Abstract
Free full text

High-Mobility Group Chromatin Proteins 1 and 2 Functionally Interact with Steroid Hormone Receptors To Enhance Their DNA Binding In Vitro and Transcriptional Activity in Mammalian Cells
Abstract
We previously reported that the chromatin high-mobility group protein 1 (HMG-1) enhances the sequence-specific DNA binding activity of progesterone receptor (PR) in vitro, thus providing the first evidence that HMG-1 may have a coregulatory role in steroid receptor-mediated gene transcription. Here we show that HMG-1 and the highly related HMG-2 stimulate DNA binding by other steroid receptors, including estrogen, androgen, and glucocorticoid receptors, but have no effect on DNA binding by several nonsteroid nuclear receptors, including retinoid acid receptor (RAR), retinoic X receptor (RXR), and vitamin D receptor (VDR). As highly purified recombinant full-length proteins, all steroid receptors tested exhibited weak binding affinity for their optimal palindromic hormone response elements (HREs), and the addition of purified HMG-1 or -2 substantially increased their affinity for HREs. Purified RAR, RXR, and VDR also exhibited little to no detectable binding to their cognate direct repeat HREs but, in contrast to results with steroid receptors, the addition of HMG-1 or HMG-2 had no stimulatory effect. Instead, the addition of purified RXR enhanced RAR and VDR DNA binding through a heterodimerization mechanism and HMG-1 or HMG-2 had no further effect on DNA binding by RXR-RAR or RXR-VDR heterodimers. HMG-1 and HMG-2 (HMG-1/-2) themselves do not bind to progesterone response elements, but in the presence of PR they were detected as part of an HMG-PR-DNA ternary complex. HMG-1/-2 can also interact transiently in vitro with PR in the absence of DNA; however, no direct protein interaction was detected with VDR. These results, taken together with the fact that PR can bend its target DNA and that HMG-1/-2 are non-sequence-specific DNA binding proteins that recognize DNA structure, suggest that HMG-1/-2 are recruited to the PR-DNA complex by the combined effect of transient protein interaction and DNA bending. In transient-transfection assays, coexpression of HMG-1 or HMG-2 increased PR-mediated transcription in mammalian cells by as much as 7- to 10-fold without altering the basal promoter activity of target reporter genes. This increase in PR-mediated gene activation by coexpression of HMG-1/-2 was observed in different cell types and with different target promoters, suggesting a generality to the functional interaction between HMG-1/-2 and PR in vivo. Cotransfection of HMG-1 also increased reporter gene activation mediated by other steroid receptors, including glucocorticoid and androgen receptors, but it had a minimal influence on VDR-dependent transcription in vivo. These results support the conclusion that HMG-1/-2 are coregulatory proteins that increase the DNA binding and transcriptional activity of the steroid hormone class of receptors but that do not functionally interact with certain nonsteroid classes of nuclear receptors.
Steroid hormone receptors are members of a superfamily of ligand-dependent transcriptional activators which direct the expression of specific gene networks involved in regulating the differentiation and growth of reproductive tissues, as well as other metabolic processes. Receptors for steroid hormones are a subgroup of the nuclear receptor supergene family which have distinctive properties. In the absence of hormone, these receptors associate with heat shock proteins (HSPs) that serve as protein folding chaperones to maintain an active receptor conformation capable of receiving and responding to the hormonal signal (55). Steroid binding induces a series of changes in the receptor that leads to transcriptional activation, including a conformational change, dissociation from the oligomeric HSP complex, dimerization, and binding to hormone response elements (HREs) of target genes (see review in reference 62). Consensus HREs for steroid receptors are inverted palindromes arranged as hexanucleotide core motifs separated by 3 bp of undesignated sequence (61, 75). Steroid receptors bind preferentially as homodimers to HREs with the axis of dyad symmetry over the center of the palindromic element (19). Other members of the superfamily include receptors for nonsteroidal ligands, such as thyroid hormone receptor (TR), retinoic acid receptor (RAR), and vitamin D receptor (VDR) (35, 36). These nuclear receptors are distinguished from steroid receptors by the lack of stable interaction with HSPs (55), recognition of HREs that are arranged as direct repeats (DR) with variable half-site spacing, and binding to DR elements as heterodimers with retinoid X receptor (RXR) as the common dimer partner (29, 34, 63, 72 74).
How receptor binding to HREs enhances transcription of target genes is not well understood. Nuclear receptors are thought to stabilize the formation of a preinitiation complex at promoters through protein-protein interactions with basal transcription factors (23, 59), TATA-binding protein-associated factors (24), or a specific group of coactivators that are thought to provide a bridge between the receptor and the basal transcriptional machinery (22, 25, 26, 43, 59, 66).
Receptor interacting proteins which can either facilitate or inhibit receptor binding to target DNA sequences have also been described. A thyroid hormone receptor uncoupling protein (TRUP) was isolated that interacts with TR and RAR to block DNA binding in vitro and transactivation function within the cell (6). The calcium binding protein, calreticulin, has been reported to inhibit the binding of several different nuclear receptors to their target DNA sites in vitro as a result of interacting with conserved sequences in the DNA binding domains (DBDs) of all nuclear receptors. When overexpressed in mammalian cells, calreticulin also can inhibit receptor-dependent transcription (5, 12, 13). Additionally, several studies have reported the existence of proteins that stimulate sequence-specific DNA binding of nuclear receptors. Purification of recombinant receptors typically results in a loss of DNA binding activity that can be partially or fully restored by the addition of other protein(s). It has been well established that RXR enhances the DNA binding and transcriptional activity of a subgroup of nuclear receptors (TR, VDR, and RAR) by heterodimerization, where RXR directly contacts HREs (29, 34, 35, 72, 74). RXR has not been observed to functionally interact with any of the steroid hormone receptors. Studies have also described proteins that facilitate the binding of steroid receptor homodimers to palindromic HREs in vitro. For the most part, the protein(s) responsible for facilitating steroid receptor-DNA binding has not been identified and has been characterized as an enriched protein fraction or as an isolated protein of a specific molecular mass (9, 11, 14, 30, 37, 41, 51, 56). We previously showed that mammalian-cell nuclear extracts contain a protein(s) that substantially increases the binding of human progesterone receptor (PR) to its target DNA in vitro (15). The major nuclear factor responsible for enhancing PR-DNA binding was later identified as the chromatin nonhistone high-mobility group protein 1 (HMG-1) (42, 46). Whether HMG-1 or other accessory proteins that enhance steroid receptor-DNA binding in vitro have a physiological role in steroid receptor action in the cell has not been determined.
HMG-1 and the closely related HMG-2, referred to collectively as HMG-1/-2, are modular proteins that contain duplicated amino-terminal and centrally located DNA binding domains, termed HMG boxes, and an acidic carboxyl terminus. HMG-box motifs have been identified in several other proteins, some of which are sequence-specific transcription factors (7, 8, 31, 32). HMG-1/-2 have no known specific DNA recognition sequence; they prefer to bind to specific DNA structures, such as prebent DNA or the sharp angles at four-way junction DNA (7). HMG-1/-2 also have the ability to induce bends in DNA, and they bind in the minor groove. Based on nuclear magnetic resonance (NMR) solution structure, the tertiary structure of the HMG box from different proteins has been reported to be composed of three α helices folded into an L shape configuration believed to be important for the characteristic ability of HMG-box proteins to recognize and bind to bends or other distortions in DNA structure (49, 68). Although the physiological function of HMG-1/-2 remains poorly understood, this ability to recognize and manipulate DNA structure has led to the idea that these proteins function as architectural factors in processes that require transient manipulation of DNA structure, such as DNA repair, recombination, replication, and transcription (20, 70). As evidence that they can function as architectural transcriptional cofactors, HMG-1/-2 have been reported to facilitate the binding of several types of sequence-specific transcription factors to their target DNA sites; these factors include the octamer transcription factors Oct-1, Oct-2, and Oct-6 (77); the homeotic protein HOXD9 (73); the adenovirus major late promoter transcription factor MLTF (60, 67); p53 (25); and progesterone receptor (39, 42). In addition, transient-cotransfection assays demonstrated that HMG-1/-2 enhanced the transcriptional activity of the octamer transcription factors and HOX proteins within mammalian cells (73, 77). In a cell-free transcription assay with purified components, HMG-1/-2 were also found to be essential for activator-mediated assembly of the TFIID-TFIIA initiation complex on several promoters (54). Genetic evidence for a functional role of HMG-1/-2 in transcriptional regulation was recently provided by studies that deleted the HMG-like NHP6A and NHP6B genes in the yeast Saccharomyces cerevisiae (44). Mutant yeast cells carrying disruptions for both NHP6A and NHP6B exhibited a substantial reduction in activated transcription of a subset of yeast genes without altering basal promoter activity.
In the present study we investigated whether HMG-1 enhancement of the sequence-specific DNA activity of PR occurs with other members of the nuclear receptor superfamily. By use of cotransfection assays, we also investigated the effect of HMG-1 on the transcriptional activity of nuclear receptors within mammalian cells. We show that HMG-1 and the closely related HMG-2 increase the sequence-specific DNA binding activity in vitro and the transcriptional activity in whole cells of all of the steroid receptors tested. In contrast, HMG-1/-2 had little or no effect on the in vitro DNA binding activity of the nonsteroid class of nuclear receptors tested, including VDR, RXR, and RAR, and had minimal effect on VDR-mediated gene transcription in vivo. These results suggest that HMG-1/-2 are previously unrecognized positive coregulatory proteins for the steroid hormone subgroup of the nuclear receptor superfamily.
MATERIALS AND METHODS
Materials.
Promegestone (17α,21-dimethyl-19-norpregna-4,9-dien-3,20-dione) was obtained from NEN Life Sciences (Boston, Mass.). Other steroids (triamcinolone acetonide, 17β-estradiol, and dihydrotestosterone) were purchased from Sigma Chemical Co. (St. Louis, Mo.). 1,25-Dihydroxy-vitamin D3 [1,25-(OH)2D3] was obtained from Solvay Duphar and 9-cis-retinoic acid was purchased from Sigma. [3H]sodium acetate was from ICN Pharmaceuticals (Irvine, Calif.); coenzyme A and acetyl coenzyme A synthase were obtained from Sigma. Talon metal ion affinity resins were purchased from Clontech (Palo Alto, Calif.), and nickel chelation affinity resins were obtained from Qiagen (Chatsworth, Calif.). The radionucleotides [α-32P]dATP (3,000 Ci/mmol) and [α-32P]dCTP (3,000 Ci/mmol) were purchased from NEN Life Sciences. The following mouse monoclonal antibodies (MAbs) to steroid receptors were used: AB-52 prepared against human PR, which recognizes both the A and B receptor forms (16); h151 raised against a synthetic peptide corresponding to amino acids 287 to 300 of the hinge region of human estrogen receptor (ER) (17); and N441 raised against a synthetic peptide corresponding to amino acids 340 to 356 in the amino-terminal region of the human androgen receptor (AR; unpublished data). Polyclonal rabbit antiserum raised against human glucocorticoid receptor (GR) was provided by John Cidlowski (National Institute of Environmental Health Sciences, Research Triangle Park, N.C.). A rat MAb to VDR, 9A7, was provided by Wesley Pike (University of Cincinnati, Cincinnati, Ohio). Polyclonal antisera against human RXRα and RARγ have been previously described (58). A mouse immunoglobulin M (IgM) MAb (clone 854/E10) to calf thymus HMG-1 was prepared as described previously (47), and a rabbit polyclonal antibody specific for HMG-2 was provided by Raymond Reeves (Washington State University, Pullman, Wash.).
Expression of recombinant receptors and HMG-1 in the baculovirus system.
