Abstract
Free full text

The M2 Ectodomain Is Important for Its Incorporation into Influenza A Virions
Abstract
M2 is an integral protein of influenza A virus that functions as an ion channel. The ratio of M2 to HA in influenza A virions differs from that found on the cell surface, suggesting selective incorporation of M2 and HA into influenza virions. To examine the sequences that are important for M2 incorporation into virions, we used an incorporation assay that involves expressing M2 from a plasmid, transfecting the plasmid into recipient cells, and then infecting those cells with influenza virus. To test the importance of the different regions of the protein (extracellular, transmembrane, and cytoplasmic) in determining M2 incorporation, we created chimeric mutants of M2 and Sendai virus F proteins, exchanging corresponding extracellular, transmembrane, and cytoplasmic domains. Of the six possible chimeric mutants, only three were expressed on the cell surface. Of these three chimeric proteins, only one mutant (with the extracellular domain from M2 and the rest from F) was incorporated into influenza virions. These results suggest that the extracellular domain of M2 is important for its incorporation into virions.
The influenza A virus is an enveloped negative-strand virus with eight RNA segments encapsidated with nucleoprotein (NP) (reviewed in reference 20). Spanning the viral membrane are three proteins, hemagglutinin (HA), neuraminidase (NA), and M2. The infectious cycle of influenza virus is initiated by HA binding to sialic acid-containing receptors on the cell surface, leading to endocytosis of the virion into the cell. In the endosome, HA, triggered by the low pH in the endosome, mediates fusion, and uncoating of the virion occurs. It is at this point that the M2 protein is thought to act as an ion channel (14, 30, 34), transmitting the low pH of the endosome into the virion core, allowing M1 (matrix protein) to separate from RNP (viral RNA and its associated proteins, NP and three polymerase proteins). Dissociation of M1 from RNP is important, because only free RNP can enter the nucleus to be replicated and transcribed (23). After replication, transcription, and translation, the viral proteins are assembled at the cell surface, from which virions, incorporating viral proteins and RNA, bud out (reviewed in reference 20).
Incorporation of proteins into influenza virions appears to be a selective process, because relatively few host cell proteins, such as actin, are included in the virion (1, 20). It is apparent that selection (or exclusion) of influenza virus proteins occurs, since the HA-to-M2 ratios found on the cell surface are not mirrored in the virion. Even though M2 is expressed in fairly large amounts on the cell surface, relatively few copies (20 to 60 molecules/virion) are incorporated into virions (21, 40). By contrast, HA is present in large amounts both on the cell surface (12) and in the virion (approximately 500 molecules/virion) (7, 16). Some mechanism must exist to selectively include or exclude these proteins. What determines which proteins are included in the virion and how much of each? Are there any specific sequences in these proteins that determine whether they will be incorporated into the virion? The transmembrane region of HA has been found to be essential for the incorporation of HA into virions (17, 26). For NA, the incorporation sequences have not been localized. However, neither the cytoplasmic tail of NA nor that of HA is essential for incorporation, though they seem to play important roles in virion morphology (11, 17, 18, 25). Very little is known about the sequences important for M2 incorporation into virions.
The M2 gene encodes a 97-amino-acid protein that is expressed on the cell surface as a tetramer. It is composed of 24 extracellular amino acids, 19 transmembrane amino acids, and 54 cytoplasmic residues (21). Disulfide bonds link the protein through cysteines located in the extracellular region (14). The transmembrane domain, when incorporated into a lipid bilayer, can act as an ion channel, suggesting that this domain forms the pore and is responsible for the activity (9). However, the roles of the other M2 domains in virus replication remain unknown.
The goal of this study was to determine which region(s) of M2 (extracellular, transmembrane, or cytoplasmic) is responsible for its incorporation into virions. To this end, we have used a system that allows us to examine M2 incorporation into virions. In this system, the viral proteins being studied are expressed in a plasmid expression vector, under the control of the chicken β-actin promoter. This system is unlike previous incorporation systems, which used viral expression vectors, such as vaccinia virus. Interpretation of results was more difficult with these systems because many foreign or irrelevant viral proteins were also being expressed. Using our system, we have identified M2 incorporation sequences.
MATERIALS AND METHODS
Virus and cells.
COS-1 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. A/Puerto Rico/8/34 (H1N1) (PR8) and A/turkey/Minnesota/833/80 (H4N2) (Ty/MN) viruses were grown in eggs and purified (22).
Antibodies.
The antibodies used against M2 were mouse monoclonal antibody 14C2 (40) and rabbit antiserum R3C, which was raised against a peptide spanning the last 15 amino acids of PR8 M2, AVDADDGHFVSIELE. The M1 monoclonal antibodies used were a pool of anti-WSN 174/4 and anti-MEM/2/85 E2-1. The anti-F monoclonal antibody pool was composed of M33/1, M38/1, and M16/1 (31).
Construction of plasmids.
