Abstract
Free full text

Differential Early Interactions between Salmonella enterica Serovar Typhi and Two Other Pathogenic Salmonella Serovars with Intestinal Epithelial Cells
Abstract
Salmonella enterica serovar Typhi (hereafter referred to as S. typhi) is a host-restricted pathogen that adheres to and invades the distal ileum and subsequently disseminates to cause typhoid fever in humans. However, S. typhi appears to be avirulent in small animals. In contrast, other pathogenic salmonellae, such as S. enterica serovars Typhimurium and Dublin (S. typhimurium and S. dublin, respectively), typically cause localized gastroenteritis in humans but have been used as models for typhoid fever because these organisms cause a disease in susceptible rodents that resembles human typhoid. In vivo, S. typhi has been demonstrated to attach to and invade murine M cells but is rapidly cleared from the Peyer’s patches without destruction of the M cells. In contrast, invasion of M cells by S. typhimurium is accompanied by destruction of these M cells and subsequently sloughing of the epithelium. These data have furthered our view that the early steps in the pathogenesis of typhoidal and nontyphoidal Salmonella serovars are distinct. To extend this concept, we have utilized an in vitro model to evaluate three parameters of initial host-pathogen interactions: adherence of three Salmonella serovars to human and murine small intestinal epithelial cell (IEC) lines, the capacity of these salmonellae to invade IECs, and the ability of the bacteria to induce interleukin-6 (IL-6) in these cell lines as a measure of host cell activation and the host acute-phase response. The results demonstrate that S. typhi adheres to and invades human small IECs better than either S. typhimurium or S. dublin. Interestingly, invA and invE null mutants of S. typhi are able neither to adhere to nor to invade IECs, unlike S. typhimurium invA and invE mutants, which adhere to but cannot invade IECs. S. typhi also induces significantly greater quantities of IL-6 in human small IEC lines than either of the other two Salmonella serovars. These findings suggest that differential host cytokine responses to bacterial pathogens may play an important role in the pathological sequelae that follow infection. Importantly, S. typhimurium did not induce IL-6 in murine IECs. Since S. typhimurium infection in mice is often used as a model of typhoid fever, these findings suggest that, at least in this case, the mouse model does not reflect the human disease. Taken together, our studies indicate that (i) marked differences occur in the initial steps of S. typhi, S. typhimurium, and S. dublin pathogenesis, and (ii) conclusions about S. typhi pathogenesis that have been drawn from the mouse model of typhoid fever should be interpreted conservatively.
Salmonella enterica serovar Typhi (hereafter referred to as S. typhi) is a human pathogen that causes typhoid fever. After being ingested in contaminated food or water, S. typhi adheres to and invades microfold (M) and epithelial cells in the distal small intestine (17, 18, 23, 37, 39). Subsequently, the organisms disseminate throughout the reticuloendothelial system, and symptoms ensue (17, 18). Other Salmonella serovars, specifically, S. enterica serovar Typhimurium, S. enterica serovar Dublin, and S. enterica serovar Enteritidis (referred to as S. typhimurium, S. dublin, and S. enteritidis, respectively), usually do not cause a disseminated, systemic disease in humans but clinically manifest as gastroenteritis and diarrhea in humans (10, 20).
The studies of S. typhi pathogenesis have been limited mostly to in vitro systems, since this organism fails to establish disease or significant pathology in small laboratory animal models (2, 9, 30, 35). Wild-type S. typhi, isolated from humans with clinical disease, is a host-restricted pathogen and is avirulent in susceptible strains of mice (Itys) (50% lethal dose [LD50] = >109 organisms orally) (2, 30, 35). In contrast to S. typhi, wild-type S. typhimurium and S. dublin are lethal in Itys mice at low doses (oral LD50 of 104 organisms [40] and intraperitoneal LD50s of <50 S. typhimurium and <10 S. dublin organisms [7, 40]). This lethal murine disease has been likened to human typhoid (3, 7, 10, 36, 40).
Several studies have suggested that the clinical and pathological sequelae associated with specific serovars of Salmonella may be the result of differences in the early steps of pathogenesis. Recently, Pascopella et al. concluded that S. typhi invades the murine intestinal epithelium via M cells but that invasion does not destroy the M cells and the organisms are quickly found in phagocytic cell vacuoles beneath the follicle-associated epithelium (39). Previous studies have demonstrated that S. typhimurium also preferentially invades M cells (2a, 20, 39) but, more importantly, that invasion results in generalized destruction of the follicle-associated epithelium and is accompanied by replication within the Peyer’s patches (20, 39). Moreover, McCormick et al. demonstrated that Salmonella serovars that cause gastroenteritis show transepithelial signaling to neutrophils across polarized human intestinal epithelial cell (IEC) monolayers, whereas Salmonella serovars that elicit human enteric fever do not elicit this response (29). Finally, Kops et al. (24) showed that S. typhi migrates through polarized human IECs significantly better than S. typhimurium. Taken together, these studies suggest that differences which dictate the subsequent clinical sequelae may exist among Salmonella serovars in the initial steps of pathogenesis in humans.
Recent studies in our laboratory have focused on the early essential steps of S. typhi pathogenesis, namely, the adherence to and invasion of the human IECs by these bacteria. We demonstrated that S. typhi can induce the proinflammatory cytokine interleukin-6 (IL-6) in human small and large IEC lines (47). In this report, the initial steps of S. typhi pathogenesis were compared with those of S. typhimurium and S. dublin. Our results demonstrate that the early interactions between S. typhi and small IECs differ quantitatively and qualitatively from the initial interactions of other Salmonella serovars with host IECs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacteria used in this study are listed in Table Table1.1. S. typhi H553 (invE) was constructed by P22 transduction of S. typhi ISP1820 by the procedure previously described by Miller (31). Luria-Bertani (LB) broth or Sambrook agar (41) with 0.3 M NaCl was used for routine growth of bacteria.
TABLE 1
Bacterial strains used in this study
study
| Serovar | Relevant genotype | Source (reference[s]) |
|---|---|---|
| S. typhi | ||
 ISP1820 ISP1820 | Wild type | Center for Vaccine Development |
 SB130 SB130 | invA ISP1820 | Jorge Galán (13) |
 H553 H553 | invE ISP1820 | This paper |
| S. typhimurium | ||
 TML TML | Wild type | Alison O’Brien |
 SR-11 SR-11 | Wild type | Jorge Galán |
 C5 C5 | Wild type | Alison O’Brien |
 SB147 SB147 | invA SR-11 | Jorge Galán (12) |
 SB109 SB109 | invE SR-11 | Jorge Galán (12, 14) |
| S. dublin Lane | Wild type | Roy Curtiss |
Cell lines and culture conditions.