Recombinant baculovirus transfer vectors for human PR-A and PR-B were constructed by inserting the cDNAs for each receptor isoform into BamHI sites of pBlueBacHis-2 (InvitroGen, San Diego, Calif.). This placed PR coding sequences in frame with amino-terminal plasmid sequences that contain an ATG translation start site, six sequential histidine residues, and an enterokinase cleavage site. A polyhistidine-tagged human GR baculovirus transfer vector was constructed by inserting a BamHI-XhoI fragment from pI9 (18) encoding amino acids 9 to 777 of human GR in frame into pBlueBacHis-2 (C) restricted with BamHI and HindIII. The XhoI and HindIII ends of GR cDNA were blunted by filling in before joining. The pI9 plasmid was provided by Ron Evans (Salk Institute, San Diego, Calif.). A 2.8-kb full-length human androgen receptor (hAR) cDNA, plus sequences coding for one extra amino-terminal alanine and six histidine residues, was cloned into a 9.1-kb pAcC4 baculovirus transfer vector (Cetus Corp.) that resulted in the following amino-terminal sequence: Met-Ala-His6-Glu-Val. The hAR recombinant baculovirus transfer vector was constructed by PCR amplification with a primer that placed an NcoI site at the starting methionine of human AR, making it coincident with the position of the starting methionine of the polyhedron protein. A triple-ligation reaction was performed as previously described (71). A recombinant baculovirus transfer vector for HMG-1 was constructed by inserting rat HMG cDNA from pBS-rHMG-1 into EcoRI and SalI sites of pBlueBacHis-2(B). The rat HMG coding region was placed in frame with amino-terminal sequences of the plasmid containing an ATG translation start site, six sequential histidine residues, and an enterokinase cleavage site. A recombinant baculovirus transfer vector for steroid receptor coactivator 1 (SRC-1) (43) was constructed by inserting SRC-1 cDNA (amino acids 1 to 1140) from pBK-CMVSRC-1 (provided by Bert O’Malley, Ming-Jer Tsai, and Sergio Oñate, Baylor College of Medicine) into BamHI and PstI sites of the baculovirus transfer plasmid, pBlueBacHis-2(C). The SRC-1 coding region was also inserted in frame with amino-terminal plasmid sequences containing an ATG translation start site, six sequential histidine residues, and an enterokinase cleavage site. Cloning junctures in all transfer plasmids were sequenced (Sequenase 2.0; U.S. Biochemicals), and inserts were determined to be correctly oriented and in frame with the starting ATG codon and the six sequential histidine residues.
To construct recombinant viruses encoding the above proteins, Spodoptera frugiperda insect cells (Sf9) were cotransfected with baculovirus transfer plasmid and wild-type Autographa californica nuclear polyhedrosis virus baculovirus DNA as previously described (10). Recombinant viruses were identified by visual inspection under a reversed-phase light microscope, and the individual viruses were plaque purified. Viruses were screened by Western blot analysis of infected Sf9 cells for their ability to express protein. PR, AR, and GR proteins were produced in Sf9 cells in a stirred oxygenated 5-liter bioreactor (Applikon, Inc.). Cells were grown in Grace’s insect medium (Gibco-BRL) supplemented with lactalbumin hydrolysate, yeastolate, 0.1% Pluronic F68, 10% heat-inactivated fetal bovine serum (HyClone Laboratories, Inc.) and 50 μg of gentamicin per ml. Cells were grown at 27°C in the bioreactor to a density of 1.5 × 106 to 1.8 × 106 cells/ml; cultures were then inoculated with virus at a multiplicity of infection of 1.0 and allowed to grow for an additional 32 to 36 h at 27°C. Cells typically reached a density of 2 × 106 to 2.2 × 106 cells/ml at the time of harvest. Hormone was added to the bioreactors for the last 6 to 8 h of infection as follows: R5020 (500 nM) for PR, dihydrotestosterone (500 nM) for AR, and triamcinolone acetonide (500 nM) for GR. Cells were harvested by centrifugation at 1,500 rpm for 15 min in 50-ml aliquots, washed once in TG buffer (10 mM Tris-HCl [pH 8.0], 10% glycerol), and frozen as pellets at −80°C.
Expression of recombinant VDR, RXRα, and RARγ in yeast.
Human VDR, RXRα, and RARγ were expressed as polyhistidine-tagged proteins in the protease-deficient yeast strain BJ2168 from a copper-inducible promoter on 2μm plasmids as described previously (3). Briefly, transformed yeast were grown in 2-liter volumes to an A600 of 1.0 in minimal selection medium, and expression was stimulated by addition of 100 μM copper sulfate for 16 h at 30°C. VDR cultures were also induced by addition of 1 μM 1,25-(OH)2 D3. Cells were then harvested and lysed with acid-washed glass beads in the following buffer: 10 mM sodium phosphate, pH 8.0; 0.4 M KCl; 0.5 mM phenylmethylsulfonyl fluoride (PMSF); aprotinin, 1 μg/ml; and 5 mM β-mercaptoethanol. Lysates were centrifuged at 100,000 × g for 45 min at 4°C to yield a soluble supernatant.
Purification of polyhistidine-tagged nuclear receptors with metal ion affinity columns.
For polyhistidine-tagged steroid receptors expressed from baculovirus vectors, Sf9 insect cells were lysed in the following buffer: 20 mM Tris-HCl, pH 8.0; 350 mM NaCl; 10 mM imidazole; 5% glycerol; and a cocktail of protease inhibitors (16). All procedures were done at 0 to 4°C. Cell lysates were centrifuged at 100,000 × g for 30 min, and the supernatant was taken as a soluble whole-cell extract. Whole-cell extract was passed once over Talon resins in a column at a flow rate of 1 to 2 ml/min. The resins were washed with cell lysis buffer until the optical density at 280 nm returned to the buffer baseline. Bound receptors were eluted under nondenaturing conditions by competition with 100 mM imidazole. Receptors were eluted into siliconized tubes to prevent binding to surfaces, and dithiothreitol (DTT; 1 mM), zinc chloride (1 μM), and MgCl2 (1 mM) were added immediately to stabilize receptor DNA and steroid binding activity. Samples were stored at −80°C in aliquots and remained stable for DNA binding activity for several months if not repeatedly frozen and thawed.
Baculovirus-expressed human estrogen receptor was a gift from Angelo Notides (Rochester University) and was prepared as previously described (39). Briefly, Sf9 insect cells containing expressed ER bound to 17β-estradiol were lysed by freeze-thawing in TDEE buffer (40 mM Tris-Cl, pH 7.4; 1 mM EDTA; 1 mM DTT; 1 mM EGTA), and the cell lysates were centrifuged at 28,000 × g followed by centrifugation at 143,000 × g to obtain soluble cytosol. ER was precipitated with 40% saturated ammonium sulfate, and the pellet was redissolved in TDEE buffer and circulated over a DNA affinity column constructed with an estrogen response element (ERE) oligonucleotide. The column was washed several times in TDEE buffer containing 200 mM KCl, and bound ER was eluted with a linear 200 to 800 mM KCl gradient.
Purification of recombinant polyhistidine-tagged human VDR and human RXRα from soluble yeast cell lysates was done with nickel chelation affinity columns (Qiagen) according to the manufacturer’s recommended protocol. Yeast whole-cell extracts were bound to resins in a column at 4°C, and the column was equilibrated and washed in the yeast cell lysis buffer described above with the addition of 10% glycerol. Bound receptors were eluted in the cell lysis buffer with a pH 4.0 to 6.0 linear gradient. Collected fractions were immediately neutralized with 1 M Tris base (pH 8.5). Polyhistidine-tagged human RARγ was purified by using Talon resins as described above for steroid receptors except that the lysates were loaded onto the Talon resins and the resins were washed in the lysis buffer containing 2 mM imidazole. The eluted receptor was then diluted to reduce imidazole and was repurified on fresh Talon resins. VDR was bound to 1,25-(OH)2 D3 in yeast cells prior to extraction and purification. RXRα and RARγ were purified in their unliganded state.
Purification of HMG-1 and HMG-2.
A non-acid extraction method was used to purify HMG-1 and HMG-2 from calf thymus by a modification of the method originally defined by Adachi et al. (1). Fresh calf thymus (60 g) was cut into small pieces, trimmed of connective tissue, and homogenized in a stainless-steel blender in 6 volumes (tissues to buffer) of 10 mM sodium citrate buffer containing 0.15 M NaCl and 1 mM PMSF. The homogenate was filtered through cheesecloth and centrifuged for 15 min at 2,000 × g. The pellet was then washed three times by resuspension in homogenization buffer and centrifugation at 2,000 × g. The pellet was resuspended in 180 ml of 50 mM Tris-HCl (pH 7.8) containing 3 mM PMSF and then centrifuged for 10 min at 6,000 × g. This step was repeated. The resulting pellet containing nuclei and other membrane fractions was resuspended in a buffer (10 mM Tris-HCl, pH 7.8; 0.35 M NaCl; 5 mM DTT; 1 mM PMSF) in a glass-glass Dounce homogenizer. After homogenization, samples were left on ice for 1 h with brief intermittent vortexing to release 0.35 M NaCl-extractable nuclear proteins. The samples were then centrifuged at 5,000 × g for 20 min. The salt extraction of the nuclear pellet was repeated, and the two salt nuclear extracts were combined and centrifuged at 50,000 × g for 30 min. The supernatant was dialyzed overnight at 4°C against 10 mM Tris-HCl (pH 7.8) containing 1 mM DTT. The dialyzed salt nuclear extract was then passed over a PBE94 chromatofocusing column with a peristaltic pump at a flow rate of 1 to 2 ml/min. The column was washed and eluted with a linear 0 to 1.5 M NaCl gradient (in 10 mM Tris-HCl, pH 7.8; 5 mM DTT), and 1.0-ml fractions were collected and analyzed by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining for the presence of HMG-1/-2. The peak fractions containing HMG-1/-2 were pooled, dialyzed against 10 mM Tris-HCl (pH 7.8)–1 mM DTT, and rechromatographed on a second PBE94 column. The fractions containing HMG-1 and HMG-2 were pooled and stored at −80°C.
Coimmunoprecipitation assay.
Purified calf thymus HMG-2 (1,000 ng) was incubated with purified PR-B (200 ng) for 1 h at 4°C. The samples were then incubated for 3 h at 4°C on an end-over-end rotator with a 100-μl suspension of protein A-Sepharose that was prebound with a rabbit polyclonal antibody (≈2 μg) to HMG-2. As controls to determine the extent of nonspecific binding, blank protein A-Sepharose-coated beads and anti-HMG-2 antibody-coated beads were incubated with PR-B (200 ng) in the absence of HMG-2. Beads were washed four times with TEDGN100 (10 mM Tris base, pH 7.4; 1 mM EDTA; 1 mM DTT; 100 mM NaCl; 10% glycerol), transferred to a new tube, and washed twice more. Bound PR-B was eluted with SDS-sample buffer and analyzed by Western blotting with the PR-specific MAb 1294. The same coimmunoprecipitation assay was performed except that 200 ng of purified human VDR was used in place of PR-B and protein A-Sepharose eluates were analyzed for VDR by Western blotting with a rat MAb (9A7) to human VDR.
Immobilized metal affinity chromatography pull-down assays.
Baculovirus-expressed polyhistidine-tagged HMG-1 (10 μg) was incubated with non-histidine-tagged PR-B (8 μg) that was expressed in baculovirus and purified by MAb affinity chromatography as described in earlier studies (42, 46). Samples were incubated for 30 min at 4°C and then added to Talon resins in suspension and incubated for another 1 h at 4°C. Talon resins were washed four times in 20 mM Tris-HCl (pH 8.0)–10% glycerol–100 mM NaCl–15 mM imidazole, transferred to a new tube, and washed twice more. Bound proteins were eluted with SDS-sample buffer, and PR-B was detected by Western blotting with the PR-specific MAb 1294.
EMSA.