All constructs were made in pCAGGS/MCS, which was adapted from pCAGGS (27) by adding a multicloning site at the EcoRI/BglII site, using the primer 5′-AATCGAGCTCATCGATGCATGGTACCCGGGCATGCTCGAGCTAGCAG-3′. pCAGGS contains the eukaryotic chicken β-actin promoter. For PR8 M2, the wild-type M2 gene was generated by PCR (33) from pUCT3PRM (the PR8 M gene cloned in pUC19 [5]) and cloned into the EcoRI and SmaI sites of pCAGGS/MCS. The Sendai virus F gene was excised from pTF1-SVF (4) as an EcoRI/SmaI fragment and placed into pCAGGS/MCS. We named the resulting construct pSVF.
All subsequent chimeric constructs were made, with pTF1-SVF and wild-type PR8 M2 as templates, by gene splicing through overlap extension by PCR (15) with Pfu polymerase (Stratagene) under conditions recommended by the manufacturer.
The PR8 M2 mutant lacking its cytoplasmic tail [M2 (no-tail)] was created by PCR with the wild-type PR8 M2 plasmid as a template. All of the constructs were sequenced to ensure that unwanted mutations were not present.
Fluorescence-activated cell sorting (FACS).
Twenty-four hours after transfection, cells were washed three times with phosphate-buffered saline (PBS), pH 7.2, and detached from the plates by trypsinization at 37°C. They were then passed through 100-μm-pore-size Spectra/mesh (Spectrum) to remove large clumps. Trypsin was removed by spinning down the cells, washing them twice, and then resuspending them in PBS with 5% newborn calf serum (NCS-PBS). The cells were incubated for 30 min at 4°C with an appropriate primary mouse monoclonal antibody. After three washes, the cells were treated with a 1/20 dilution of anti-mouse immunoglobulin G (IgG)-conjugated fluorescein isothiocyanate-labeled antibody (Boehringer Mannheim Biochemicals) and incubated for another 30 min at 4°C. The cells were then washed with NCS-PBS and analyzed on a FACScan (Becton Dickinson Instruments). Propidium iodide was added before analysis to aid in detection of intact cells, thus decreasing the likelihood of false-positive signals.
Incorporation assay.
Approximately 10 μg of plasmid DNA was electroporated into COS-1 cells (as previously described [2] except that 60-mm-diameter plates were used in the final step). Twenty-four hours later, the electroporated cells were infected with Ty/MN virus (at a multiplicity of infection of 1). Twenty-four hours after infection, the supernatant was spun to remove debris and purified through a five-step sucrose gradient (25, 40, 47.5, 55, and 70%) over 2.5 h at 50,000 × g and 4°C. Fractions (0.3 ml) were collected, after a hole was pierced in the bottom of the tube, and each fraction was assayed by hemagglutination for the presence of virus. The fractions that contained virus were pooled and spun down at 50,000 × g for 1 h at 4°C and resuspended in 20 μl of lysis buffer (0.6 M KCl, 50 mM Tris-Cl [pH 7.5], 0.5% Triton X-100).
Western immunoblot analysis.
The viral lysates from the incorporation assay were examined by Western immunoblot analysis. Lysates were run on 10% Tris-glycine gels in sodium dodecyl sulfate (SDS)–Tris-glycine buffer. The gel was incubated in transfer buffer (48 mM Tris, 39 mM glycine, 0.0375% SDS, 20% methanol) for 30 min and then transferred to a nylon membrane (Nytran; Schleicher and Schuell) by using a semidry transblotter (Bio-Rad) for 1.5 h at 12 V. The membrane was probed with the appropriate antibody and then with either the anti-mouse antibody conjugated to horseradish peroxidase (Bio-Rad) at a 1/3,000 dilution or a primary anti-rabbit horseradish peroxidase-linked antibody from donkeys, provided in the ECL kit (Amersham). The proteins were visualized by using the Amersham ECL kit according to the manufacturer’s instructions. Antibodies were removed by incubating the membrane with a solution containing 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl (pH 6.7) at 50°C for 30 min.
Immunoprecipitation.
The cells were lysed with lysis buffer (1% Nonidet P-40, 20 mM Tris [pH 8], 0.15 M NaCl, 2 mM EDTA) at 4°C for 10 min and spun down to remove debris. Protein A-Sepharose beads (50 μl) (Fast Flow immobilized rProtein A; Repligen) were added to the lysate and incubated for 1 h at 4°C to remove proteins that bound nonspecifically to the beads. After being spun, the supernatant was incubated with the anti-M2 monoclonal antibody 14C2 overnight. Protein A-Sepharose beads (50 μl) coated with rabbit anti-mouse antibody (Sigma) were then added and incubated for 1 h. The beads were washed with ice-cold lysis buffer (with 0.02% SDS) four times and heated at 95°C in 40 μl of sample buffer (Novex) with 5% β-mercaptoethanol for 5 min to detach the proteins from the beads. The tubes were spun down once more, and the supernatant was run on a gel.