The culture system was established with the human embryonic small IEC line Intestine 407 (16) (Int407; ATCC CCL6; American Type Culture Collection, Rockville, Md.) and the murine small IEC cell line MODE-K (46). The Int407 cell line was grown and maintained in minimal essential medium (MEM; Gibco/BRL Life Technologies, Inc., Gaithersburg, Md.) to which 10% heat-inactivated fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah) and 2 mM glutamine (Gibco/BRL) were added. The MODE-K cell line was grown and maintained in Dulbecco’s modified Eagle’s medium with 4.5 g of glucose (BioWhittaker, Walkersville, Md.) per liter, to which 10% heat-inactivated fetal bovine serum (HyClone) and 2 mM glutamine were added. Stock IEC cultures were maintained in 75-cm2 culture flasks (Becton Dickinson Labware, Lincoln Park, N.J.) at 37°C in a 6% CO2 atmosphere and split weekly. Monolayers for adherence and invasion assays were prepared by seeding either 2.5 × 105 cells in 1 ml of growth medium in each well of a 24-well tissue culture plate (Costar Corp., Cambridge, Mass.) or 1.3 × 106 cells in 2.5 ml of growth medium in each well of a six-well tissue culture plate (Costar Corp.). Confluent monolayers were obtained after 24 h. Immediately before adherence and invasion assays, the medium was removed from each well of the 24- or 6-well tissue culture plates and replaced with 0.3 or 1.5 ml of fresh growth medium, respectively.
Quantitation of cell-associated bacteria.
The assays used in the present studies were a modification of the procedures developed by Tartera and Metcalf (44) and Weinstein et al. (47). Briefly, bacteria were grown overnight at 37°C in a rotary shaking water bath (200 rpm) and then subcultured by diluting them 1:100 in 10 ml of fresh LB medium containing 0.3 M NaCl, unless otherwise stated. The bacteria were grown to an A600 of 0.5 (mid- to late logarithmic phase), and 1 ml of each culture was centrifuged at 5,000 × g for 10 min. The pellets were resuspended in an equal volume of MEM supplemented with 10% fetal calf serum and 2 mM l-glutamine. For the 24-well plate assay, 25 μl of this suspension was added to each of three wells of IEC monolayers, representing an initial inoculum of 2.5 × 106 to 6 × 106 CFU per well. In the six-well plate assay, each of six wells of epithelial cell monolayers was inoculated with 130 μl of a bacterial suspension (1.3 × 107 to 2.6 × 107 CFU per well). All microtiter plates were centrifuged at 2,000 × g for 10 min to permit optimal interaction of bacteria with the cell monolayers.
For quantitation of cell-associated bacteria, bacteria were incubated with the monolayers of IECs for 90 min at 37°C in 5% CO2, unless otherwise stated, and then each well was rinsed six times with Earle’s balanced salt solution (EBSS; Gibco/BRL). The cell-associated bacteria were released with 1% Triton X-100 (Sigma) in phosphate-buffered saline (PBS). The CFU were quantified by plating the appropriate dilutions on LB agar.
For the invasion assays, bacteria were incubated with monolayers of IECs for 90 min at 37°C in a CO2 incubator and then washed three times with EBSS. One (for 24-well plates) or 2.5 (for 6-well plates) ml of prewarmed MEM containing 100 μg of gentamicin per ml was added per well and incubated for an additional 90 min to kill extracellular bacteria (44). The supernatant from each well was collected into 1.5-ml microcentrifuge tubes, centrifuged (8,000 × g for 10 min), and/or filter-sterilized through a 0.22-μm-pore-size low-protein binding filter (Millipore, Bedford, Mass.) to remove bacteria and cell debris. The supernatants were frozen at −20°C until assayed for IL-6. Subsequently, the wells were washed three times with EBSS, and intracellular bacteria were released by lysis of the monolayer with 1% Triton X-100 in PBS. The cell lysate was then diluted in saline and plated on LB agar to determine viable bacterial counts.
For mRNA isolation, the monolayers from six-well plates were infected as described above. After the wells were washed with EBSS, the cells were lysed and resuspended in 1 ml of RNA-STAT60 (Tel-Test “B,” Friendswood, Tex.). These suspensions were immediately frozen at −70°C until the mRNA could be extracted.
Microscopy.
For staining of the IEC monolayers, 8 × 105 epithelial cells (semiconfluent) were seeded into six-well plates, and these cells were infected with a multiplicity of infection (MOI) of approximately 20 or 400 bacteria per cell by the standard protocol described above, unless otherwise stated. The bacteria were incubated with the IECs for 90 min at 37°C in 5% CO2, and then each well was rinsed six times with EBSS. For the Leukostat stain, the Fisher Leukostat stain kit (Fisher Scientific, Pittsburgh, Pa.) was used to fix and stain the monolayers and the bacteria, according to the procedure described by the manufacturer. After staining, the wells were rinsed with water. After air drying, 2 drops of Gel/Mount (Biomeda, Foster City, Calif.) were placed in the center of each stained well and then carefully overlaid with a 1.5-mm coverslip. The monolayers were photographed under the oil immersion objective (×100 magnification) of an Olympus BX50 microscope with Ektachrome 100 Plus film (Eastman Kodak Co., Rochester, N.Y.). For the lipopolysaccharide-immunofluorescence staining, the infected monolayers were fixed with 3% formalin diluted in PBS overnight at 4°C. The monolayers were then washed three times with PBS and permeabilized with 0.1% Triton X-100 in PBS for 4 min. The wells were then treated with blocking reagent (5% nonfat dry milk [Super G, Inc., Landover, Md.] and 3% gelatin [Bio-Rad Laboratories, Hercules, Calif.] in TBS [0.14 M sodium chloride, 0.025 M Tris (pH 8.0), 2.7 mM potassium chloride]) at 37°C for 30 min. Following the blocking step, the S. typhi- and S. dublin-infected monolayers were incubated with anti-O:9 antigen-specific serum (Difco, Detroit, Mich.), and the S. typhimurium-infected monolayers were incubated with anti-O:4 antigen-specific serum (Difco) for 1 h at 37°C. Both antisera were diluted 50-fold in blocking reagent prior to use. After this incubation, the monolayers were washed three times with PBS and then incubated for 30 min at 37°C with fluorescein-labeled, affinity-purified goat anti-rabbit immunoglobulin G (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) diluted 50-fold in blocking reagent. The monolayers were washed three times with PBS, and then 2 drops of Slow Fade (Molecular Probes, Eugene, Oreg.) was placed in the center of each stained well and then carefully overlaid with a 1.5-mm coverslip. The plates were left in the dark overnight, and then the monolayers were photographed under the oil immersion objective (×100 magnification) of an Olympus BX50 microscope with the WIB filter with Kodak Elite II ASA400 film (Eastman Kodak Co.).
