Abstract
Free full text

Open Reading Frame 1a-Encoded Subunits of the Arterivirus Replicase Induce Endoplasmic Reticulum-Derived Double-Membrane Vesicles Which Carry the Viral Replication Complex
Abstract
The replicase of equine arteritis virus (EAV; family Arteriviridae, order Nidovirales) is expressed in the form of two polyproteins (the open reading frame 1a [ORF1a] and ORF1ab proteins). Three viral proteases cleave these precursors into 12 nonstructural proteins, which direct both genome replication and subgenomic mRNA transcription. Immunofluorescence assays showed that most EAV replicase subunits localize to membranes in the perinuclear region of the infected cell. Using replicase-specific antibodies and cryoimmunoelectron microscopy, unusual double-membrane vesicles (DMVs) were identified as the probable site of EAV RNA synthesis. These DMVs were previously observed in cells infected with different arteriviruses but were never implicated in viral RNA synthesis. Extensive electron microscopic analysis showed that they appear to be derived from paired endoplasmic reticulum membranes and that they are most likely formed by protrusion and detachment of vesicular structures with a double membrane. Interestingly, very similar membrane rearrangements were observed upon expression of ORF1a-encoded replicase subunits nsp2 to nsp7 from an alphavirus-based expression vector. Apparently, the formation of a membrane-bound scaffold for the replication complex is a distinct step in the arterivirus life cycle, which is directed by the ORF1a protein and does not depend on other viral proteins and/or EAV-specific RNA synthesis.
Equine arteritis virus (EAV) (20) is the prototype of the family Arteriviridae, a novel family of positive-stranded RNA viruses (for reviews, see references 35 and 45) which also includes lactate dehydrogenase-elevating virus, porcine reproductive and respiratory syndrome virus, and simian hemorrhagic fever virus. Together with the coronaviruses, the arteriviruses belong to the recently established order Nidovirales (10). Despite remarkable differences in virion architecture and genome size (13 to 16 kb for arteriviruses against 27 to 32 kb for coronaviruses), the two virus families were united on the basis of their striking similarities in polycistronic genome organization and expression strategy. The latter includes the discontinuous transcription of a nested set of subgenomic mRNAs to express the mostly structural genes in the 3′ end of the genome (10, 17, 46). Comparative sequence analysis strongly suggested that the genes encoding the arterivirus and coronavirus replicases are related by common ancestry (15, 46).
As in all nidoviruses, the EAV replicase gene is comprised of two large open reading frames (ORFs), ORF1a and -1b, which are both expressed from the genomic RNA. Following ORF1a translation, a −1 ribosomal frameshift into ORF1b occurs with an estimated efficiency of 15 to 20% (15). This expression strategy leads to the generation of two multidomain precursor proteins: the 1,727-amino-acid ORF1a protein and the 3,175-amino acid ORF1ab protein. Both polyproteins are cleaved extensively by three ORF1a-encoded proteases (47, 49, 50). Our current understanding of EAV replicase processing is summarized in Fig. Fig.1.1. The ORF1a protein can be cleaved at seven sites (48, 50, 61), yielding a number of processing intermediates and eight end products, nonstructural protein 1 (nsp1) to nsp8. The N-terminal cleavage products nsp1 and nsp2 are rapidly liberated by internal cysteine autoprotease activities (47, 49). The remaining nsp3-8 intermediate (96 kDa) is processed by the nsp4 serine protease (SP) (50, 61), which also cleaves the ORF1b-encoded polyprotein three times (58, 59). The latter part of the replicase contains a set of highly conserved functions which are essential for viral RNA replication and mRNA transcription (15, 57). Among its processing products (nsp9 to nsp12) are the replicase subunits that carry the putative viral RNA polymerase (nsp9) and helicase (nsp10) activities (58, 59).
