during the fecal-oral transmission route. Further data are needed to establish the actual role and prevalence of the plasmid and whether the DNA probe developed in this study would provide a useful tool in such investigations.
ACKNOWLEDGMENTS
We are indebted to Beatrix Henkel and Barbara Plaschke for excellent technical assistance and Werner Brunder for helpful communications.
This work was supported by grant Ka 217/3 from the Deutsche Forschungsgemeinschaft.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.36.1.24-29.1998
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/36/1/24.full.pdf
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/full/36/1/24
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/36/1/24.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/36/1/24
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.36.1.24-29.1998
Article citations
Escherichia coli O157:H7 tir 255 T > A allele strains differ in chromosomal and plasmid composition.
Front Microbiol, 14:1303387, 15 Dec 2023
Cited by: 1 article | PMID: 38169669 | PMCID: PMC10758439
Alteration of a Shiga toxin-encoding phage associated with a change in toxin production level and disease severity in Escherichia coli.
Microb Genom, 9(2), 01 Feb 2023
Cited by: 5 articles | PMID: 36821793 | PMCID: PMC9997748
Chronological set of E. coli O157:H7 bovine strains establishes a role for repeat sequences and mobile genetic elements in genome diversification.
BMC Genomics, 21(1):562, 17 Aug 2020
Cited by: 2 articles | PMID: 32807088 | PMCID: PMC7430833
Isolation and characterization of two novel groups of kanamycin-resistance ColE1-like plasmids in Salmonella enterica serotypes from food animals.
PLoS One, 13(3):e0193435, 07 Mar 2018
Cited by: 4 articles | PMID: 29513730 | PMCID: PMC5841774
Design of synthetic epigenetic circuits featuring memory effects and reversible switching based on DNA methylation.
Nat Commun, 8:15336, 24 May 2017
Cited by: 15 articles | PMID: 28537256 | PMCID: PMC5458116
Go to all (21) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - Y10412
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Colicinogeny of O157:H7 enterohemorrhagic Escherichia coli and the shielding of colicin and phage receptors by their O-antigenic side chains.
Can J Microbiol, 37(2):97-104, 01 Feb 1991
Cited by: 19 articles | PMID: 1711924
Sorbitol-Fermenting Enterohemorrhagic Escherichia coli O157:H- Isolates from Czech Patients with Novel Plasmid Composition Not Previously Seen in German Isolates.
Appl Environ Microbiol, 83(23):e01454-17, 16 Nov 2017
Cited by: 1 article | PMID: 28970221 | PMCID: PMC5691429
A 3.3-kb plasmid of enterohemorrhagic Escherichia coli O157:H7 is closely related to the core region of the Salmonella typhimurium antibiotic resistance plasmid NTP16.
Plasmid, 39(2):134-140, 01 Jan 1998
Cited by: 6 articles | PMID: 9514711
The role of virulence factors in enterohemorrhagic Escherichia coli (EHEC)--associated hemolytic-uremic syndrome.
Semin Thromb Hemost, 27(3):207-213, 01 Jun 2001
Cited by: 30 articles | PMID: 11446654
Review