RESULTS
Assay for M2 incorporation into influenza virions.
To define the sequences in M2 that are important for its incorporation into virions, we first established an assay system. In this system, M2 protein is expressed in COS-1 cells by using a plasmid expression vector. The same cells are then infected with influenza virus. Since cell surface expression of a protein is a prerequisite for its incorporation, wild-type PR8 M2 expression on the cell surface was ascertained by FACS analysis (Fig. (Fig.1A)1A) with 14C2 (a monoclonal antibody that recognizes the extracellular domain of PR8 M2). M2 was detected on the cell surface, confirming its expression.
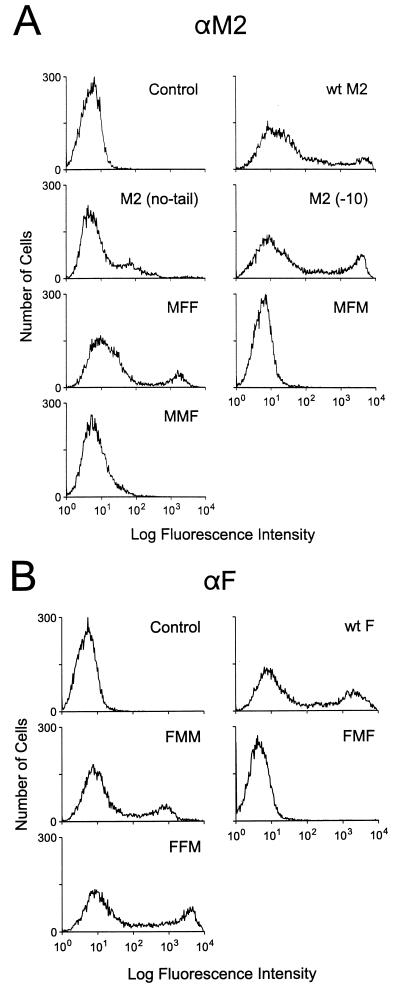
Cell surface expression of wild-type and chimeric M2 and F mutants. The wild-type and chimeric constructs described in the text and in Fig. Fig.66 were transfected into COS-1 cells and examined 24 h later by FACS analysis for cell surface expression, as described in Materials and Methods. Anti-M2 (αM2) monoclonal antibody 14C2 (A) and a pool of anti-F (αF) monoclonal antibodies (B) were used to label the cells.
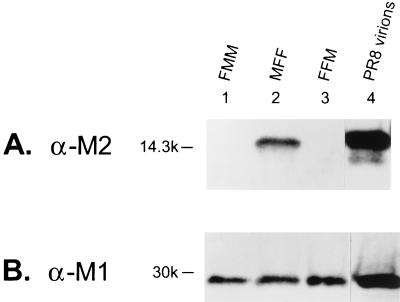
We first tested whether the wild-type PR8 M2 was incorporated into Ty/MN virions. Ty/MN was chosen as a target virus for M2 incorporation because the 14C2 M2 monoclonal antibody binds to PR8 M2 but not to Ty/MN M2. The PR8 M2-transfected cells were infected with Ty/MN virus. The virus was purified from the supernatant by fractionation on a sucrose step gradient, and viral proteins were examined by Western blot analysis. The monoclonal antibody detected a band corresponding to an estimated molecular mass of approximately 14 kDa, thereby demonstrating the presence of PR8 M2 in the Ty/MN virions (Fig. (Fig.2A,2A, lane 3). The lower band in this sample is probably a proteolytically cleaved form of M2, as reported by others (18, 40). The membrane was stripped and reprobed for M1, another influenza virus protein, to ensure that the M2 detected by Western blot analysis was indeed incorporated into virions (Fig. (Fig.2B,2B, lane 3).
Wild-type PR8 M2 is incorporated into Ty/MN virions. The incorporation assay was performed with a wild-type PR8 M2 plasmid (lanes 3) and vector alone (lanes 1) as described in Materials and Methods. Briefly, the plasmids were electroporated into COS-1 cells, which were then infected with Ty/MN virus. Virus in the culture supernatant was purified and examined by Western blot analysis. As another control for the assay, a wild-type M2 plasmid was transfected into cells, which were then subjected to a mock infection (lanes 2). Lanes 4 show the incorporation assay for the M2 mutant lacking the last 10 cytoplasmic residues [M2 (−10)]. The Western blot was probed with the anti-M2 (α-M2) monoclonal antibody 14C2 (A); the membrane was then stripped and reprobed with the anti-M1 (α-M1) monoclonal antibody pool (B).