DNA binding was determined by electrophoretic mobility shift assay (EMSA) for PR and AR under the same conditions as described previously (42, 46, 47). Briefly, receptors in whole-cell or purified extracts of Sf9 cells (amounts per assay are indicated in the figure legends) were incubated for 1 h at 0 to 4°C with 0.3 ng of a 32P-end-labeled glucocorticoid response element-progesterone response element (GRE-PRE) oligonucleotide (specific activity, 100,000 to 300,000 cpm/ng) in a DNA binding buffer containing 10 mM Tris-base (pH 7.5), 50 mM NaCl, 5 mM DTT, 2 mM MgCl2, 1 mM EDTA, and 5% glycerol. The oligonucleotide was end labeled by filling in of 5′ single-stranded ends with the Klenow fragment of DNA polymerase by using [α-32P]dATP and [α-32P]dCTP. The binding reactions also contained 1 μg of gelatin (or albumin) as a carrier protein and 80 to 100 ng of competitor poly(dA-dT)-poly(dA-dT). All components of the binding reaction were preincubated for 30 min at 4°C prior to addition of the [32P]DNA probe. After 1 h, DNA binding reactions (25 μl) were electrophoresed on 5% polyacrylamide (40:1 acrylamide/bisacrylamide ratio) gels in 0.5× TAE buffer (0.02 M Tris-acetate, pH 8.0; 0.5 mM EDTA) with cooling to maintain the gel temperature at 4°C. Gels were dried under vacuum and autoradiographed, and free [32P]DNA and [32P]DNA complexes were quantitated by direct scanning of the gels for radioactivity by a series 400 Molecular Dynamics PhosphorImager. DNA binding as determined by EMSA for other nuclear receptors was accomplished by similar methods with the following minor modifications. A different binding buffer that included 10 mM HEPES (pH 7.8), 50 mM KCl, 4 mM MgCl2, and 12% glycerol was used with GR, ER, VDR, RARγ, and RXRα. Additionally, 2.5% glycerol was added to the polyacrylamide gel, and the electrophoresis buffer was 0.25× TBE (0.02 M Tris-borate, pH 8.0; 0.02 M boric acid; 0.5 M EDTA).
A palindromic GRE oligonucleotide was used as the DNA probe for PR, AR, and GR: 5′-gatcTTTGAGAACAAACTGTTCTTAAAACGAG-3′ and 3′-AAACTCTTGTTTGACAAGAATTTTGCTCctag-5′. The DNA probe for ER binding was a palindromic ERE oligonucleotide: 5′-gatcTCTTTGATCAGGTCACTGTGACCTGACTTTG-3′ and 3′-AGAAACTAGTCCAGTGACACTGGACTGAAACctag-5′. For RXR-RXR homodimer binding the probe was a DR-1 oligonucleotide: 5′-tcgaCAGGTCAGAGGTCAGTCGA-3′ and 3′-GTCCAGTCTCCAGTCAGCTgatc-5′. For VDR-RXRα heterodimer binding the probe was a DR-3 oligonucleotide: 5′-tcgaCAGGTCAAGGAGGTCAG-3′ and 3′-GTCCAGTTCCTCCAGTCgatc-5′. For RXRα-RARγ heterodimer binding the probe was a DR-5 oligonucleotide: 5′-tcgaCAGGTCACCAGGAGGTCAG-3′ and 3′-GTCCAGTGGTCCTCCAGTCagct-5′. In some experiments PR binding was done with a longer, 43-bp oligonucleotide containing a single GRE-PRE and flanking sequences from the mouse mammary tumor virus: 5′-gatcGGGTTTAAATAAGTTTATGGTTACAAACTGTTCTTAAAACAAG3′ and 3′-CCCAAATTTATTCAAATACCAATGTTTGACAAGAATTTTGTTCctag-5′.
SDS-PAGE and Western blots.
Receptors or HMG-1/-2 were subjected to SDS-PAGE on 12, 10, or 7.5% polyacrylamide gels as previously described (42, 46, 47), and protein bands were detected by staining with Coomassie blue or silver (21). Separated proteins were transferred to nitrocellulose membranes and incubated with the appropriate receptor-specific antibody overnight at 4°C. For mouse MAbs, Western blot detection was done with a secondary goat anti-mouse–horseradish peroxidase colorimetric method or by using rabbit anti-mouse IgG and 35S-labeled protein A followed by autoradiography. With rabbit polyclonal antibodies, goat anti-rabbit–horseradish peroxidase (Cappel) was used for detection. For the rat 9A7 MAb to human VDR, the detection method was with rabbit anti-rat IgG followed by the addition of 35S-labeled protein A and autoradiography.
Expression plasmids and reporter gene constructs.
The expression plasmids pHMG-1 and pHMG-2 contain genomic clones of mouse HMG-1 and HMG-2, respectively, under the control of their own constitutive promoters that were then cloned into the EcoRI site of pBluescript KS(+) as previously described (73). pBluescript KS(+) from Stratagene was used as an empty vector control. A mammalian-cell expression plasmid for human PR-B (pPR-B) was provided by Donald McDonnell (Duke University, Durham, N.C.) (64), an expression plasmid for human AR (p5HBL-AR-A) was provided by Elizabeth Wilson (University of North Carolina, Chapel Hill, N.C.), and an expression plasmid for GR (pSVGR) was provided by Ron Evans (Salk Institute). Human VDR expression plasmid (pRShVDR) for mammalian cells was under the control of the Rous sarcoma virus promoter (76).
Two progesterone-responsive reporter plasmids were used in the transfection assays. DHRE-E1b-CAT contains two optimal GRE-PRE elements linked to the TATA box of the adenovirus E1b gene and the chloramphenicol acetyltransferase (CAT) gene. PRE2tk-LUC contains two PREs linked to the herpes simplex virus thymidine kinase promoter and the luciferase (LUC) reporter gene as previously described (64). Three VDRE (vitamin D-responsive element) LUC reporters were used. These included the human 25-hydroxyvitamin D3–24-hydroxylase [24 (OH)ase] promoter (−1177 to −22), which contains two VDREs linked to LUC as previously described (76), a synthetic VDRE subcloned into a mouse mammary tumor virus (MMTV)-LUC vector (VDRE-1ΔMTV-LUC) that has the GREs deleted (76), and a synthetic DR-3 oligonucleotide cloned into a minimal tk-LUC reporter vector (DR3-tk-LUC).
Cell culture and transient transfections.
COS1 cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (HyClone) and were plated in multiwell dishes (six-well dishes; Falcon Plastics) at a density of 175,000 to 200,000 cells/well. HeLa cells were plated at the same density in DMEM supplemented with 5% fetal bovine serum. Cells were transiently transfected by an adenovirus-mediated method described previously (4). Purified defective adenovirus particles, covalently coupled to poly-l-lysine (provided by Nancy Weigel, Baylor College of Medicine, Houston, Tex.), were mixed with plasmid DNA for 30 min at room temperature in 20 mM HEPES buffer (pH 7.8) followed by the addition of poly-l-lysine at 2.4 μg/ml for an additional 30 min at room temperature. Just prior to transfection, culture medium was removed from the wells and replaced with 3 ml of serum-free DMEM per well, and the adenovirus-plasmid mixture was added directly to the medium at a multiplicity of infection of 250 to 400 viral particles/cell. Cells were then incubated for 2 h at 37°C, and then 3 ml of DMEM supplemented with 10% fetal bovine serum was added per well to bring the final serum concentration to 5%. The cells were then incubated for another 24 h at 37°C. After 24 h the cells were treated for another 24 h with and without hormone. At 48 h after transfection, cell monolayers were rinsed with CAT-LUC wash buffer (40 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA), and the cells were lysed in the well by the addition of 300 μl of cell lysis buffer (20 mM potassium phosphate, pH 7.4; 5 mM MgCl2; 0.5% Triton X-100) per well; the lysates were then measured for CAT or LUC activity. To estimate the percentage of transfected cells, parallel cells in six-well dishes were transfected with a cytomegalovirus (CMV)–β-galactosidase (β-Gal) reporter gene. At 48 h after transfection, cells were fixed in the well with 0.5% glutaraldehyde in phosphate-buffered saline and incubated with a 2% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining solution (4). The percentage of blue-stained cells averaged 15% for HeLa cells and 30% for COS1 cells. To control for variation in transfection efficiency, the cells were routinely cotransfected with a CMV–β-Gal internal control plasmid (2 ng/well). Cotransfection cultures with receptors and HMG-1/-2 expression plasmids also contained appropriate amounts of empty control vectors so that all cultures received the same amount of total plasmid DNA.
CAT and LUC assays.
CAT enzyme activity was measured by a radiometric phase-extraction method as previously described (38). Enzyme activity was calculated as the counts per minute of 3H-labeled acetyl coenzyme A converted per μg of protein in the cell lysate. LUC assays were done with a Monolight luminometer 2001 or 2010. Aliquots of cell lysates (5 to 50 μl) were added to 0.35 ml of LUC assay buffer (100 mM potassium phosphate, pH 7.8; 15 mM magnesium sulfate, 5 mM ATP; 1 mM DTT), and light output was measured for 10 s with a built-in 2-s delay after injection of 100 μl of 1 mM luciferin. The protein concentration was measured by Bradford assay, and equal amounts of protein (10 to 30 μg) were added to the CAT assays. Specific CAT and LUC activities were determined by subtraction of the assay background obtained from lysates of mock-transfected cells that received no plasmids. β-Gal activity from internal CMV–β-Gal control plasmids was measured in a luminometer by a chemiluminescent method according to the manufacturer’s instructions (Tropix, Inc., Bedford, Mass.), and CAT and LUC activities were corrected for variations in β-Gal activity. Cell transfections were all done in duplicate culture wells, and reporter gene assays were performed in duplicate with each lysate. Therefore, all CAT and LUC values are averages from four assay determinations.
RESULTS
Purification of recombinant nuclear receptors and HMG-1/-2 proteins for in vitro DNA binding studies.
Because our previous studies dealt only with PR (42, 46), it was used in this study as a standard for comparison. Human PR is expressed as two isoforms: an amino-terminal truncated PR-A and a full-length PR-B. The two PR isoforms are identical in their centrally located DNA binding domains and carboxyl-terminal ligand-binding domains (28). PR-A and PR-B have indistinguishable DNA binding properties in vitro (10), and HMG-1 was previously observed to stimulate the binding of both PR-A and PR-B to target DNA sites in a similar manner (42, 46). All DNA binding results in this study are shown with PR-A; indistinguishable results were obtained with PR-B (data not shown). In order to have a common purification procedure, all nuclear receptors in this study were expressed as recombinant proteins with an amino-terminal polyhistidine tag for use in metal ion affinity chromatography under nondenaturing conditions. The exception was human ER, which was expressed as a nonfusion protein and purified by using sequence-specific DNA affinity columns as previously described (39). Steroid receptors, including human PR, GR, AR, and ER, were expressed in Sf9 insect cells by using the baculovirus system and were bound to their cognate hormonal ligands during expression. Nonsteroid nuclear receptors, including human VDR, RARγ, and RXRα, were expressed in yeast cells.
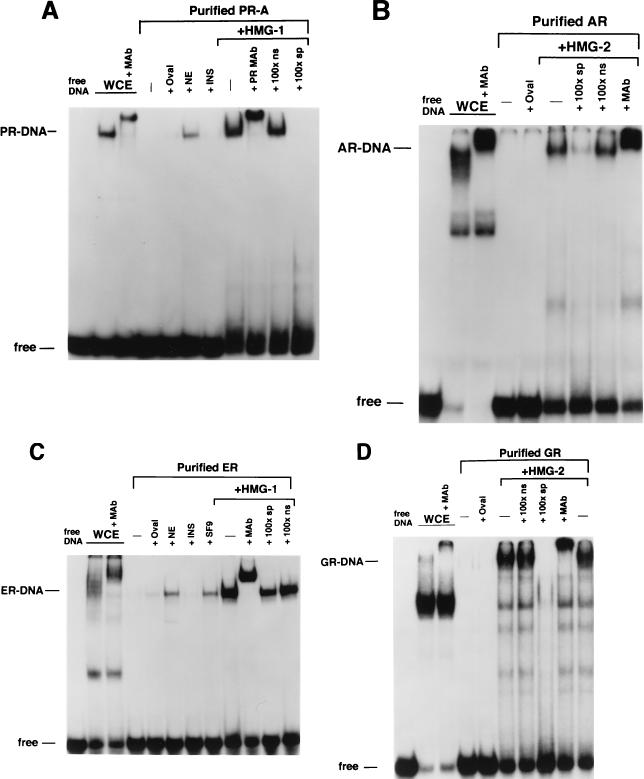
Sf9 insect cells expressing steroid receptors were lysed in aqueous buffer containing 350 mM NaCl and a low concentration of imidazole (10 to 15 mM) to reduce the binding of nonspecific proteins to the resins. The lysates were passed over a column containing a metal ion affinity resin (Talon; Clontech), and bound receptors were eluted by competition with 100 mM imidazole. As judged by silver-stain SDS-PAGE and Western blot analyses, this resulted in purification of intact full-length PR-A to greater than 95% (Fig. (Fig.1A).1A). This single-step purification also yielded intact GR (~100 kDa) at >90% purity (Fig. (Fig.1B)1B) and intact AR (~118 kDa) at approximately 40% purity. The AR preparation contained another protein band of ~100 kDa that was not reactive by Western blot with an AR-specific MAb (Fig. (Fig.1B).1B). Human ER purified by DNA affinity chromatography showed a major protein band at ~66 kDa, the size of intact hER, and another band of slightly faster mobility (Fig. (Fig.1B).1B).