Cytokine assay.
To quantify levels of IL-6 in the supernatants of S. typhi-stimulated epithelial cell cultures, a modification of a standard IL-6 bioassay was employed as described previously (34, 47). For IL-6 determinations, the IL-6-dependent cell line B9 (5) was used (kindly provided by Philip Morrissey, Immunex, Seattle, Wash.). B9 cells were maintained in Dulbecco’s MEM (Bio Whittaker), supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1% penicillin-streptomycin, and 50 ng of recombinant human IL-6 (Genzyme Diagnostics, Cambridge, Mass.). Recombinant murine IL-6 was also purchased from R&D Systems. The significance of the data was evaluated by Student’s t test.
To confirm that the biologically active IL-6 secreted by the MODE-K-derived supernatants was specific IL-6, five 50% effective doses of supernatant from S. typhi ISP1820-stimulated MODE-K cells were incubated with a range of concentrations of the polyclonal goat anti-mouse IL-6 neutralizing antibody AB406-NA, as previously described (47). A total of 3.3 μg of the anti-mouse IL-6 antibody per ml neutralized the B9-stimulating activity to the level of the background control. In contrast, the murine anti-human IL-6 monoclonal antibody MAB206 did not inhibit any of the S. typhi-induced IL-6 activity derived from the MODE-K cells at any concentration of antibody tested (data not shown).
RNA extraction and RT-PCR detection of cytokine mRNA.
RNA was extracted from IEC cultures prepared as described above, with an RNA-STAT60 kit (Tel-Test “B”), according to the directions supplied by the manufacturer. Reverse transcriptase PCR (RT-PCR) was performed by a modification of the procedure previously described by our laboratory (47) to determine the quantities of mRNA for hypoxanthine phosphoribosyltransferase (HPRT) and IL-6.
The primers and probes for human and murine HPRT and IL-6 were prepared on a DNA synthesizer (Applied Biosystems, Foster City, Calif.) and have been previously described (33). Specific cytokine gene cDNA transcripts were amplified by PCR by a procedure previously described (47). The number of PCR cycles selected for each cytokine was 26 for HPRT and 30 for IL-6. The relative expression of HPRT in each experimental sample was compared to that of the HPRT for uninfected samples to normalize for RNA in the reverse transcription reaction. Expression of a given RNA by uninfected epithelial cell cultures was arbitrarily assigned a value of 1, and the expression of IL-6 RNA by IEC cultures in other experimental groups was expressed relative to this baseline, as previously described (47).
RESULTS
Differential interaction of three Salmonella serovars with human and murine small IEC lines.
Early studies by Collins and Carter (3) showed that, in orally infected germfree mice, S. typhi appeared in the Peyer’s patches and mesenteric lymph node cultures in numbers equivalent to that of a Salmonella species that is virulent in mice, such as S. enteritidis. Subsequently, both S. typhi and S. typhimurium were shown to be able to invade murine IEC and M cells (20–23, 39). Taken together, these studies implied that S. typhi could multiply in the lumen of the murine intestinal tract and penetrate the intestinal epithelium. To test this possibility in another infection model, compare the findings with two other Salmonella serovars, and evaluate the response of the host to each organism, the capacity of S. typhi, S. typhimurium, and S. dublin strains to infect and stimulate murine and human small IECs, with the murine small IEC line MODE-K (46) and the human small IEC line Int407, was evaluated. Confluent monolayers of MODE-K or Int407 cells were exposed to fresh medium or to medium containing S. typhi ISP1820, S. typhimurium TML, S. typhimurium SR-11, or S. dublin Lane. After washing, the monolayers were lysed to determine percent cell-associated bacteria. Alternatively, medium containing gentamicin was added to kill extracellular organisms and then the monolayers were lysed to determine the percent invasion. The results in Table Table22 demonstrate that S. typhi ISP1820 could adhere to and invade both murine and human IECs more efficiently than either S. typhimurium TML or SR-11 or S. dublin Lane. Since previous studies from our laboratory suggested that adherence of S. typhi is sufficient to induce IL-6 secretion by IECs (47), we also measured IL-6 induction in the supernatants after coculture with the three Salmonella serovars. The results in Table Table22 also show that S. typhi ISP1820 induces both murine and human small IEC lines to secrete quantitatively more biologically active IL-6 than either S. typhimurium TML or SR-11 or S. dublin Lane. Steady-state levels of IL-6 mRNA in S. typhi ISP1820-, S. typhimurium TML-, and S. dublin Lane-infected IECs were also analyzed. RT-PCR was performed on the RNA isolated from uninfected and infected Int407 cells, and the results in Fig. Fig.11 demonstrate that S. typhi induced greater quantities of IL-6-specific mRNA in Int407 cells than did either S. typhimurium or S. dublin. Two other wild-type S. typhi strains, Ty2 and Quailes, were also found to adhere to, invade, and induce secretion of IL-6 in MODE-K murine IECs (data not shown). When other IEC lines, IEC-6 (a rat small IEC line), and Caco-2 (a human colonic IEC line), were analyzed, similar results were obtained (data not shown). Collectively, these data demonstrate that S. typhi can stimulate both human and murine IECs to secrete IL-6, whereas S. typhimurium and S. dublin induce only minimal levels of IL-6 in human or murine IECs. These data suggest that different pathogens can stimulate distinct host responses that may be related to differential clinical pathologies. In addition, these data suggest that results from the murine model of typhoid fever should be interpreted with caution since S. typhimurium does not induce secretion of IL-6 in murine IECs but S. typhi does induce secretion of IL-6 in human IECs.