As a negative control, we followed the same procedure as for the M2 incorporation assay except that we transfected the vector plasmid instead of that containing the M2 gene. Even though the fraction analyzed contained M1 protein (Fig. (Fig.2B,2B, lane 1), we did not observe any signal corresponding to M2 in our Western blot analysis (Fig. (Fig.2A,2A, lane 1), confirming the lack of 14C2 M2 monoclonal antibody cross-reactivity with Ty/MN M2. We also tested whether the viral purification procedures successfully removed contaminants, such as cellular debris. When cells transfected with the wild-type PR8 M2 were mock infected, no M2 was found in the fractions whose sucrose density, measured by refractometer, corresponded to the virus-containing fractions (Fig. (Fig.2A,2A, lane 2). These controls indicate that the procedures we used allow examination of PR8 M2 protein incorporation into Ty/MN influenza virions in the absence of contaminating cellular debris.
Wild-type PR8 M2 does not oligomerize with Ty/MN M2.
To exclude the possibility that M2 incorporation was due to hetero-oligomerization with Ty/MN M2, wild-type M2 was transfected into COS-1 cells, which were then infected with Ty/MN as described above. The cell lysates were immunoprecipitated with the M2 monoclonal antibody and then examined by Western blot analysis with a polyclonal antibody (R3C) that recognizes the cytoplasmic tail of M2. The antiserum to the M2 cytoplasmic tail recognizes the PR8 M2 and the Ty/MN M2 with equal sensitivity (see below). The 14C2 monoclonal antibody is specific for the ectodomain of PR8 M2 and, therefore, should immunoprecipitate PR8 M2 but not Ty/MN M2. If these two M2 proteins hetero-oligomerize, we should detect them both on the Western blot with the antiserum to the M2 cytoplasmic tail after immunoprecipitation with the 14C2 monoclonal antibody. The mobilities of Ty/MN M2 and PR8 M2 on a gel differ (Fig. (Fig.3,3, lanes 3 and 4), allowing us to distinguish between the two proteins. Western blot analysis with antiserum to the M2 cytoplasmic tail, after immunoprecipitation of the lysate with 14C2 M2 monoclonal antibody, detected the PR8 M2 but not Ty/MN M2 (Fig. (Fig.3,3, lane 2), demonstrating that the wild-type M2 does not hetero-oligomerize with Ty/MN M2. The 22-kDa band seen in the two left lanes of Fig. Fig.3,3, which contain immunoprecipitated samples, is probably the mouse IgG light chain cross-reacting with the anti-rabbit IgG used as a secondary antibody and is visible due to the sensitive chemiluminescence method of detection.
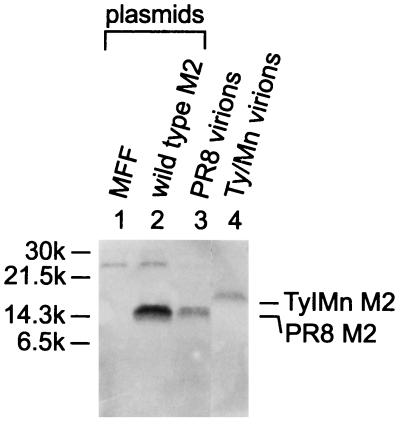
Neither the wild-type PR8 M2 nor the chimeric protein between M2 and Sendai F proteins (MFF) hetero-oligomerizes with Ty/MN M2. (Lanes 1 and 2) Each plasmid indicated was transfected into COS-1 cells, which were then infected with Ty/MN. The viral lysate was immunoprecipitated with the anti-M2 monoclonal antibody 14C2 and then incubated with anti-mouse secondary antibody-coated protein A-Sepharose beads. The immunoprecipitated product was examined by a Western blot analysis. (Lanes 3 and 4) The blot was probed with the polyclonal antibody R3C, which recognizes the cytoplasmic tails of both PR8 M2 and Ty/MN M2. PR8 and Ty/MN virions were fractionated by SDS-PAGE without immunoprecipitation and then probed with the R3C antibody as controls.
M2 C-terminal deletion mutants are incorporated into influenza virions.
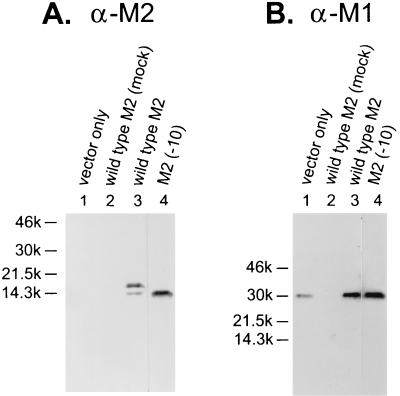
We next investigated which M2 regions are important for its incorporation into virions. Previously, we generated an influenza virus containing M2 that lacked a single carboxy-terminal residue, but we were unable to generate recombinant viruses containing M2 that lack 5 or 10 C-terminal residues (5). These data suggested to us that the C-terminal residues may play an important role in virus replication. To examine whether our failure to generate virus with M2 lacking the C-terminal 10 amino acids was due to the inability of mutant M2 to be incorporated into the virions, we tested its incorporation using our system. An M2 mutant lacking 10 amino acids of its cytoplasmic tail, referred to as M2 (−10), was efficiently expressed on the cell surface (Fig. (Fig.1A)1A) and incorporated in virions (Fig. (Fig.2A,2A, lane 4). This result shows that the C-terminal 10 amino acids are not essential for M2 incorporation into virions.