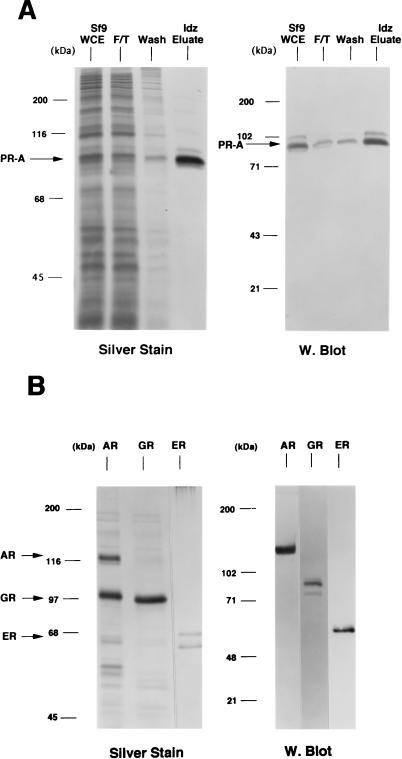
Purification of baculovirus-expressed steroid receptors. Human PR-A, AR, and GR were expressed as polyhistidine-tagged proteins in baculovirus and were purified by metal ion affinity resins (Talon). Human ER was expressed as a nonfusion protein in baculovirus and purified by using a DNA affinity column. (A) (Left panel) Silver-stained SDS–7.5% PAGE of PR-A purification fractions. Lanes: 1, Sf9 whole-cell extract (WCE; 5 μl); 2, Talon resin flowthrough (F/T; 5 μl); 3, column wash (Wash; 50 μl); and 4, 100 mM imidazole (Idz) eluate (10 μl). (Right panel) Western blot of the same PR-A purification fractions (5 μl) with the AB-52 MAb. (B) (Left panel) Silver-stained SDS-PAGE of purified AR and GR eluted from Talon resins by 100 mM imidazole. ER is an NaCl eluate of a DNA affinity resin. (Right panel) Western blots of purified AR, GR, and ER were done with the following receptor specific antibodies: N441, an MAb to AR; a polyclonal antibody to GR; and h151, an MAb to human ER.
Polyhistidine-tagged human VDR, RARγ, and RXRα were expressed in yeast cells and purified either by nickel affinity chelation resins (VDR and RXRα) or by Talon resins (RARγ). VDR and RXRα were eluted by a pH gradient followed by neutralization, while RARγ was eluted by competition with 100 mM imidazole. Silver-stained SDS-polyacrylamide gels and Western blot analysis of purified VDR, RARγ, and RXRα are shown in Fig. Fig.2.2. Each purified product exhibited a major protein band of the expected size and appropriate immunoreactivity with receptor-specific antibodies (58). All purified nuclear receptor preparations appeared to be free of contaminating HMG-1/-2, as determined by lack of Western blot reactivity with an MAb to HMG-1/-2 and a rabbit polyclonal antibody to HMG-2 (data not shown).
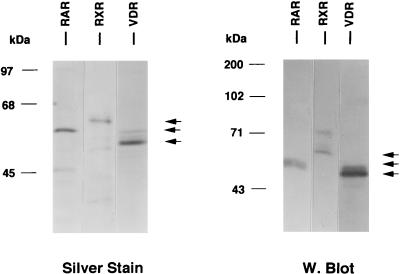
Purification of nonsteroid nuclear receptors. Human RXRα, RARγ, and VDR were expressed in yeast cells as polyhistidine-tagged proteins and were purified by using metal ion affinity resins. Purified products were analyzed by SDS-PAGE and silver staining (left panel) and by Western blotting with receptor-specific antibodies (right panel).
Separate genes encode HMG-1 and HMG-2, but the two proteins are closely related, with 82% amino acid sequence identity. HMG-1 and HMG-2 differ primarily in the carboxyl-terminal acidic domain (HMG-2 has a shorter C-terminal tail) and in a few amino acid sequences in the HMG-box DNA binding domains (7). HMG-1 and HMG-2 were purified from 0.35 M NaCl nuclear extracts from calf thymus by chromatofocusing on PBE94 resins. Eluted fractions were analyzed by SDS-PAGE and Western blotting and then pooled. As shown in Fig. Fig.3A,3A, pool 1 (lane 1) contains highly purified HMG-2 (~25 kDa), pool 2 (lane 2) contains a mixture of purified HMG-1 and HMG-2, and pool 3 (lane 3) contains only HMG-1 (~27 kDa). Also shown is recombinant baculovirus-expressed HMG-1 with a polyhistidine tag at the amino terminus that was purified by using Talon resins (Fig. (Fig.3A,3A, lane 4). It should be noted that recombinant HMG-1 is larger than native HMG-1 by ~4 kDa due to the six sequential histidine and enterokinase cleavage sequences. The purified HMG proteins shown in Fig. Fig.3A3A were stained with Coomassie blue. Upon silver staining of the same material, calf thymus and recombinant HMG are purified to near homogeneity (data not shown). In our previous studies, calf thymus HMG-1 purified by similar methods was determined by overloaded silver-stained SDS-polyacrylamide gels to be apparently homogeneous (42, 46). Because HMG actually stains more efficiently with Coomassie blue than with silver, we have switched to Coomassie blue staining of purified HMG.
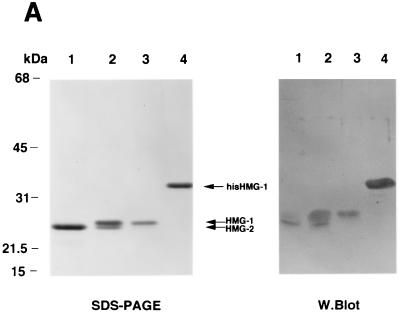
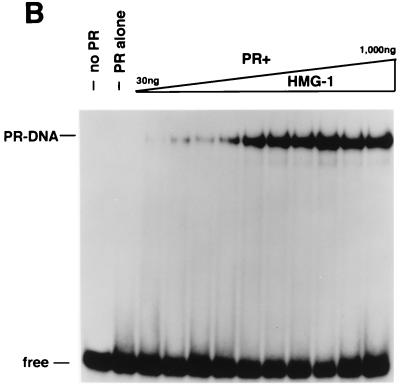
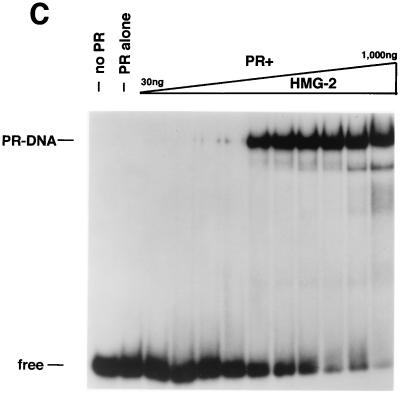
HMG-1 and HMG-2 purification and effects on PR-DNA binding in vitro. (A) HMG-1 and HMG-2 from calf thymus were purified and separated on a PBE94 chromatofocusing column as described in Materials and Methods. Coomassie blue-stained SDS-polyacrylamide gels (12%) and Western blot analysis of purified HMG-1/-2 fractions were prepared as follows: lane 1 (pool 1), HMG-2; lane 2 (pool 2), HMG-1 and HMG-2; lane 3 (pool 3), HMG-1; lane 4, purified recombinant polyhistidine-tagged HMG-1. Western blots were done with an IgM MAb (854/E10) that recognizes both HMG-1 and HMG-2. (B and C) The influence of purified calf thymus HMG-1 (pool 3) and HMG-2 (pool 1) on the binding of purified PR-A to a 32-bp PRE oligonucleotide probe as assessed by EMSA. A constant amount (30 nM) of purified PR-A was incubated with increasing amounts of purified HMG-1 (B) or HMG-2 (C). The amounts of HMG-1 and HMG-2 used were 30, 50, 70, 100, 200, 300, 400, 500, 700, and 1,000 μg/assay in lanes 3 to 12, respectively.
HMG-1 and the closely related HMG-2 facilitate the sequence-specific DNA-binding activity of recombinant purified human PR.
Our previous studies showing that HMG-1 increased the DNA binding activity of PR were done with a nonfusion baculovirus-expressed PR purified by MAb affinity chromatography (42, 46). Therefore, before analyzing the effects of HMG-1 on other nuclear receptors, we determined whether polyhistidine-tagged PR purified by metal affinity resins would respond in a similar manner to HMG-1 as had antibody-purified PR. Crude and highly purified preparations of baculovirus expressing polyhistidine-tagged PR-A (Fig. (Fig.1A)1A) were analyzed by EMSA for binding to a PRE oligonucleotide. As with our results with nonfusion MAb-purified PR (42, 46), polyhistidine-tagged PR-A in crude extracts of Sf9 insect cells bound to the PRE probe, whereas an equal amount of purified polyhistidine-tagged receptor exhibited no DNA binding activity (Fig. (Fig.4A).4A). The addition of calf thymus nuclear extracts or purified calf thymus HMG-1 stimulated the formation of PR-DNA complexes (Fig. (Fig.4A).4A). As controls for the general effect of other proteins on the stability of purified PR, the addition of unrelated proteins such as albumin or insulin had no effect on PR-DNA binding (Fig. (Fig.4A).4A). The DNA complex stimulated by HMG-1 is specific, as demonstrated by supershifts with a MAb to PR and by competition with unlabeled homologous PRE oligonucleotide but not by an ERE oligonucleotide (Fig. (Fig.4A).4A). Purified recombinant baculovirus-expressed HMG-1 exhibited stimulatory activity similar to that of native HMG-1 purified from calf thymus, indicating that minor contaminants in calf thymus preparations are not responsible for stimulating PR-DNA binding activity (data not shown). In previous studies we observed that bacterially expressed and purified HMG-1 stimulated PR-DNA binding (42). The fact that one source of purified cellular HMG-1 and two sources of purified recombinant HMG-1 all have similar activities strongly indicates that the stimulation of PR-DNA binding is an intrinsic property of HMG-1/-2 and is not due to contaminating proteins.
HMG-1/-2 stimulate the sequence-specific DNA binding activity of all steroid receptors tested. (A) Equal amounts (30 nM) of polyhistidine-tagged PR-A in Sf9 whole-cell extract (WCE) or affinity-purified PR-A were analyzed for binding to a 32P-labeled palindromic GRE-PRE oligonucleotide by EMSA. Lanes contain Sf9 whole-cell extract (WCE), purified PR alone (−), or purified PR plus 1 μg of ovalbumin (+Oval), 1 μg of nuclear extract (+NE) from calf thymus, 1 μg of insulin (+INS), or 300 ng of purified calf thymus HMG-1 (+HMG-1). The HMG-1-stimulated PR-DNA complexes were supershifted by the PR-specific MAb, AB-52, and were competed by a 100-fold molar excess of the PRE oligonucleotide (100×sp) but not by an ERE oligonucleotide (100×ns). (B) Equal amounts (70 nM) of baculovirus-expressed AR in WCE and affinity-purified AR were analyzed for binding to the GRE-PRE oligonucleotide by EMSA as described for panel A. (C) Equal amounts (10 nM) of baculovirus-expressed ER in WCE and affinity-purified ER were analyzed for binding to an ERE oligonucleotide by EMSA as described for panel A with or without the addition of 300 ng of purified calf thymus HMG-1. (D) Equal amounts (200 nM) of baculovirus-expressed GR in WCE and affinity-purified GR were analyzed for binding to the GRE-PRE by EMSA as described for panel A. All receptors were bound to their cognate (see Materials and Methods) hormonal ligands in Sf9 cells during expression.