TABLE 2
S. typhi is better able to adhere to, invade, and induce IL-6 in human (Int407) and murine (MODE-K) small IEC lines than other Salmonella serovars
serovars
| Serovar used for infectiona | Int407
| MODE-K
| ||||
|---|---|---|---|---|---|---|
| Cell-associated bacteria (%)b | Invasion (%)c | IL-6 (pg/ml)d | Cell-associated bacteria (%)b | Invasion (%)c | IL-6 (pg/ml)d | |
| Expt 1 | ||||||
 None (medium) None (medium) | NAe | NA | 230 | NA | NA | 60 |
 S. typhi ISP1820 S. typhi ISP1820 | 128 | 119 | 10,150 | 99 | 105 | 8,200 |
 S. typhimurium TML S. typhimurium TML | 22 | 15 | 1,520 | 29 | 18 | 1,490 |
 S. dublin Lane S. dublin Lane | 21 | 37 | 2,460 | 31 | 51 | 2,160 |
| Expt 2 | ||||||
 None (medium) None (medium) | NA | NA | 170 | NA | NA | 0 |
 S. typhi ISP1820 S. typhi ISP1820 | 96 | 141 | 10,270 | 79 | 73 | 6,020 |
 S. typhimurium SR-11 S. typhimurium SR-11 | 32 | 14 | 840 | 33 | 21 | 410 |
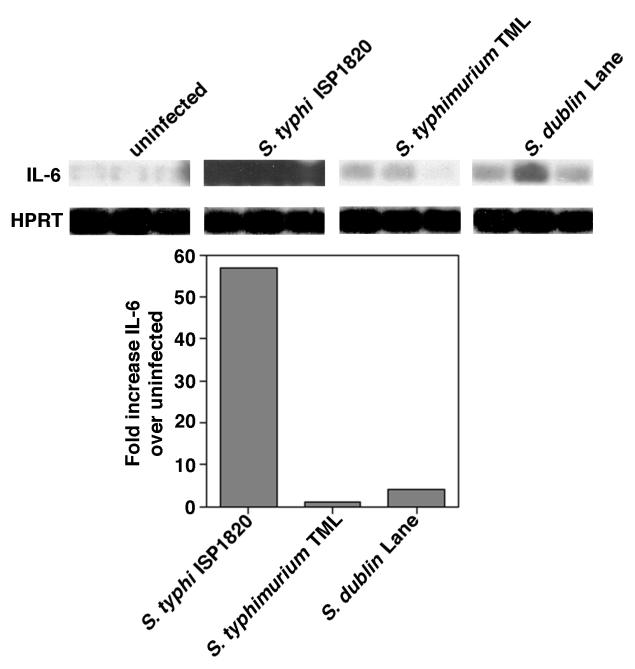
Cytokine gene expression by Int407 stimulated with S. typhi ISP1820, S. typhimurium TML, and S. dublin Lane. RNA was prepared from 2.6 × 106 uninfected Int407 cells or cells stimulated with S. typhi ISP1820, S. typhimurium TML, or S. dublin Lane at MOIs of approximately 20 bacteria per cell in a standard invasion assay. cDNA was prepared from 1 μg of each mRNA sample, and RT-PCR was performed with primers specific for human IL-6 and the housekeeping gene HPRT. Blots were hybridized with enhanced chemiluminescence-labeled oligonucleotide probes specific for the corresponding genes. Autoradiographs were scanned, and the fold increase of the IL-6-specific cytokine gene was calculated by comparison of the signals elicited by bacteria with those elicited by medium alone. Each lane of the autoradiograph represents the RT-PCR product from a single well. These results are representative of three separate experiments.
Analysis of entry requirements and ability to induce IEC-derived IL-6 among Salmonella serovars.
The results in Table Table22 suggested that both S. typhimurium and S. dublin associate with and invade murine and human IECs less well than does S. typhi. Therefore, it was possible that the lower production of IL-6 in S. typhimurium- and S. dublin-stimulated IECs was the consequence of a decreased capacity to associate with IECs. To ensure that the differential induction of IL-6 was not due simply to a shift in the kinetics of induction, adherence, invasion, and IL-6 induction in Int407 cells for the other two Salmonella species were evaluated after a 90-min adherence step followed by a 90-, 120-, 180-, or 240-min gentamicin killing step. The adherence step was not extended beyond 90 min because the bacterial multiplication in the well overwhelms the culture. The data shown in Fig. Fig.22 demonstrate that, over longer incubation periods, the IL-6 concentration in the supernatant of S. typhimurium TML- or S. dublin Lane-infected IECs increased slightly, although the maximum levels of IL-6 were still significantly less than those induced by S. typhi ISP1820. The slight reduction of intracellular S. typhi observed at time points later than 180 min postinfection was a result of decreased viability of the epithelial cell cultures, as determined by trypan blue exclusion (data not shown). Taken together, neither the percent cell association nor the ability to induce IEC-derived IL-6 was increased by longer incubations of S. typhimurium TML or S. dublin Lane with IECs. In contrast, in the IEC cultures infected with S. typhi ISP1820, the IL-6 levels continued to rise for the duration of the experiment.
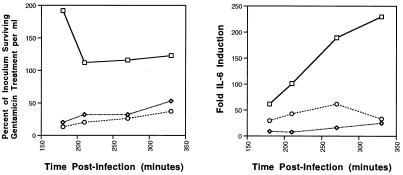
The effects of extending the incubation periods postinfection on invasion and IL-6 induction of Salmonella-infected Int407 cells. Monolayers of Int407 cells (2.5 × 105 cells per well of a 24-well tissue culture plate) were infected with an MOI of 20 S. typhi ISP1820 (squares), S. typhimurium TML (diamonds), or S. dublin Lane (circles) bacteria per cell and incubated for 90 min to allow the bacteria to adhere and invade. After removal of the extracellular bacteria by washing, the cultures were further incubated in the presence of gentamicin for an additional 90, 120, 180, or 240 min. At the end of the culture period, the supernatants were removed, and the concentration of IL-6 in each supernatant was determined by bioassay. The Int407 cells were lysed, and the number of bacteria surviving gentamicin treatment was determined. These results are representative of two independent experiments.