To further address the importance of the cytoplasmic tail for incorporation, we tested a mutant that lacked the entire cytoplasmic tail [M2 (no-tail)]. The M2 (no-tail) mutant was incorporated into virions but at a drastically reduced level compared to the incorporation of wild-type M2 (Fig. (Fig.4,4, lanes 1). However, since cell surface expression of the M2 (no-tail) mutant was also greatly reduced (Fig. (Fig.1A),1A), it is difficult to determine where the defect lies, and no firm conclusions can be drawn about the importance of the tail in incorporation. We tried varying the temperature, the amount of DNA transfected, and the hours of incubation after transfection in attempts to match the level of cell surface expression (represented by the FACS profile) of the no-tail mutant to that of the wild type, but no satisfactory solution was found. Therefore, we can only conclude that the cytoplasmic tail is not an absolute requirement for incorporation.
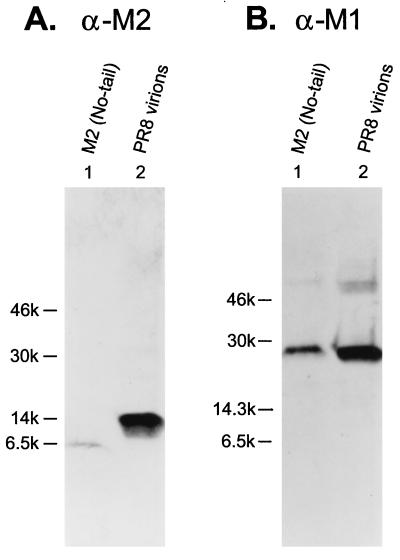
M2 lacking its cytoplasmic tail is incorporated into virions at very low levels. (Lanes 1) The M2 (no-tail) mutant was tested for its incorporation into virions. (Lanes 2) Purified PR8 virions were analyzed as a control. (A) The blot was probed with the anti-M2 (α-M2) monoclonal antibody 14C2. (B) The M2 monoclonal antibody was stripped from the membrane, which was then reprobed with the anti-M1 (α-M1) antibody pool.
Sendai virus F protein is excluded from influenza virions.
Another way to examine virion incorporation determinants is to find M2 sequences that allow incorporation of non-influenza virus proteins, or portions of these proteins, that are usually not incorporated into influenza virions. Sendai virus F was chosen because it is a paramyxovirus, a close cousin of orthomyxoviruses. The F protein, a trimeric class I membrane protein, is involved in the fusion between the viral envelope and the cellular membrane. It comprises 499 extracellular amino acids, 24 transmembrane amino acids, and 42 cytoplasmic amino acids (3). We tested whether the Sendai virus F protein is incorporated into influenza virions by placing its gene in the same vector we used for other constructs and expressing the F protein in COS-1 cells. FACS analysis demonstrated cell surface expression of F protein (Fig. (Fig.1B);1B); however, the protein was not detected in influenza virions by Western blot analysis (Fig. (Fig.5A,5A, lane 2) upon superinfection of F-expressing cells with Ty/MN virus. This finding indicates that the F protein is excluded from influenza virions.
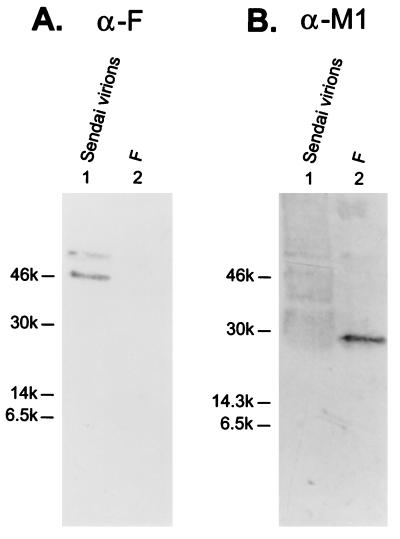
Sendai virus F protein is not incorporated into Ty/MN virions. The Sendai virus F gene in pCAGGS/MCS was transfected into COS-1 cells, and the incorporation assay was performed (lanes 2). Purified Sendai virions (lanes 1) were included as a control. (A) The Western blot was probed with the anti-F (α-F) monoclonal antibody pool. (B) The anti-F monoclonal antibodies were stripped from the membrane, which was then reprobed with a pool of anti-M1 (α-M1) monoclonal antibodies.
Incorporation of the M2-F chimeric proteins into influenza virions.