In our previous studies, only HMG-1 was analyzed (42). To compare the activity of HMG-1 and the closely related HMG-2, PR-DNA binding assays were performed with a constant amount of purified PR-A and various amounts of purified HMG-1 or HMG-2 (see Fig. Fig.3A).3A). Both HMG-1 (Fig. (Fig.3B)3B) and HMG-2 (Fig. (Fig.3C)3C) stimulated the formation of specific PR-DNA complexes in a dose-dependent manner. Maximal stimulation was observed with the addition of between 200 and 400 ng for both HMG-1 and HMG-2, which is approximately a 5- to 10-fold molar excess over PR. HMG-1 and HMG-2 exhibited slight differences in activity in two respects, however. First, HMG-2 was consistently observed to stimulate a higher maximal level of PR-DNA complexes (Fig. (Fig.3B3B and C). Second, HMG-2 appeared to stimulate cooperative binding of PR to DNA, a finding that was not evident with HMG-1. The dose-dependent increase in PR-DNA binding caused by the addition of HMG-1 occurred gradually over the entire range of HMG-1 concentrations. In contrast, nearly all of the stimulation of PR-DNA complexes produced by addition of HMG-2 was obtained over a much narrower concentration range, i.e., between 100 and 200 ng of HMG-2 per assay (Fig. (Fig.33C).
HMG-1/-2 facilitate the sequence-specific DNA binding activity of all steroid receptors tested.
We next analyzed the influence of HMG-1/-2 on the DNA binding activities of other steroid receptors. Because PR, GR, and AR all recognize a common GRE, these receptors were assayed for binding to the same palindromic GRE oligonucleotide probe. ER binding was assessed with a palindromic ERE probe. As with the results with PR, recombinant baculovirus-expressed AR, GR, and ER in crude Sf9 cell extracts bound specifically to their target DNA probes. Equal amounts of purified preparations of these same receptors exhibited a loss of DNA binding activity that was restored by the addition of either HMG-1 or HMG-2 but not by the addition of other unrelated proteins (Fig. (Fig.4B4B to D). The DNA complexes stimulated by HMG-1 or HMG-2 were specific, as demonstrated by supershifts with the appropriate receptor-specific antibody and by competition with homologous unlabeled DNA probes (Fig. (Fig.4B4B to D). Similar to results with PR, HMG-1 and HMG-2 were found to be interchangeable with respect to stimulating the DNA binding activity of other purified steroid receptors (data not shown).
In previous studies with PR purified by MAb affinity chromatography, we observed that receptor alone was able to bind to 32P-labeled GRE-PRE but with low affinity requiring a high receptor concentration for detection by EMSA. The addition of purified HMG-1 increased the apparent affinity of PR for its target DNA sequences and thus was most effective in stimulating DNA binding activity at low receptor concentrations (42, 46). To determine whether HMG-1/-2 also increased the binding affinity of other steroid receptors for target DNA, saturation DNA binding assays were performed with purified GR, AR, and ER in the presence or absence of HMG-1/-2. Purified polyhistidine-tagged PR-A was used as a reference. These experiments were done by varying the concentration of purified receptors against a constant amount of [32P]DNA probe under equilibrium binding conditions. Specific receptor-DNA complexes at each receptor concentration were quantitated by phosphorimage scanning of the gels, and the data were plotted as the percent upshifted DNA relative to total DNA. Figure Figure55 (A to D) shows that each purified steroid receptor bound to specific DNA with low affinity, generating complexes only at high receptor concentrations. Addition of either HMG-1 or HMG-2 but not an unrelated protein such as ovalbumin caused a substantial leftward shift of the binding curves for all purified steroid receptors. The relative DNA binding affinity and the fold increase caused by the addition of HMG-1/-2 varied for each receptor. This could be due to differences in intrinsic binding affinities of each receptor for target DNA or to differences in the estimated concentration of each purified receptor. Interestingly, AR showed no detectable binding in the absence of HMG-1/-2 at any of the concentrations tested, whereas other steroid receptors produced low levels of DNA binding in the absence of HMG-1/-2. Thus, the fold increase in the apparent DNA binding affinity caused by the addition of HMG-1 or HMG-2 was greatest with AR. We also observed that the concentrations of AR and GR in the presence of HMG-1/-2 required to saturate the DNA binding sites were higher than that required for PR and ER and that ER achieved saturation at the lowest concentration. This suggests that AR and GR have lower binding affinities than PR for the same target DNA and that ER has the highest affinity for target DNA. Alternatively, variation in the percentage of purified receptors that are active could also contribute to these concentration differences. Nevertheless, all four purified steroid receptor preparations exhibited a substantial increase in affinity for target DNA in response to the addition of HMG-1 or HMG-2.
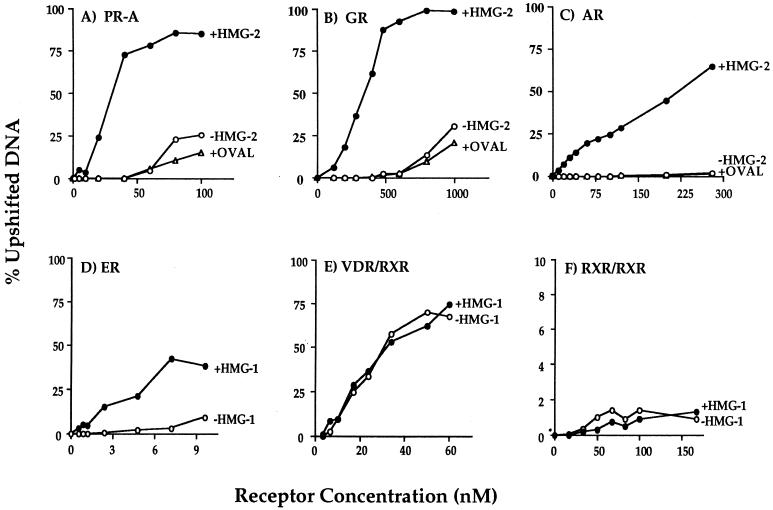
Saturation DNA binding analysis of purified nuclear receptors in the presence or absence of HMG-1/-2. The concentrations of purified receptors indicated in the figure were varied in DNA binding reactions against a constant amount (0.3 ng) of 32P-labeled oligonucleotide probe, and DNA binding was analyzed by EMSA. Each receptor was assayed alone (−HMG-1 or −HMG-2) or with the addition of 150 to 300 ng of HMG-1, HMG-2, or ovalbumin (+OVAL). The oligonucleotide probe for PR-A (panel A), GR (panel B), and AR (panel C) was a palindromic GRE-PRE. A palindromic ERE oligonucleotide was used as the probe for ER (panel D), a DR-3 oligonucleotide was used for the VDR-RXR heterodimers (panel E), and a DR-1 probe was used for the RXR homodimers (panel F). Purified receptors were quantitated by protein Bradford assay and by comparing silver-stained receptor band intensities with those of known amounts of purified molecular weight protein standards. The specific DNA complexes at each concentration of receptor were quantitated by phosphorimage analysis and plotted as the percentage of total DNA.
HMG-1/-2 have no effect on the binding of nonsteroid nuclear receptors to direct-repeat target DNA sequences.
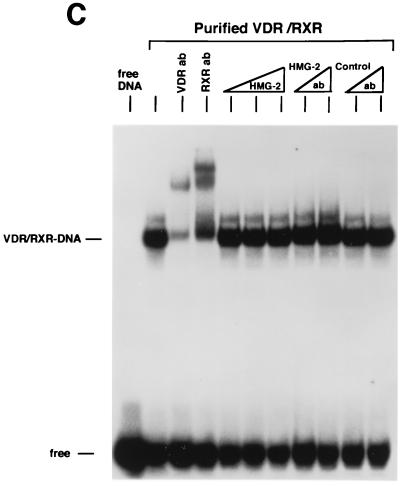
We next examined the influence of HMG-1/-2 on binding of human VDR and RARγ to DR target DNA sequences. EMSA showed that purified VDR did not bind to a DR-3 probe (Fig. (Fig.6A)6A) and that purified RARγ failed to interact with a DR-5 probe (Fig. (Fig.6B).6B). In contrast to the results with steroid receptors, the addition of HMG-1 or HMG-2 had no effect on the DNA binding activities of VDR and RARγ (Fig. (Fig.6).6). As expected, the addition of purified RXR stimulated the DNA binding activity of purified VDR and RAR by a heterodimerization mechanism, as shown by supershifts with RXR-, VDR-, and RAR-specific antibodies (Fig. (Fig.6).6). Competition with excess unlabeled homologous and unrelated oligonucleotides demonstrates that the RXR-induced complexes are specific (Fig. (Fig.6A6A and B). The addition of HMG-1 or HMG-2 also had no effect on the DNA binding activity of RXR-VDR and RXR-RAR heterodimers (Fig. (Fig.6A6A and B). To determine whether HMG-1/-2 might in fact stimulate this class of nonsteroid nuclear receptors but simply require a higher concentration than is needed to stimulate steroid receptors, increasing amounts of HMG-2 were added to VDR-RXR heterodimer DNA binding reactions. As shown in Fig. Fig.6C,6C, at concentrations three times higher (900 ng) than are required to maximally stimulate PR-DNA binding, HMG-2 had no effect on the sequence-specific DNA binding activity of VDR-RXR heterodimers (Fig. (Fig.6C).6C). Also, the HMG-2 antibody did not supershift VDR-RXR DNA complexes in the presence of large amounts of HMG-2, suggesting that HMG-2 is not recruited to the complex (Fig. (Fig.6C).6C).
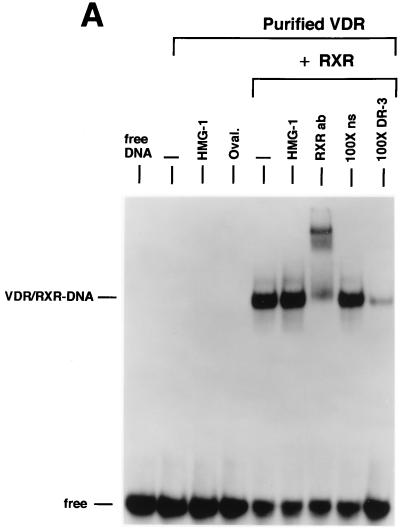
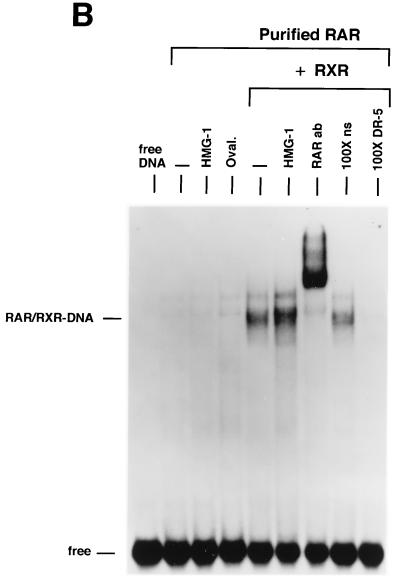
HMG-1/-2 has no influence on sequence-specific DNA binding activity of purified human VDR and RARγ. (A) Human VDR expressed and purified from yeast cells was incubated for 1 h at 4°C with 0.3 ng of a [32P]DR-3 probe, and samples were analyzed by EMSA. A single concentration of purified VDR (60 nM) was incubated alone (−) or with the addition of 150 ng of purified HMG-1, 1 μg of ovalbumin (Oval), 50 ng of purified RXRα, or 50 ng of RXRα plus 150 ng of purified HMG-1. A supershift of RXR-induced DNA complexes with an antibody to RXRα (RXRab) shows that RXR is a component of the stimulated DNA complex. The specificity of the DNA complexes is shown by the lack of competition with a 100-fold molar excess of an unlabeled nonspecific DNA (100×ns) compared to an unlabeled DR-3 (100×DR-3). (B) Purified human RARγ (60 nM) expressed and purified from yeast cells was incubated with 0.3 ng of a [32P]DR-5 probe, and samples were analyzed by EMSA as for panel A. (C) Purified VDR and RXR were incubated with the DR-3 probe as for panel A. Supershifts of the RXR-VDR heterodimer-DNA complex are shown with antibodies to VDR (VDR ab) and RXR (RXR ab). Increasing amounts of purified HMG-2 (300, 600, and 900 ng) were added to the VDR-RXR heterodimer-DNA binding reaction. At the intermediate amount of HMG-2 (i.e., 600 ng), DNA binding reactions were incubated with two concentrations of HMG-2 antibody (HMG-2 ab) or a control unrelated antibody (Control ab).