The data thus far suggest a correlation between percent cell association or invasion and IL-6 induction. The following series of experiments were conducted to address this possibility. Wild-type strain S. typhi ISP1820 was grown in media with osmolarities suboptimal for its adherence to IECs (as previously shown [44, 45]) so that the percent cell-associated bacteria for the S. typhi strain would be similar to the percent cell-associated bacteria for S. typhimurium and S. dublin. The results shown in Table Table33 demonstrated that S. typhi ISP1820 grown in low-osmolarity medium (LB, 0.06 M NaCl) exhibited a percentage of cell-associated bacteria and bacteria that had invaded cells that was comparable to that exhibited by S. typhimurium and S. dublin. Nonetheless, S. typhi ISP1820 still induced statistically significantly more IL-6 in Int407 cells than did the other two Salmonella strains. These data suggest that the capacity of S. typhi ISP1820 to induce more IL-6 in IECs than either S. typhimurium or S. dublin is not totally a result of the increased ability of S. typhi to adhere to and invade host cells.
TABLE 3
Differential IL-6 induction ensues even after changes in culture conditions that permit equivalent adherence and invasion of IECs by the three Salmonella pathogens
pathogens
| Bacteriuma | Mol of NaCl per liter added to LB growth medium | % Cell-associated bacteria | % Invasion | IL-6 (pg/ml) [fold increase over uninfected cells] |
|---|---|---|---|---|
| None (uninfected cells) | NAb | NA | NA |       600 600 |
| S. typhi ISP1820 | 0.30 | 167 | 221 | 22,720 [37.9] [37.9] |
| S. typhi ISP1820 | 0.06 | 25 | 27 | 9,030 [15.1] [15.1] |
| S. typhimurium TML | 0.30 | 28 | 15 | 2,330c [3.9] [3.9] |
| S. dublin Lane | 0.30 | 18 | 21 | 3,080c [5.1] [5.1] |
Previous studies from this laboratory demonstrated that the osmolarity of the bacterial growth medium, as well as the growth phase of S. typhi, significantly affects the capacity of S. typhi to adhere to and invade IECs (44). Therefore, the possibility existed that the bacterial growth conditions of S. typhimurium and S. dublin were not optimal for adherence and invasion to IECs, and as a consequence, the ability of these organisms to induce epithelial cell-derived IL-6 was underestimated. To optimize the association of S. typhimurium with IECs, we first compared the effects of centrifuging the bacteria onto the monolayers with those of not centrifuging infected monolayers. Centrifugation did not affect the association of S. typhi with IECs nor the ability of S. typhi to induce IEC-derived IL-6, but centrifugation did enhance the association of S. typhimurium with IECs (data not shown). Therefore, all IEC cultures were centrifuged after addition of bacteria in order to maximize adhesion.
A second variable that has been shown to affect adherence and invasion of IECs by Salmonella is the oxygen tension of the growth media. Lee and Falkow reported that microaerophilic growth conditions were optimal for Salmonella choleraesuis adherence to epithelial cells in culture (26). Francis et al. (11) and Schiemann and Shope (42) showed that S. typhimurium also adhered to and invaded intestinal epithelial cells more efficiently when grown in standing culture and with lower oxygen tension. To ensure that oxygen tension was not affecting our present observations, all bacterial cultures were grown under reduced oxygen tension to increase the percentage of adherence to cell monolayers. When the adherence and invasion of S. typhi ISP1820 and S. typhimurium TML grown at 37°C in a microaerophilic environment (incubation of bacteria in 1.5-ml tubes filled with LB broth with no aeration [32]) were compared with those of bacteria grown under our standard laboratory conditions (incubation of bacteria in 15-ml tubes with 10 ml of LB broth and shaking at 225 rpm [44]), no significant differences were observed in the ability to adhere to and invade IECs or the capacity to induce IL-6 in the IEC cultures (data not shown). Two possible explanations for this observation are (i) our normal growth conditions are microaerophilic, and/or (ii) the high osmolarity of our growth medium (0.3 M), perhaps combined with the low level of oxygen available in the growth tubes, is optimal for adherence and invasion by Salmonella serovars.
An additional variable that could account for the different levels of IL-6 induced by these three Salmonella pathogens is the MOI. Therefore, a study was conducted in which the MOIs of S. typhimurium (24a) and S. dublin were increased from about 20 bacteria per cell to approximately 4-fold, 16-fold, or 64-fold more bacteria per eukaryotic cell, and standard adherence and invasion assays, as well as an IL-6 induction assay, were conducted. The results in Fig. Fig.33 indicate that the actual number of adherent or internalized bacteria per cell increased as the MOI increased (24a). However, the quantity of IL-6 induced in the Int407 cells increased only with the MOIs of S. typhi ISP1820 and did not increase with the increasing MOIs of S. typhimurium TML or S. dublin Lane. The actual quantity of IL-6 induced by S. typhimurium or S. dublin at the most efficient MOI (240 or 1,200 bacteria per cell, respectively) remained greater than eightfold less than the amount of IL-6 induced by the optimal MOI for S. typhi ISP1820 (approximately 20 bacteria per cell). Collectively, these results indicate that S. typhi infection induces IECs to secrete IL-6 via a mechanism that is much more efficient in the case of S. typhi-host interactions and is directly correlated with the MOI. Moreover, adherence and invasion of S. typhimurium and S. dublin with IECs appear to be dissociable from IL-6 induction.
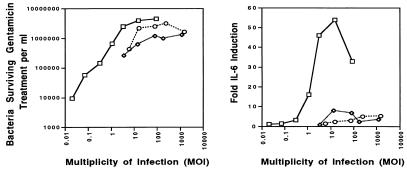
Effects of increasing the MOI on invasion and IL-6 induction of Salmonella-infected Int407 cells. Monolayers of Int407 cells (2.5 × 105 cells per well of a 24-well tissue culture plate) were infected with various doses of S. typhi ISP1820 (squares), S. typhimurium TML (circles), or S. dublin Lane (diamonds) and incubated for 90 min to allow the bacteria to adhere and invade. After removal of the extracellular bacteria by washing, the cultures were further incubated in the presence of gentamicin for an additional 90 min. At the end of the culture period, the supernatants were removed, and the concentration of IL-6 in each supernatant was determined by bioassay. The Int407 cells were lysed, and the number of bacteria surviving gentamicin treatment was determined. These results are representative of four independent experiments. The values plotted for S. typhi ISP1820 are taken from the work of Weinstein et al. (47).