To identify the M2 regions that are important for virion incorporation, we made chimeric mutants of M2 and Sendai virus F proteins. Basically, the extracellular, transmembrane, or cytoplasmic domain of M2 was replaced with the corresponding region of the Sendai virus F protein and vice versa. Six different chimeric mutants were constructed, as depicted in Fig. Fig.6,6, and tested for their cell surface expression. Of the six chimeric protein constructs, only three—FMM (with the M2 extracellular domain replaced with the corresponding F domain), MFF (with the F extracellular domain replaced with the corresponding M2 domain), and FFM (with the F cytoplasmic domain replaced with the M2 cytoplasmic domain)—were expressed on the cell surface, as evidenced by FACS analysis (Fig. (Fig.1).1). The first set of FACS data was obtained with the 14C2 monoclonal antibody, which recognizes the ectodomain of M2, and it showed expression of MFF at the cell surface at high levels, although not as high as those for wild-type M2. The second set was obtained with a pool of anti-F monoclonal antibodies (M16/1, M33/1, and M38/1), and it showed high expression of FMM and FFM, with FFM being expressed at levels equivalent to those for wild-type F.
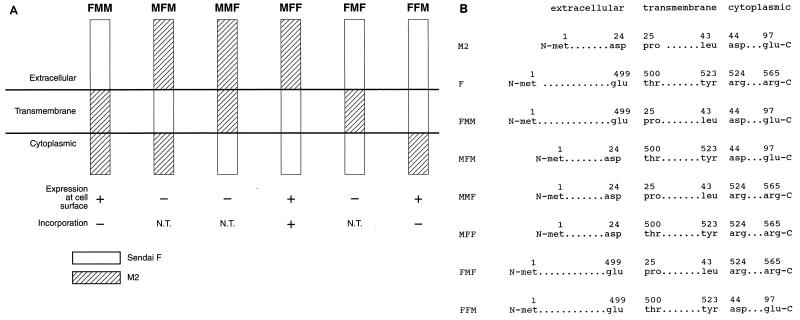
(A) Diagram of chimeric mutants of PR8 M2 and Sendai virus F. Chimeric mutants were constructed as shown, replacing a region (extracellular, transmembrane, or cytoplasmic) of one protein with its counterpart from the other protein. N.T., not tested. (B) Detailed description of chimeric mutants. The numbers above the amino acids indicate the positions of the amino acids in the wild-type protein.
We next investigated whether the three chimeric proteins that were expressed on the cell surface were also incorporated into the virions. Neither FFM nor FMM was detected in the virions by Western blot analysis (Fig. (Fig.7A);7A); however, MFF was detected in the influenza virions (Fig. (Fig.8A).8A). In each case, equivalent amounts of virus were used, as confirmed by probing the blots with M1 monoclonal antibodies (Fig. (Fig.7B7B and and8B).8B). These data indicate that the extracellular region of M2 contains sequences that are essential for its incorporation into virions, since it allowed incorporation of the transmembrane and cytoplasmic domains of F, which in its entirety is not incorporated into influenza virions, as shown in Fig. Fig.5.5.
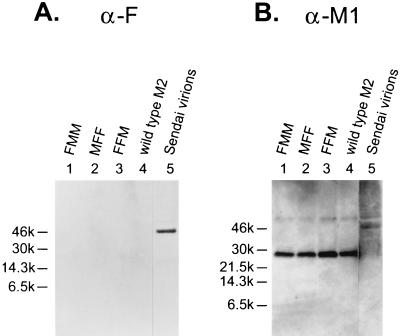
Chimeric proteins containing the M2 transmembrane and cytoplasmic domains or only the cytoplasmic domain are not incorporated into Ty/MN virions. The three chimeric constructs found to be expressed on the cell surface were assayed for incorporation as described in Materials and Methods. (A) The viral lysates were probed on a Western blot with anti-F (α-F) monoclonal antibodies. (B) The anti-F monoclonal antibodies were stripped from the membrane, which was then reprobed with a pool of anti-M1 (α-M1) monoclonal antibodies. Purified Sendai virions were loaded in the right-hand lanes as a control.
Incorporation into virions of the M2-F chimeric protein that contains only the M2 ectodomain is not due to hetero-oligomerization with the influenza virus M2.
As with wild-type PR8 M2, it is possible that MFF is incorporated into virions due to its hetero-oligomerization with the M2 of the Ty/MN virus. To exclude this possibility, an MFF-expressing plasmid was transfected into COS-1 cells, which were then infected with Ty/MN virus (Fig. (Fig.3).3). The cell lysates were immunoprecipitated with the 14C2 M2 monoclonal antibody and then probed by a Western blot analysis with the antiserum to the M2 cytoplasmic tail (R3C). 14C2 should immunoprecipitate MFF because it recognizes the extracellular domain of PR8 M2. If MFF protein hetero-oligomerizes with the M2 of the Ty/MN virus, the latter molecule should be detected on the Western blot with the antiserum to the M2 cytoplasmic tail. No band was seen at the position where the Ty/MN M2 would be (Fig. (Fig.3,3, lane 1), even though MFF expression on the cell surface was detected by immunostaining (data not shown). In addition, no MFF band was seen because R3C only recognizes the cytoplasmic tail of M2 and not the M2 ectodomain that MFF contains. Therefore, the results indicate that the chimeric protein MFF does not hetero-oligomerize with Ty/MN M2. These findings prove that incorporation of a chimeric protein containing only the M2 ectodomain and the transmembrane and cytoplasmic domains of the F protein into influenza virions occurs by virtue of the M2 extracellular sequence.