Saturation DNA binding analyses were performed with purified RXRα-VDR and RXRα-RARγ heterodimers in the presence or absence of HMG-1. The apparent binding affinity of RXRα-VDR heterodimers for a DR-3 element (Fig. (Fig.5E)5E) and the affinity of RXRα-RARγ heterodimers for a DR-5 probe (not shown) were not affected by the addition of HMG-1. Thus, HMG-1 and HMG-2 are not able to functionally substitute for RXR as coregulators of VDR and RAR DNA binding, and these two proteins did not influence the DNA binding activity of RXR-VDR or RXR-RAR heterodimers.
RXR is capable of binding as a homodimer to DR-1 elements. However, this binding is weaker than that of RXR heterodimers for their cognate DR sites. Therefore, we also asked whether HMG-1/-2 were capable of stimulating RXR homodimer binding to DNA. As shown by the saturation DNA binding analysis in Fig. Fig.5F,5F, HMG-1 had no influence on the low binding activity of purified RXR for a DR-1 probe. Thus, based on an analysis of four different steroid receptors and three nonsteroid nuclear receptors, it appears that HMG-1/-2 are capable of stimulating only the sequence-specific DNA binding activity of steroid receptors.
HMG-1 and HMG-2 transiently interact with PR and are recruited by PR to the DNA complex.
To further investigate the mechanism by which HMG-1/-2 stimulate the apparent binding affinity of steroid receptors for HREs, we focused our analysis on PR. Using short PRE oligonucleotide probes, we previously observed that stimulation of PR-DNA binding by HMG-1 was not accompanied by a decrease in electrophoretic mobility of the DNA complex, nor were stimulated complexes supershifted by the addition of antibodies to HMG-1 (42, 46). This suggested that HMG-1 is a less-stable component of the DNA complex than PR and that it dissociates during electrophoresis. However, in the context of a longer DNA fragment containing a single PRE, the addition of HMG-1 did result in reduced mobility of some of the stimulated PR-DNA complexes, which was further supershifted by anti-HMG-1 MAb (47). To test whether HMG-2 can also associate with the PR-DNA complex, we used a longer PRE oligonucleotide probe than that used in Fig. Fig.33 to to55 (43 bp versus 32 bp). As shown in Fig. Fig.7A,7A, purified HMG-2 alone did not bind to the 43-bp PRE probe at any of the concentrations tested, nor were complexes formed with purified PR alone at a submaximal receptor concentration. The addition of purified PR and HMG-2 together resulted in a synergistic stimulation of DNA complexes that contained both HMG-2 and PR, as evidenced by supershifts with PR (AB-52)- or HMG-2-specific antibodies (Fig. (Fig.7A).7A). As a control, an antibody (h151) to ER had no effect on the electrophoretic mobility of the complex. These results show that HMG-2 can associate with the PR-DNA complex. Furthermore, the inability of HMG-2 to associate with the PRE until receptor was added indicates that HMG-2 is recruited to the complex by PR.
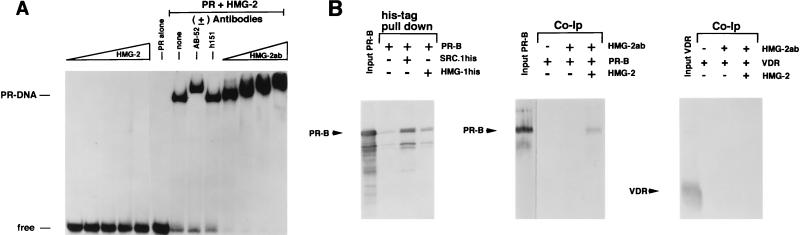
Association of HMG-2 with the PR-DNA complex and direct binding of HMG-1/-2 to PR in the absence of DNA. (A) HMG-2 is recruited to the PR-DNA complex. A 43-bp oligonucleotide containing the distal-most GRE-PRE and flanking sequences of MMTV was used as the DNA probe for EMSA. The DNA probe (0.3 ng) was incubated with increasing amounts of purified HMG-2 alone (100 to 300 ng), with purified PR-A alone (30 nM), or with purified PR-A (30 nM) plus 300 ng of HMG-2, either without (none) or with antibody to PR (AB-52), ER (h151), or HMG-2 (HMG-2ab; rabbit antisera added in increasing amounts). (B) (Left panel) Baculovirus-expressed polyhistidine-tagged HMG-1 was incubated with nonfusion recombinant PR-B, and the samples were bound to Talon resins. Resins were washed six times in a buffer containing 100 mM NaCl and 15 mM imidazole, and bound proteins were eluted with SDS-sample buffer and analyzed by Western blotting with the 1294 MAb to PR. As a control for nonspecific binding, PR-B was incubated with Talon resins in the absence of polyhistidine-tagged HMG-1. As a positive control for a known interacting protein, PR-B was incubated with polyhistidine-tagged SRC-1. Purified PR-B (middle panel) or VDR (right panel) was incubated with calf thymus HMG-2, and samples were immunoprecipitated with a rabbit antibody to HMG-2 by using protein A-Sepharose as an absorbent. The protein A-Sepharose beads were washed six times in a buffer containing 100 mM NaCl, and bound proteins were eluted with SDS-sample buffer and analyzed by Western blotting with the 1294 MAb to PR (middle panel) or the 9A7 MAb to human VDR (right panel). The leftmost lane in each of these two panels is the assay input of PR-B or VDR.
We next asked whether HMG-1/-2 can interact directly with PR in the absence of DNA. This was tested by a pulling down of the PR with histidine-tagged HMG-1 and by coimmunoprecipitation of PR and HMG-2 with an anti-HMG-2 antibody. As shown in Fig. Fig.7B,7B, a weak but specific association of PR-B with immobilized polyhistidine–HMG-1 was detected. As a positive control for a known receptor-interacting protein, Fig. Fig.7B7B also shows PR-B association with immobilized polyhistidine-tagged steroid receptor coactivator (SRC-1–his). With a rabbit polyclonal antibody specific to HMG-2, PR-B was coimmunoprecipitated with HMG-2 in a specific manner when the two proteins were added together. PR was not immunoprecipitated by the HMG-2 antibody in the absence of HMG-2 (Fig. (Fig.7B).7B). Interestingly, HMG-2 did not interact with VDR as indicated by coimmunoprecipitation assay (Fig. (Fig.7B),7B), nor did it affect VDR-DNA binding activity, suggesting that the transient interaction of HMG-1/-2 with PR may be meaningful.
HMG-1/-2 stimulate PR-mediated gene activation in mammalian cells.
To determine whether HMG-1/-2 can affect the transcriptional activity of PR in vivo, COS1 cells were cotransfected with expression plasmids for hPR-B, HMG-1, or HMG-2, and a progesterone-responsive reporter construct containing two GREs-PREs fused to the TATA box of E1b and the CAT reporter gene (DHRE-E1b-CAT). PR-B in general is a stronger transcriptional activator than PR-A; however, this differential activity is both cell-type and promoter context specific (64, 69). With this simple GRE-PRE promoter, PR-B in COS1 cells is a much stronger activator than PR-A (not shown). Thus, our studies have focused on the effect of HMG-1/-2 on the activity of PR-B. When COS1 cells were transfected with phPR-B alone, addition of the synthetic progestin R5020 resulted in an ~10-fold stimulation of CAT activity (Fig. (Fig.8A).8A). Upon cotransfection with phPR-B and increasing amounts of the HMG-1 expression plasmid (pHMG-1), R5020 stimulation of CAT activity was increased in a dose-dependent manner by another sevenfold at the highest amount of transfected pHMG-1 (7.03-fold ± 1.9 standard error of the mean [SEM], n = 11). Cotransfection with an empty vector control had minimal effect on R5020 stimulation of the DHRE-E1b-CAT reporter gene (Fig. (Fig.8A).8A). In the absence of hormone small increases in reporter gene activity were observed when cotransfecting cells with phPR-B and pHMG-1. The increase in R5020 induction by cotransfected HMG-1 is PR dependent and is not due to an effect on the basal promoter activity of the reporter gene. This is shown by the lack of stimulation of CAT activity when cells were transfected with pHMG-1 in the absence of hPR-B (Fig. (Fig.8A).8A). These results demonstrate that cotransfection of HMG-1 substantially increases hormone-dependent PR-mediated transactivation without affecting the basal promoter activity of the reporter gene.
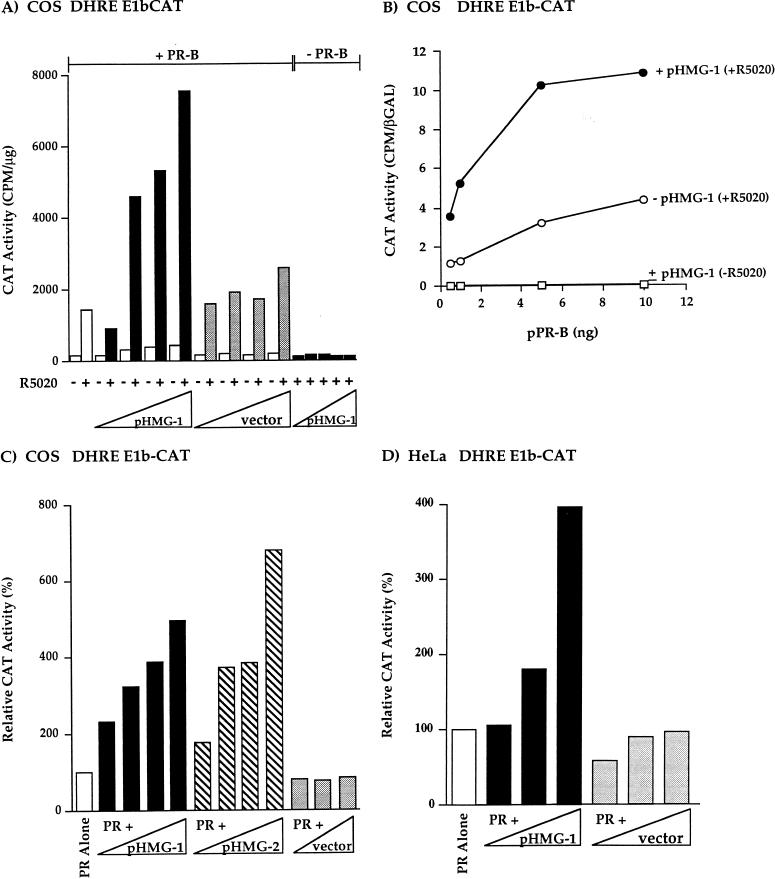
Ectopically expressed HMG-1 or HMG-2 stimulates the transcriptional activity of human PR in mammalian cells. (A) COS1 cells were transfected with a PR-B expression plasmid (phPR-B; 0.5 ng/well), increasing amounts of an HMG-1 expression plasmid (pHMG-1; 1 to 20 ng/well) or an empty vector control (1 to 20 ng/well), and the DHRE-E1b-CAT reporter plasmid (500 ng/well). Cells were treated with or without R5020 (100 nM) for the last 24 h of transfection. In some wells, cells were cotransfected with increasing amounts of pHMG-1 (1 to 20 ng/well) and no PR-B (−PR-B). CAT activity was calculated as counts per minute of 3H-labeled acetyl coenzyme A converted per microgram of total protein, and the values shown are averages from three independent experiments. (B) COS1 cells were transfected with various amounts of phPR-B expression plasmid in the presence or absence of pHMG-1 (20-fold molar excess over phPR-B). At 24 h after transfection, cells were treated for the next 24 h with or without R5020 (100 nM), and CAT activity was measured and calculated as the ratio of CAT activity to β-Gal activity of the internal control plasmid. The results are average values from duplicate transfection wells from a single experiment and are representative of two independent experiments. (C) COS1 cells were transfected with phPR-B (1 ng/well), DHRE-E1b-CAT reporter (500 ng/well), various amounts of pHMG-1 and pHMG-2, or an empty control vector (0.5- to 20-fold molar excess over the PR expression plasmid). The CAT activity in each treatment group was calculated as the fold R5020 induction over that with no hormone. The data were expressed as relative CAT activity, setting the hormone-induced level obtained in cells transfected with PR alone as the 100% value. The values are averages from four independent experiments. (D) HeLa cells were transfected with phPR-B (10 ng/well), increasing amounts of either pHMG-1 or the empty control vector (1.0- to 20-fold molar excess over phPR-B), and the DHRE-E1b-CAT (500 ng/well). The relative CAT activity was calculated as for panel C. The values are averages from duplicate transfections from a single experiment that is representative of three independent experiments.