Microscopic comparison of the adherence and invasion profiles of S. typhi ISP1820, S. typhimurium TML, and S. dublin Lane.
The data presented above suggested that, under optimal conditions, S. typhi adhered to, invaded, and induced IL-6 secretion in small IECs more effectively than the other two Salmonella serovars analyzed. In a previous study, adherence of S. typhi to epithelial cells was shown to be sufficient for IL-6 induction, and approximately 20 bacteria per cell was the optimal MOI for IL-6 induction (47). However, those quantitative assays did not distinguish between the possibilities that either all IECs are infected, but with smaller numbers of bacteria, or that some IECs remain uninfected. To address these alternatives, the number of bacteria per cell present during a standard quantitation of cell-associated bacteria and invasion assay was assessed by light and fluorescence microscopy. Int407 monolayers were infected with S. typhi ISP1820, S. typhimurium TML, and S. dublin Lane at MOIs of about 20 or 400 bacteria per cell. The infected monolayers were fixed and stained with either the Fisher Leukostat staining kit or O-antigen-specific immunofluorescence. The number of bacteria per cell was quantified in at least 100 cells per well of the O-antigen-labeled samples. As shown in Fig. Fig.44 and Table Table4,4, at the 20:1 MOI, almost all of the Int407 cells in the S. typhi-infected monolayers were infected with at least 15 bacteria per cell, while significant numbers of the Int407 cells in the S. typhimurium- and S. dublin-infected cultures remained uninfected. Moreover, of those Int407 cells that were infected by S. typhimurium and S. dublin, the majority of the epithelial cells had fewer than 15 cell-associated bacteria. At the MOI of 400 bacteria per cell, all of the cells in the S. typhi-infected monolayers had greater than 15 bacteria. It is interesting to note that, in the Int407 monolayers infected with an MOI for S. typhi ISP1820 of 400:1, there appeared to be more bacteria per cell than at 20:1. This observation suggests that, at the MOI of 20:1, the Int407 cells were not saturated with the maximum number of bacteria. In the S. typhimurium TML and S. dublin Lane monolayers infected at the higher MOI, most of the cells were infected with greater than 15 bacteria. Nevertheless, even at this higher MOI, neither S. typhimurium TML nor S. dublin Lane induced appreciable levels of IL-6 (Fig. (Fig.3).3). Taken together, these results suggest that the higher level of IEC-derived IL-6 induced by S. typhi than by S. typhimurium and S. dublin may reflect an inductive event in the IECs that is unique to S. typhi.
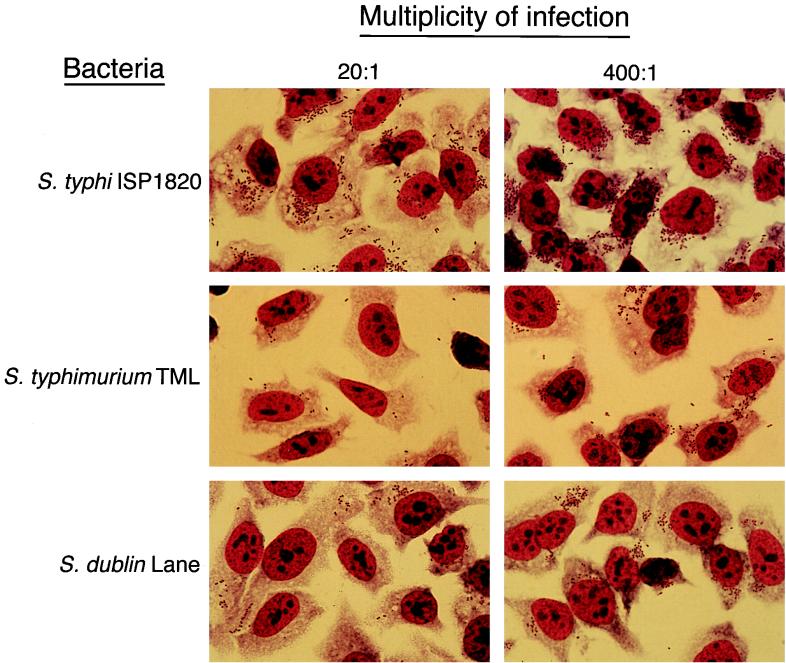
Microscopic comparison of the adherence and invasion profiles of S. typhi ISP1820-, S. typhimurium TML-, and S. dublin Lane-infected Int407 cells. Semiconfluent monolayers of Int407 cells (8 × 105 cells per well of a six-well tissue culture plate) were overlaid with medium (uninfected control) or infected at a bacterium-to-cell ratio of 20:1 or 400:1 with S. typhi ISP1820, S. typhimurium TML, or S. dublin Lane and incubated for 90 min to allow the bacteria to adhere and invade. Cultures were washed, fixed, and then stained with Fisher Leukostat staining reagents. The monolayers were photographed under the oil immersion objective (×100 magnification) of an Olympus BX50 microscope. These results are representative of three independent experiments.
TABLE 4
Quantitation of cell-associated bacteria when Int407 cells are infected with S. typhi, S. typhimurium, and S. dublin at two different MOIs
MOIs
| Bacterium | MOIb | % of cells with no. of bacteria per cella:
| ||||
|---|---|---|---|---|---|---|
| 0 | 1–5 | 6–10 | 11–15 | >15 | ||
| S. typhi ISP1820 | 20 | 0 | 0 | 5 | 6 | 90 |
| 400 | 0 | 0 | 0 | 0 | 100 | |
| S. typhimurium TML | 20 | 7 | 26 | 17 | 22 | 28 |
| 400 | 1 | 5 | 8 | 8 | 78 | |
| S. dublin Lane | 20 | 15 | 32 | 17 | 17 | 16 |
| 400 | 1 | 3 | 5 | 5 | 84 | |
Invasion mutants of S. typhi and S. typhimurium indicate differences in the earliest host-bacterium interactions.