DISCUSSION
We have used a method to assay the incorporation of protein into virions that makes use of a plasmid expression vector rather than viral vectors, which produce additional proteins that may affect the experiment. Using our system, we demonstrate that the extracellular region of M2 is sufficient for its incorporation into influenza virions. In the other systems studied, the cytoplasmic region (vesicular stomatitis virus [VSV] G protein [28, 38] and alphavirus spike protein [35]) or the transmembrane region (Rous sarcoma virus envelope protein [8] and influenza virus HA [17, 26]) has been found to contain sequences important for incorporation. To our knowledge, this study represents the first documented case of a virion incorporation determinant residing in the extracellular domain.
What mechanism would involve incorporation sequences in the extracellular domain? Any proposed mechanism must account for the inefficient incorporation of foreign proteins into influenza virions, as well as the selective incorporation of viral proteins such that the ratio of membrane proteins in the virion is low for M2 and high for HA. It must also explain the presence of incorporation determinants in the extracellular region of M2 versus determinants in the transmembrane region of HA. To satisfy these requirements, we propose a model (Fig. (Fig.9)9) that involves protein-protein interactions occurring outside the cell, between the ectodomains of the proteins expressed at the cell surface, such as HA, NA, and M2. M2 may contain sequences in the extracellular domain that allow it to interact with other viral proteins in an “incorporation complex.” This complex would contain the viral membrane proteins, bound by extracellular interactions, that would assemble at the cell surface just before budding. A lack of interactions between foreign proteins and viral membrane proteins would lead to exclusion, accounting for the inefficient incorporation of foreign proteins, whereas correct interactions among the viral membrane proteins at the cell surface would lead to incorporation.
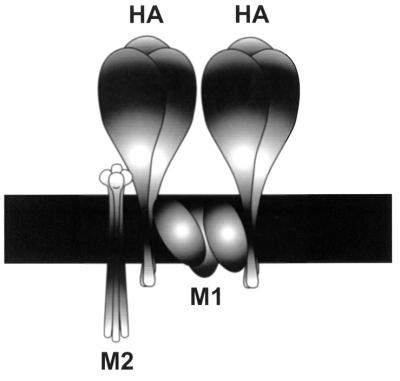
Schematic model of M2 incorporation into influenza A virions. As a part of the postulated incorporation complex, the M2 proteins interact with HA (and possibly with NA as well) in the extracellular region, whereas HA trimers interact with each other in the transmembrane region, possibly through M1.
The M2/HA ratio in the virion is less than that on the cell surface. Therefore, it is possible that only a few M2 molecules are included in the incorporation complex to interact with other viral membrane proteins. Those molecules that do not get the opportunity to interact with other viral membrane proteins are not included. It is likely that the presence of too many M2 molecules is disadvantageous for the virus because an overabundance of M2 may cause too rapid and too much acidification of the virus in the endosome, which could disrupt the M1-RNP complex before it is required.
Because the transmembrane region of HA is important for determining its incorporation (17, 26), it is reasonable to speculate that the HA interactions that are important for incorporation (e.g., those with other HA trimers or with other viral proteins) occur through its transmembrane domain. In contrast, incorporation determinants for M2 were found in the extracellular region and, therefore, interactions important for M2 incorporation must occur there. To elaborate, it is possible that in the incorporation complex, M2 and HA (Fig. (Fig.9),9), and maybe NA also, interact in the extracellular domain, whereas HA trimers interact with other trimers through contacts in the transmembrane region. This interaction between HA trimers may be mediated by M1, which is found in the membrane as well as in the cytoplasmic region. Our model (Fig. (Fig.9)9) is supported by the fact that the presence of M1 in the membrane has been demonstrated by various fractionation studies and labeling experiments (13, 29). M1, NA, and HA interactions are corroborated by studies that show that M1 protein association with the membrane is stimulated by HA and NA expression (10), although Zhang and Lamb (41), as well as Kretzschmar et al. (19), obtained opposite results for unknown reasons. There is evidence in the literature of cytoplasmic protein and transmembrane protein interactions serving important functions in assembly, as is the case for alphaviruses (35). In Semliki Forest virus, the interaction of the cytoplasmic capsid protein with the cytoplasmic region of the transmembrane spike protein is thought to drive budding during assembly (42). Thus, a precedent has been set for the proposed interaction of M1 with HA, except that this interaction is suggested to exist in the transmembrane domain, as opposed to the cytoplasmic domain for Semliki Forest virus. For NA, the locations of sequences essential for incorporation have not been identified (11, 25); therefore, it is difficult to speculate as to whether the extracellular or the transmembrane domain is involved in its incorporation. Nonetheless, important interactions for incorporation may be taking place in both the extracellular and transmembrane regions.