As shown in Fig. Fig.8A,8A, cells were transfected with a suboptimal level of phPR-B (0.5 ng/well). To determine whether the stimulatory effect of HMG-1 on PR transactivation occurs only with subthreshold levels of receptor or whether it can also affect the activity of higher levels of receptor, COS1 cells were cotransfected with a constant ratio of pHMG-1 (20-fold molar excess over PR plasmid) and increasing amounts of phPR-B. As shown in Fig. Fig.8B,8B, pHMG-1 increased R5020 induction of DHRE-E1b-CAT at all of the amounts of transfected phPR-B tested, indicating that HMG-1 stimulates the maximal transcriptional activity of PR.
Since HMG-1 and HMG-2 are interchangeable with respect to stimulating PR binding to PREs in vitro, we also tested whether coexpression of HMG-2 affects the transcriptional activity of PR in whole cells. As shown in Fig. Fig.8C,8C, R5020 induction of the DHRE-E1b-CAT reporter gene was stimulated in a similar dose-dependent manner by cotransfection of COS1 cells with phPR-B and increasing amounts of either pHMG-1 or pHMG-2. R5020-dependent, PR-mediated transcription was increased fivefold by the highest amount of pHMG-1 and sevenfold (7.27 ± 2.8 SEM, n = 4) by pHMG-2 (Fig. (Fig.8C).8C). Thus, consistent with the in vitro DNA binding results, both HMG-1 and HMG-2 are capable of increasing the transcriptional activity of PR in vivo, with HMG-2 tending to have a greater effect.
To determine whether the effect of HMG-1/-2 on PR activity in vivo extends to other cell types and target promoters, cotransfection experiments were conducted in HeLa cells and with another progestin-inducible reporter construct (PRE2-tk-LUC). In HeLa cells, PR-mediated activation of DHRE-E1b-CAT was stimulated in a dose-dependent manner with increasing amounts of cotransfected pHMG-1 (Fig. (Fig.8D).8D). In multiple independent experiments, it was found that the largest amount of pHMG-1 stimulated PR activity in HeLa cells by ca. fourfold (3.5 ± 0.044 SEM, n = 3). In COS1 cells, cotransfected pHMG-1 stimulated R5020-dependent PR-mediated activation of PRE2-tk-LUC by a factor of ca. 10-fold (10.7 ± 2.3 SEM, n = 9). These results provide strong evidence that HMG-1/-2 have a general role in PR function in vivo that does not appear to be cell type or target gene restricted.
HMG-1 stimulates transcriptional activity of other steroid receptors (GR and AR) in vivo but has minimal effect on VDR.
We also tested, by use of transient-transfection assays, the effect of HMG-1 on the transcriptional activity of other nuclear receptors in vivo. Because PR, GR, and AR can all bind and mediate functional responses through the same consensus hormone response element, the PRE2-tk-LUC reporter vector was used to assess the effect of HMG-1 on transcriptional activation mediated by GR and AR. Both AR- and GR-mediated activation of PRE2-tk-LUC was increased substantially in cells that were cotransfected with pHMG-1 compared to cells cotransfected with receptor alone or with receptor and an empty vector control (Fig. (Fig.9A).9A). The only difference from results obtained with PR was a greater stimulation of ligand-independent activation of AR and GR by HMG-1 (Fig. (Fig.9A).9A). To determine whether HMG-1 affects the activity in vivo of a receptor from the nonsteroid class of nuclear receptors, COS1 cells were cotransfected with a VDR expression plasmid and increasing amounts of pHMG-1, and the ability of vitamin D3 to induce expression of VDRE-promoter constructs was assessed. Since COS1 cells express endogenous RXR, these assays measured the transcriptional activity of the RXR-VDR heterodimers. At the two lowest amounts of cotransfected plasmids, pHMG-1 had no effect on vitamin D3-induced, VDR-dependent activation of the VDRE-24(OH)ase-LUC reporter gene (Fig. (Fig.9B).9B). However, at the highest amount of cotransfected plasmid, pHMG-1 stimulated VDR-mediated gene activation by 2.6-fold (2.6 ± 0.73 SEM, n = 7). Within the same experiments, cotransfection with pHMG-1 increased R5020-dependent, PR-mediated activation of the PRE-tk-LUC reporter gene by 10-fold (Fig. (Fig.9B).9B). Because VDRE-24(OH)ase-LUC is a complex promoter with potential sites for other transactivators that could be responsible for the 2.6-fold enhancement of VDR activity by pHMG-1, we have also analyzed the effect of HMG-1 on the VDR-dependent activation of two other reporters, including VDRE-1ΔMTV-LUC and DR3-tk-LUC. Ectopically expressed HMG-1 had no stimulatory effect on VDR transactivation of either of these vitamin D3-responsive reporter constructs, even with amounts of cotransfected pHMG-1 10 times greater than that required to maximally enhance PR activity in vivo (Fig. (Fig.9C9C and D). Again, coexpressed HMG-1, as an internal positive control, stimulated PR activity within the same transient transfection assays by ca. 10-fold (Fig. (Fig.9C9C and D). Thus, consistent with the effects of HMG-1/-2 on receptor-DNA binding in vitro, ectopically expressed HMG-1/-2 substantially increased the transcriptional activity of steroid receptors in vivo but had minimal [24(OH)ase promoter] or no (VDRE-1ΔMTV-LUC and DR3-tk-LUC) effect on the activity of at least one other class of nuclear receptor.
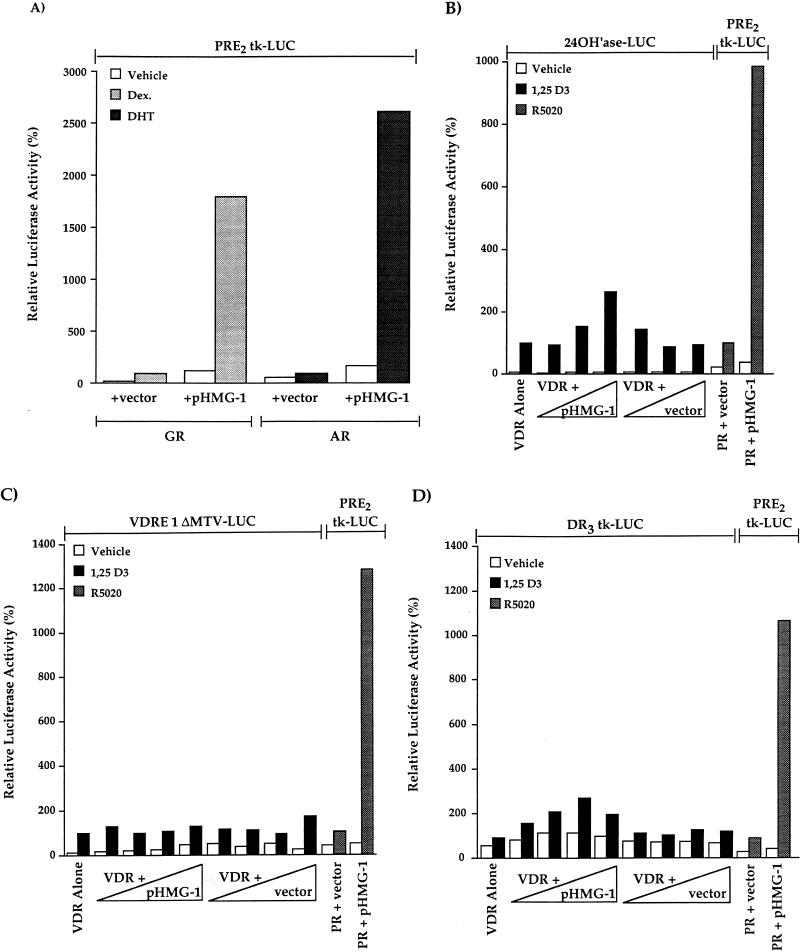
HMG-1 stimulates transcriptional activity of other steroid receptors in vivo but has minimal effects on VDR-mediated transcription. COS1 cells were cotransfected with human AR (p5HBL-AR-A) or GR (pSVGR) expression plasmids (10 ng/well), pHMG-1 (400 ng/well) or an empty vector control, and the PRE2-tk-LUC (200 ng/well) reporter. Cells were incubated for 24 h prior to harvest with vehicle, dexamethasone (Dex.; 10−6 M) or dihydrotestosterone (DHT) (10−7 M). LUC was measured and calculated as relative activity, setting the hormone-induced levels in cells transfected with receptor alone (plus empty vector control) at 100%. The values are averages from three independent experiments for AR and from four independent experiments for GR. (B) COS1 cells were transfected with a human VDR expression plasmid (1 ng/well), increasing amounts of pHMG-1 or an empty vector control (4- to 40-fold excess over VDR), and the VDRE-24(OH)ase-LUC reporter (200 ng/well). At 24 h after transfection, cells were treated for another 24 h without and with 1,25-(OH)2 D3 (10 nM). Within the same experiments, COS1 cells were cotransfected with phPR-B (1 ng/well), pHMG-1 or empty vector (40 ng/well), and PRE2-tk-LUC (200 ng/well). At 24 h after transfection, the cells were treated for another 24 h without and with R5020 (10 nM). LUC activity was measured and calculated as the relative activity as for panel A. Values are averages from seven independent experiments. (C and D) COS1 cells were cotransfected with VDR expression plasmid (1 ng/well), increasing amounts of pHMG-1, or empty control vector (4- to 400-fold excess over VDR) and VDRE-1ΔMTV-LUC (200 ng/well) or DR3-tk-LUC (200 ng/well) reporter plasmids. Cells were treated for the last 24 h prior to harvest with 10 mM 1,25-(OH)2 D3, and luciferase was assayed and calculated as the relative activity as for panels A and B. Within the same transfections, the effect of pHMG-1 on the PR-mediated activation of PRE2-tk-LUC was assessed as for panel B. The values are averages from four independent experiments for VDRE-1ΔMTV-LUC and from two independent experiments for DR3-tk-LUC.
DISCUSSION
In previous studies we showed that HMG-1 markedly and specifically increased the binding affinity of PR for target DNA in vitro (42, 46). We now show by transient-cotransfection assays that HMG-1 and the closely related HMG-2 can also stimulate the transcriptional activity of PR in mammalian cells. This increase in PR activity was observed in two different cell types and with two different target promoters, suggesting a generality to the in vivo functional interaction between HMG-1/-2 and PR. The major effect of ectopically expressed HMG-1 and HMG-2 was to increase the transcriptional activity of the receptor-hormone complex without affecting the basal promoter activity of the reporter gene (Fig. (Fig.8A).8A). It also appears that coexpression of HMG-1/-2 directly stimulates PR-mediated transcription as opposed to altering the cellular levels of expressed PR or the copy number of the reporter gene plasmid. As determined by Western blots and whole-cell-binding assays, cotransfection of COS1 cells with pHMG-1 and phPR-B did not increase PR expression levels, nor did it increase expression of the β-Gal reporter gene used as the internal control for assessing transfection efficiency (data not shown). In a more direct analysis of the question of whether HMG-1 can affect stability or replication of transfected plasmids, Aizawa et al. (2) reported that HMG-1 coexpression did not increase the copy number of simian virus 40 (SV40) enhancer-controlled β-Gal reporter plasmid. HMG-1/-2 are ubiquitous proteins expressed in much higher amounts in the cell (7) than are the steroid receptors, which raises the question of how cotransfected pHMG-1/-2 can stimulate PR activity on a cellular background of endogenous HMG-1/-2. One possible explanation is that free HMG-1/-2 may be a limiting factor under the conditions of our transfection assays. It has been estimated that there is one molecule of HMG-1/-2 per 10 to 15 nucleosomes. Because HMG-1/-2 is actively engaged in multiple transactions (7), introducing exogenous HMG-1/-2 may increase the cellular pool of free protein available to interact with the PR-DNA complex. In COS1 cells transfected with pHMG-1, a two- to threefold increase in the levels of HMG-1 protein was detected by Western blot (data not shown). This may actually represent a six- to eightfold increase, since an average of 30% of the cells were transfected by the method used in this study. Alternatively, in transient transfections in which proteins are overexpressed on a per-cell basis, receptors may require increased cellular levels of HMG proteins to achieve maximal activity.