We previously showed that cytochalasin D blocked invasion but not adherence of S. typhi to Int407 cells (47). More importantly, IL-6 induction was not reduced in S. typhi-infected cultures that had been treated with cytochalasin D. These data suggested that adherence of S. typhi to IECs was sufficient for IL-6 secretion. Since previous studies have shown that invA mutants of S. typhi and invA and invE null mutants of S. typhimurium do not invade (13, 14), we tested invA and invE mutants of S. typhi ISP1820, as well as invA and invE mutants of S. typhimurium SR-11, for their capacity to induce IL-6 secretion in Int407 cells. The results in Fig. Fig.55 show that the S. typhi invA and invE mutants were able neither to adhere to nor to invade human Int407 IECs and were unable to induce significant quantities of IL-6 in IECs. Consistent with this finding is a recent study from our laboratory in which a panel of invasion-defective S. typhi mutants were isolated and none of these mutants adhered to or invaded IECs (25). In contrast, Galán and coworkers (13, 14) showed that invA and invE mutants of S. typhimurium SR-11 adhered to the cell monolayers as well as the wild-type parent strain but invaded significantly less well. Figure Figure55 also demonstrates that the S. typhimurium wild-type parent and the invasion mutants secreted similarly low levels of IL-6. Because the amount of IL-6 induced by the parent strain of S. typhimurium is already low, it may be difficult to detect a significant decrease in that induced by the mutants. An S. dublin Lane invA mutant adhered to the Int407 cells, although only about a third as well as the parent, but the IL-6 level induced by this organism was also low due to low levels of IL-6 induced by the wild-type parent (data not shown). These data indicate additional distinctions among the three Salmonella serovars in the earliest steps of pathogenesis.
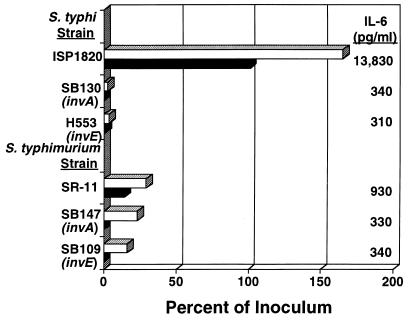
Effects of invA and invE mutations on the adherence, invasion, and IL-6 induction of S. typhi ISP1820 and S. typhimurium TML on Int407 cells. Monolayers of Int407 cells (2.5 × 105 cells per well of a 24-well tissue culture plate) were overlaid with medium (uninfected control) or infected at a bacterium-to-cell ratio of 20:1 with S. typhi ISP1820, S. typhi SB130 (ISP1820 invA), S. typhi H553 (ISP1820 invE), S. typhimurium SR-11, S. typhimurium SB147 (SR-11 invA), or S. typhimurium SB109 (SR-11 invE) and incubated for 90 min to allow the bacteria to adhere and invade. Cultures were washed and incubated in the presence of gentamicin for an additional 90 min to eliminate extracellular bacteria. At the end of the culture period, the supernatants were removed, and the concentration of IL-6 in each supernatant was determined by B9 bioassay. The IECs were lysed, and the number of viable intracellular bacteria was determined. These results are representative of two independent experiments. Top and bottom bars for each strain represent percent cell association and percent invasion, respectively.
DISCUSSION
These studies were undertaken to characterize the initial steps in S. typhi pathogenesis compared to that of other pathogenic Salmonella serovars. Our findings demonstrate that S. typhi adheres to and invades human small IECs more effectively than do two other serovars of Salmonella, S. typhimurium and S. dublin. Moreover, even when the conditions were optimized for S. typhimurium and S. dublin adherence and invasion, S. typhi induced significantly more IL-6 than the other Salmonella strains. In addition, the present studies demonstrate that, although S. typhimurium also adheres to and invades murine small IECs, this organism does not induce IL-6 even in mouse IECs, whereas S. typhi does. Finally, our findings show that S. typhi invasion mutants do not adhere to or invade IECs whereas S. typhimurium invasion mutants adhere at levels similar to those of the wild type. Taken together, these data indicate marked differences in the early, small intestine-associated steps in the pathogenesis of different Salmonella serovars.
In the present study, S. typhi was significantly more effective at inducing IEC-derived IL-6 than either S. typhimurium or S. dublin. Since we had previously shown that adherence of S. typhi to IECs appeared to be sufficient for these bacteria to induce IL-6, we wanted to ensure that the other bacteria were also cultured in conditions that were optimal for adherence to the epithelial cells. Other investigators have cultured all Salmonella serovars in either Luria broth (0.6 M NaCl) or LB broth (0.17 M NaCl) (41) and have demonstrated that, under these conditions, S. typhi and S. typhimurium adhere to and invade cultured IECs in equivalent quantities. Tartera and Metcalf (44) clearly demonstrated that S. typhi adherence was osmoregulated, with a concentration of 0.3 M NaCl in LB broth being the optimal condition for S. typhi adherence. Thus, in the studies presented here, the optimal adherence and invasion conditions (0.3 M NaCl in LB broth) were used for all bacteria. We have also previously shown that an MOI of 20 S. typhi organisms per IEC was optimal for adherence to and invasion of IECs, and this MOI also resulted in the optimal induction of IEC-derived IL-6 (47). In the present study, a higher MOI (approximately 400 bacteria per epithelial cell) for S. typhimurium and S. dublin resulted in optimal numbers of bacteria adhering to and invading IECs, but this higher MOI did not induce significantly more IEC-derived IL-6, for either strain. Thus, even when the parameters for adherence and invasion were optimized for all three Salmonella pathogens, S. typhi remained the most significant inducer of IL-6.
Recent studies from several laboratories have demonstrated that stimulation of a panel of both large and small IEC lines by a variety of bacteria, including S. typhi (46a), induced mRNA synthesis and secretion of IL-8 (1, 4, 6, 8, 19, 21, 27, 43). In addition, Weinstein et al. also showed that significant quantities of IL-6 are induced by both Caco-2 and other small IEC lines after stimulation with S. typhi (47). In contrast, neither Eckmann et al. (8) nor Hedges et al. (15) were able to demonstrate IL-6 induction in the Caco-2 IEC line after exposure to either S. dublin or Escherichia coli, respectively. One possible explanation for the different observations is that S. typhi attaches to and invades IECs significantly better than S. dublin. In addition, while S. typhi is a highly virulent human pathogen, S. dublin is most commonly found in bovine disease and causes a systemic disease only in humans with other underlying illnesses (10). Since the cell lines used in both this study and that of Eckmann et al. (8) were of human origin, it is possible that the different results reflect the human specificity of S. typhi or the lack of specificity of S. dublin.