In the phenotypic mixing experiments, Zawada and Rosenbergova (39) coinfected cells with fowl plague influenza virus and VSV. They observed VSV particles which could be neutralized by anti-influenza virus serum and concluded that the virions contained influenza membrane proteins. This is not surprising in view of the numerous reports of VSV phenotypically mixing with a variety of other viral glycoproteins (24, 37). However, no influenza virus particles neutralizable with anti-VSV serum were observed. On the basis of our results and our model, we can explain the lack of influenza virions containing VSV transmembrane proteins. Influenza membrane proteins can be incorporated into VSV because VSV operates through a different mechanism to incorporate influenza proteins, whereas influenza virions cannot incorporate VSV membrane proteins because they may fail to participate in the influenza virus incorporation complex.
The coinfection experiments illustrate incorporation mechanisms which can explain why influenza virus membrane proteins can be incorporated into VSV but not vice versa. A mechanism employed by VSV and retroviruses makes use of proteins that direct assembly and budding, such as the retroviral Gag and the VSV M proteins. This mechanism allows incorporation, albeit at a lower efficiency (36), of many foreign proteins that are expressed on the cell surface (6, 24, 32). This is in contrast to viruses like influenza virus, which lack proteins that single-handedly direct assembly and budding. Instead, these viruses probably require a concerted interplay of interactions among many proteins at the cell surface to form an incorporation complex that gives rise to assembly and budding. Specific interactions with several proteins are required to create the postulated incorporation complex; therefore, foreign proteins are more likely to be excluded from incorporation into influenza virions.
An influenza virus containing the M2 mutant that lacks only the carboxy-terminal residue but not 10 residues of the 54-amino-acid cytoplasmic tail has been generated by reverse genetics (5). Because the last 10 amino acids of the cytoplasmic tail were not necessary for M2 incorporation into virions, the defect in the M2 (−10) mutant must be at other steps during the virus replication cycle. Therefore, the function of the M2 cytoplasmic tail remains unknown.
ACKNOWLEDGMENTS
We gratefully acknowledge Robert A. Lamb for the hybridoma cell for 14C2 monoclonal antibody, Brian Murphy for its production, Robert G. Webster for the M1 monoclonal antibody pool, and Susan Watson for editing the manuscript.
Support for this work came from National Institute of Allergy and Infectious Diseases Public Health Service research grant AI-29599, Cancer Center Support (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.72.3.2449-2455.1998
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/72/3/2449.full.pdf
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/72/3/2449
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/72/3/2449
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/content/full/72/3/2449
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.72.3.2449-2455.1998
Article citations
Antiviral Approaches against Influenza Virus.
Clin Microbiol Rev, 36(1):e0004022, 16 Jan 2023
Cited by: 21 articles | PMID: 36645300 | PMCID: PMC10035319
Review Free full text in Europe PMC
Influenza Viruses: Harnessing the Crucial Role of the M2 Ion-Channel and Neuraminidase toward Inhibitor Design.
Molecules, 26(4):880, 07 Feb 2021
Cited by: 13 articles | PMID: 33562349 | PMCID: PMC7916051
Review Free full text in Europe PMC
A Novel Mechanism Underlying Antiviral Activity of an Influenza Virus M2-Specific Antibody.
J Virol, 95(1):e01277-20, 09 Dec 2020
Cited by: 6 articles | PMID: 33055251 | PMCID: PMC7737731
X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59(4):627-634, 13 Jan 2020
Cited by: 13 articles | PMID: 31894969 | PMCID: PMC7224692
Viroporins in the Influenza Virus.
Cells, 8(7):E654, 29 Jun 2019
Cited by: 22 articles | PMID: 31261944 | PMCID: PMC6679168
Review Free full text in Europe PMC
Go to all (49) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding.
J Virol, 82(20):10059-10070, 13 Aug 2008
Cited by: 169 articles | PMID: 18701586 | PMCID: PMC2566248
Amphipathic Helices of Cellular Proteins Can Replace the Helix in M2 of Influenza A Virus with Only Small Effects on Virus Replication.
J Virol, 94(3):e01605-19, 17 Jan 2020
Cited by: 7 articles | PMID: 31694941 | PMCID: PMC7000973
Influenza A virus M2 ion channel protein: a structure-function analysis.
J Virol, 68(3):1551-1563, 01 Mar 1994
Cited by: 163 articles | PMID: 7508997 | PMCID: PMC236612
[Structure and function of the influenza virus M2 ion channel protein].
Nihon Rinsho, 55(10):2587-2592, 01 Oct 1997
Cited by: 1 article | PMID: 9360376
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA021765
Grant ID: CA-21765
NIAID NIH HHS (1)
Grant ID: AI-29599