An additional finding of this study was that HMG-1/-2 functionally interact in vitro and in vivo with all steroid hormone receptors examined but not with certain other classes of nuclear receptors. HMG-1 and HMG-2 stimulated the sequence-specific DNA binding activity of highly purified preparations of PR, ER, GR, and AR, but they had no effect on DNA binding by purified VDR, RARγ, or RXRα. As previously established, RXR is required for high-affinity DNA binding of VDR and RAR through a heterodimerization mechanism by which RXR directly contacts one half-site of a DR element (29, 34, 63, 72, 74). We also found that HMG-1/-2 had no effect on the DNA binding activities of VDR-RXR and RAR-RXR heterodimers. In transient transfection assays, HMG-1 overexpression had a marked stimulatory effect on the transcriptional activity of three different steroid receptors (PR, AR, and GR), but it had minimal or no effect on the activity of VDR in vivo (Fig. (Fig.9B9B to D). Since completion of our studies, Verrier et al. (65) reported that HMG-1 stimulated binding of partially purified recombinant ER to an ERE and that ER-dependent transcription in vitro was enhanced by the combined effects of the TATA-binding protein-associated factor TAFII30 and HMG-1. Bianchi and coworkers (73) examined the influence of HMG-1/-2 on RARα; this was done as part of a study on the interaction of HMG-1 with HOX proteins. RARα-mediated transcription of a retinoic acid response element-controlled promoter was reported to be unaffected by the coexpression of HMG-1 under conditions in which HMG-1 substantially increased the transcriptional activity of HOX9D (73). Thus, a good correlation has been observed between the ability of HMG-1/-2 to stimulate sequence-specific DNA binding activity of nuclear receptors in vitro and their ability to increase the transcription activity of receptors in vivo. These results, when taken together, support the notion that HMG-1 and HMG-2 are coregulatory proteins that positively modulate the DNA binding and transcriptional activity of the steroid class of receptors but not certain other members of the nuclear receptor family.
Whether the closely related HMG-1 and HMG-2 are redundant gene products or whether they have distinct functional properties is not known. With respect to stimulating DNA binding and the transcriptional activity of steroid receptors, HMG-1 and HMG-2 were found to be interchangeable (Fig. (Fig.33 and and8C),8C), with HMG-2 exhibiting a slightly higher activity in receptor-DNA binding assays. With other sequence-specific transcriptional activators, it also appears that HMG-1 and HMG-2 are functionally interchangeable. Both HMG-1 and HMG-2 were reported to enhance the sequence-specific DNA binding affinity and transcriptional activity of the octamer transcription factors (73), MLTF/USF (60, 67), and p53 (25). Additionally, HMG-1 and HMG-2 were reported to be functionally equivalent as factors required for activator initiation of purified TFIID-TFIIA complexes in vitro (54). At least one study, however, has suggested that HMG-1 and HMG-2 have different functions. When mammalian cells were stably transfected to overproduce HMG-1 or HMG-2, only HMG-1 was observed to stimulate transcription from reporter plasmids driven by the SV40 promoter, while HMG-2 overexpression was associated with increased proliferation (2, 40). However, it is difficult to conclude from these studies whether HMG-1 and HMG-2 have different effects on activator-mediated transcription, since SV40 reporter plasmids are constitutively active.
Although HMG-1/-2 are ubiquitous proteins and have been implicated as regulatory cofactors for several different classes of transcriptional activators, it appears they are not cofactors for all families of sequence-specific transcription factors. The studies presented here indicate that HMG-1/-2 selectively stimulate only the activity of the steroid class of nuclear receptors and not that of other nonsteroid nuclear receptors. Additionally, disruption of the yeast HMG-like proteins (NHP6A and NHP6B) was reported to reduce or abolish only a subset of the yeast transcriptional activators examined (44). The rule for which transcriptional activators are positively modulated by HMG-1/-2 is unknown. One possibility for why HMG-1/-2 stimulated the activity of steroid receptors but did not affect the nonsteroid nuclear receptors tested in this study is suggested by the fundamental differences in the structure of the DBDs of the two subgroups of nuclear receptors and how they recognize DNA. The core DBDs of GR and ER contain two α helices, a recognition helix that makes basic specific contacts in the major groove of target DNA and a second helix that is positioned above DNA perpendicular to the recognition helix (52, 75). The core DBDs of RXR and TR have the same two perpendicular α helices folded into a single domain but also contain a third α helix missing in the steroid receptors that is located immediately C terminal to the second zinc binding module (33, 48). From molecular modeling and biochemical studies with wild-type and mutant RXR DBDs, helix-3 is predicted to make contacts in the minor groove, providing additional interactive surfaces required for high-affinity binding to DNA (33). In the crystal structure of the TR-RXR heterodimer complexed to a DR-4 element, helix-3 from TR projects across the minor groove to make extensive interactions over the half-site and spacer nucleotides (48). Thus, as a result of helix-3 interaction with the minor groove, the TR DBD appears to make more-extensive DNA contacts than do the DBDs of ER and GR (33, 48). Because HMG-1/-2 are known to bind to the minor groove, it is tempting to speculate that HMG-1/-2 association with the steroid receptor-DNA complex may functionally substitute for helix-3 in the C-terminal extension. The presence of helix-3 may sterically interfere with the ability of HMG-1/-2 to interact with the DBDs of nonsteroid nuclear receptors and may explain the lack of effect of HMG-1/-2 on VDR-RXR and RAR-RXR heterodimer-DNA binding. In support of this idea, HMG-1/-2 can stimulate the DNA binding activity of the expressed DBDs of ER (50) and PR (15a), indicating that the DBD is the minimal region capable of functional interaction with HMG. Testing this hypothesis more directly will require an analysis of the effects of HMG-1/-2 on the DBDs of nonsteroid receptors bearing deletions or other structure-disrupting mutations of helix-3 and an analysis of chimeras containing the core DBDs of steroid receptors linked to the C-terminal extensions from the DBDs of nonsteroid receptors.
The mechanism by which HMG-1/-2 stimulate the DNA binding affinity of steroid receptors in vitro and transcriptional activity in vivo remains unknown. Based on the ability of HMG-1/-2 to recognize and manipulate DNA structure, two general models can be considered. One possibility is that the optimal DNA binding site for steroid receptors is a combination of a nucleotide recognition sequence and a specific DNA structure. Acting as a DNA chaperone, HMG-1/-2 could transiently manipulate the structure of target DNA and hold it in an energetically unfavorable position until the receptor is stably bound (20, 70). By this mechanism, HMG-1/-2 would not be required as a component of the final receptor-DNA complex since it would dissociate upon stable receptor binding. This “hit and run” mechanism predicts an ordered binding with HMG-1/-2 interacting with PREs prior to PR binding. A second possibility is that HMG-1/-2 are recruited into the receptor-DNA complex, resulting in additional protein-DNA and/or protein-protein contacts. The present results are most consistent with this second mechanism. HMG-1/-2 themselves have no detectable binding for PREs in the absence of receptor, even at much higher concentrations than are required to enhance steroid receptor-DNA binding. HMG-1 association with PREs was detected only in the presence of PR, suggesting that PR recruits HMG into the complex. The mechanism responsible for HMG recruitment into the complex is unknown. The transient protein-protein interaction observed between HMG and PR in the absence of DNA is probably not sufficient. However, this weak interaction may be meaningful since HMG-1/-2 did not interact with two nonsteroid nuclear receptors, VDR (Fig. (Fig.7)7) and RAR (data not shown), that are unaffected functionally by HMG-1/-2. Using circular permutation and phasing gel mobility shift assays, we previously reported that purified PR alone, at high enough concentrations to bind to PREs with low affinity, can mediate a significant bend in DNA over the PRE toward the major groove (47). The addition of HMG-1 had minimal influence on the magnitude or direction of PR-mediated DNA bending (47). These results indicate that DNA bending is an intrinsic property of the receptor that does not require HMG-1/-2. Since HMG-1 has a substantially greater affinity for bent DNA than for linear DNA (7, 42), it is tempting to speculate that HMG-1/-2 are recruited into the receptor-DNA complex by the combined effects of transient protein-protein interaction and receptor-mediated bending of target DNA.
Whether the observed stimulatory effect of HMG-1/-2 on steroid receptor transcriptional activity in vivo is due solely to increasing receptor-DNA interaction is not known. As an architectural cofactor recruited to the receptor-DNA complex, HMG-1/-2 could conceivably potentiate the transcriptional activity of the receptor by altering or stabilizing the conformation of DNA to help establish the three-dimensional structure at the promoter required for assembly of higher-order nucleoprotein complexes. HMG-1/-2 recruited to the receptor-DNA complex could also contribute to modification of the chromatin structure that is associated with activated gene transcription. Indeed, HMG-1 and HMG-2 have been reported to preferentially associate with regions of transcriptionally active chromatin (7). A family of novel steroid receptor coactivators have been identified that directly interact with transcriptional activation domains of receptor and are essential for receptor activation of target promoters. These coactivators are thought to function as transcriptional signaling intermediates by coupling receptors with the general transcriptional apparatus through protein-protein interaction (22, 26, 27, 43, 57, 66). It is also conceivable that HMG-1/-2 could have a role in facilitating the assembly of steroid receptor coactivator complexes at the target gene promoters.
ACKNOWLEDGMENTS
The first three authors made equal contributions to this work.
This work was supported in part by the Tissue Culture/Monoclonal Antibody Core facility of the University of Colorado Cancer Center (Cancer Center Core grant P30 CA46934) and by Public Health Service grant NCI-CA 46938 (D.P.E.).
We thank Kurt Christensen, Lori Sherman, Suzanne Meintzer, and Nicole Flessner for expert technical assistance and Elizabeth Wilson, University of North Carolina at Chapel Hill, for providing baculovirus AR expression vectors and for helpful suggestions and critical reading of the manuscript.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.18.8.4471
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc109033?pdf=render
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/18/8/4471
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/content/full/18/8/4471
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/18/8/4471
Citations & impact
Impact metrics
Article citations
An i-motif-regulated enhancer, eRNA and adjacent lncRNA affect Lhb expression through distinct mechanisms in a sex-specific context.
Cell Mol Life Sci, 81(1):361, 19 Aug 2024
Cited by: 0 articles | PMID: 39158745 | PMCID: PMC11335282
Testosterone Exacerbates the Formation of Liver Cancer Induced by Environmental N-Nitrosamines Exposure: Potential Mechanisms and Implications for Human Health.
Onco Targets Ther, 17:395-409, 17 May 2024
Cited by: 0 articles | PMID: 38774818 | PMCID: PMC11107913
HMGB2 is a potential diagnostic marker and therapeutic target for liver fibrosis and cirrhosis.
Hepatol Commun, 7(11):e0299, 06 Nov 2023
Cited by: 1 article | PMID: 37930124 | PMCID: PMC10629741
HMGB2 upregulation promotes the progression of hepatocellular carcinoma cells through the activation of ZEB1/vimentin axis.
J Gastrointest Oncol, 14(5):2178-2191, 24 Oct 2023
Cited by: 1 article | PMID: 37969822 | PMCID: PMC10643579
Structure and Functions of HMGB2 Protein.
Int J Mol Sci, 24(9):8334, 05 May 2023
Cited by: 12 articles | PMID: 37176041 | PMCID: PMC10179549
Review Free full text in Europe PMC
Go to all (199) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences.
Mol Cell Biol, 14(5):3376-3391, 01 May 1994
Cited by: 114 articles | PMID: 8164686 | PMCID: PMC358703
Thyroid hormone receptor does not heterodimerize with the vitamin D receptor but represses vitamin D receptor-mediated transactivation.
Mol Endocrinol, 12(9):1367-1379, 01 Sep 1998
Cited by: 13 articles | PMID: 9731705
The interaction of the vitamin D receptor with nuclear receptor corepressors and coactivators.
Biochem Biophys Res Commun, 253(2):358-363, 01 Dec 1998
Cited by: 57 articles | PMID: 9878542
The vitamin D hormone and its nuclear receptor: molecular actions and disease states.
J Endocrinol, 154 Suppl:S57-73, 01 Sep 1997
Cited by: 137 articles | PMID: 9379138
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01 CA046938
Grant ID: P30 CA046934
Grant ID: P30 CA46934
Grant ID: CA 46938