It should also be noted that Jung et al. (21) and Panja et al. (38) showed that fresh human colonic epithelial cells secreted modest levels of IL-6 constitutively or after stimulation with Yersinia enterocolitica, Listeria monocytogenes, or IL-1β. Thus, it is possible that the discrepancy in results is due only to differences in the nature of the stimulating antigen. The fact that IL-6 is secreted from normal, nontransformed small IECs (Int407 and IEC-6) after stimulation (47) also lends credence to the notion that the production of IL-6 by these cells is a physiologically relevant activity of the cells that represent the host’s first line of defense against mucosal enteric pathogens.
While our findings confirm those of Pascopella et al. (39) that S. typhi and S. typhimurium have different effects on the murine intestinal epithelium, our results also extend their findings because we show (i) that S. typhi adheres to and invades both human and murine IECs better than S. typhimurium and (ii) that these two organisms induce a different physiological response in human IECs, i.e., IL-6 production. Our data show that S. typhi, the etiologic agent of typhoid fever in humans, induces IL-6 in human IECs whereas S. typhimurium does not. S. typhimurium infection in mice has been used as a model of typhoid fever in humans. However, S. typhimurium stimulation of murine IECs does not induce IL-6, as would have been expected from the human data. Therefore, these findings suggest that the data derived from the mouse model of typhoid fever should be interpreted conservatively.
One hypothesis for the differences observed in the capacity of S. typhi to induce IL-6 in IECs is that IL-6 secretion is physiologically related to restricted S. typhi-host interactions and may play a role in enhancing the systemic dissemination of the bacteria. In contrast, S. typhimurium and S. dublin, which are not normally disseminated systemically in humans, elicit a different cascade of host responses. This hypothesis is supported by the data of McCormick et al., which demonstrated that only Salmonella serovars causing human gastroenteritis, not Salmonella serovars that elicit human enteric fever, show transepithelial signaling to neutrophils across polarized IEC monolayers (28, 29). McCormick and her coworkers also suggested that this species-specific polymorphonuclear leukocyte signaling mechanism may significantly contribute to the development of gastroenteritis. Thus, those Salmonella serovars that do not elicit gastroenteritis but do elicit enteric fever may stimulate other cellular pathways involved in enteric fevers such as typhoid fever.
In conclusion, the data from this study suggest a major physiologic role for IEC-derived IL-6 in response to S. typhi infection but not to that by other Salmonella serovars. The exact role of IL-6 in either regulation of, or interactions with, other components of an immune response after S. typhi infection is not well understood, as yet. However, it is possible that a selective cascade of soluble mediators, such as cytokines and acute-phase reactants, are induced in IECs by bacteria that cause enteric fevers, whereas a distinct but potentially overlapping subset of soluble mediators is induced in IECs by bacteria that cause gastroenteritis. The data presented here also suggest that there may be important differences between the pathogenesis of S. typhi in human typhoid and the pathogenesis of S. typhimurium in the widely used murine typhoid model.
ACKNOWLEDGMENTS
We thank Stefanie Vogel, Carmen Tartera, Anne Jerse, Robin Sandlin, and Clare Schmitt for review of the manuscript and for helpful discussions. We are especially grateful to Dominique Kaiserlian for providing the MODE-K cell line and to Cindy Salkowski for sharing her expertise on RT-PCR.
This work was supported by NIH grant AI32951 to E.S.M. and USUHS grant R073FE to E.S.M.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.66.5.2310-2318.1998
Read article for free, from open access legal sources, via Unpaywall:
https://iai.asm.org/content/iai/66/5/2310.full.pdf
Free after 4 months at iai.asm.org
http://iai.asm.org/cgi/content/full/66/5/2310
Free after 4 months at iai.asm.org
http://iai.asm.org/cgi/reprint/66/5/2310
Free to read at iai.asm.org
http://iai.asm.org/cgi/content/abstract/66/5/2310
Citations & impact
Impact metrics
Citations of article over time
Article citations
Differential regulatory control of curli (csg) gene expression in Salmonella enterica serovar Typhi requires more than a functional CsgD regulator.
Sci Rep, 13(1):14905, 09 Sep 2023
Cited by: 3 articles | PMID: 37689734 | PMCID: PMC10492818
N-acyl homoserine lactonase attenuates the virulence of Salmonella typhimurium and its induction of intestinal damages in broilers.
Anim Nutr, 14:334-342, 12 Jul 2023
Cited by: 3 articles | PMID: 37635927 | PMCID: PMC10448016
Probiotic Properties of Chicken-Derived Highly Adherent Lactic Acid Bacteria and Inhibition of Enteropathogenic Bacteria in Caco-2 Cells.
Microorganisms, 10(12):2515, 19 Dec 2022
Cited by: 2 articles | PMID: 36557770 | PMCID: PMC9788042
Understanding the Mechanism of Antimicrobial Resistance and Pathogenesis of Salmonella enterica Serovar Typhi.
Microorganisms, 10(10):2006, 11 Oct 2022
Cited by: 6 articles | PMID: 36296282 | PMCID: PMC9606911
Review Free full text in Europe PMC
Salmonella enterica Serovar Typhi Induces Host Metabolic Reprogramming to Increase Glucose Availability for Intracellular Replication.
Int J Mol Sci, 22(18):10003, 16 Sep 2021
Cited by: 3 articles | PMID: 34576166 | PMCID: PMC8467381
Go to all (53) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Salmonella enterica Serovars Typhi and Paratyphi A are avirulent in newborn and infant mice even when expressing virulence plasmid genes of Salmonella Typhimurium.
J Infect Dev Ctries, 4(11):723-731, 24 Nov 2010
Cited by: 4 articles | PMID: 21252450 | PMCID: PMC4059606
Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages.
Infect Immun, 68(3):1005-1013, 01 Mar 2000
Cited by: 96 articles | PMID: 10678900 | PMCID: PMC97241
The Type III Secretion System Effector SptP of Salmonella enterica Serovar Typhi.
J Bacteriol, 199(4):e00647-16, 30 Jan 2017
Cited by: 24 articles | PMID: 27920299 | PMCID: PMC5287405
Clinical pathogenesis of typhoid fever.
J Infect Dev Ctries, 2(4):260-266, 30 Aug 2008
Cited by: 59 articles | PMID: 19741286
Review