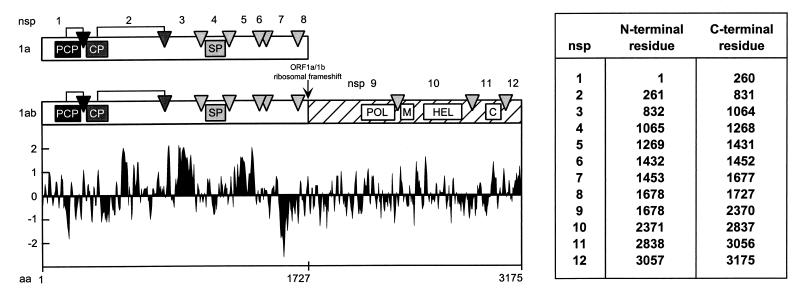
Proteolytic processing scheme, hydrophobicity plot, and subunit nomenclature of the EAV ORF1a and ORF1ab replicase polyproteins (48, 59, 61). The three EAV protease domains (papainlike cysteine protease [PCP], cysteine protease [CP], and SP) and their cleavage sites (arrows and arrowheads) are shown. In the ORF1b-encoded polypeptide (hatched), the four major domains conserved in nidoviruses are depicted: POL, putative RNA-dependent RNA polymerase; M, putative metal-binding domain; HEL, putative RNA helicase; C, conserved C-terminal domain specific for nidoviruses. The hydrophobicity plot was generated by the method of Kyte and Doolittle (29). Values above the axis indicate hydrophobicity. The table specifies the N- and C-terminal residues of each of the EAV nonstructural proteins.
Immunofluorescence studies have revealed that most ORF1a- and ORF1b-encoded replicase subunits localize to the perinuclear region of EAV-infected cells, suggesting association with intracellular membranes (56, 59). Double-label experiments showed a substantial overlap with compartments (the endoplasmic reticulum [ER] and/or the intermediate compartment [IC]) containing the cellular enzyme protein disulfide isomerase (PDI). Furthermore, metabolic RNA labeling experiments using bromouridine triphosphate (BrUTP) revealed that viral RNA synthesis colocalizes with the membrane-bound complex in which the replicase subunits accumulate. Since the ORF1b-encoded part of the replicase does not contain domains with significant hydrophobicity, ORF1a-derived cleavage products which do contain such regions (in particular, nsp2, nsp3, and nsp5) were proposed to mediate the membrane association of the arterivirus replication complex (48, 61). Biochemical analyses confirmed that a number of ORF1a-encoded replicase subunits are indeed tightly associated with membranes and behave as integral membrane proteins (22, 56).
In order to extend our characterization of the arterivirus replication complex to the ultrastructural level, we have used electron microscopy (EM) to analyze EAV-infected cells after conventional Epon embedding or cryoimmunogold labeling. Our data suggest that EAV RNA synthesis is attached to virus-induced double-membrane structures, which were previously observed in cells infected with different arteriviruses (8, 36, 52, 62, 64). Interestingly, very similar structures were induced upon expression of ORF1a-encoded replicase subunits nsp2-7 from a heterologous expression vector. This strongly suggests that the formation of a membrane-bound scaffold for the arterivirus replication complex is an important function of these ORF1a-encoded subunits.
MATERIALS AND METHODS
Cells, viruses, and BrUTP labeling.
Baby hamster kidney (BHK-21) and rabbit kidney (RK-13) cells were used for infection experiments with the EAV Bucyrus strain (20) by the protocol described by de Vries et al. (16). The newly synthesized viral RNA in infected BHK-21 cells was labeled by using BrUTP from 6.5 to 7.5 h postinfection (p.i.) as described previously (56).
Sindbis virus expression system.
The construction of Sindbis virus expression vector pSinEAV(261-1677)His, which expresses a C-terminally hexahistidine-tagged version of nsp2-7, was described by Wassenaar et al. (61). As before (61), recombinant virus particles [vSinEAV(261-1677)His] were used for high-multiplicity-of-infection (MOI) experiments with BHK-21 cells, which were fixed and processed for immunofluorescence assays or EM between 6 and 9 h p.i. A control Sindbis virus expression vector for the green fluorescent protein (pSinRep/GFP) was kindly provided by C. M. Rice, Washington University, St. Louis, Mo.
Antisera and lectins.
The EAV replicase-specific rabbit antisera used in this study have been described previously (48, 59), with the exception of a novel anti-nsp3 rabbit serum. The latter was raised by using a bovine serum albumin-coupled synthetic peptide with the sequence Tyr-Val-Thr-Gly-Thr-Thr-Arg-Leu-Tyr-Ile-Pro-Lys-Glu-Gly-Gly-Met-Val-Phe-Glu (corresponding to residues 1046 to 1064 of the EAV ORF1a protein, the C-terminal 19 residues of nsp3 [50]) and following the immunization protocol described before (48). Due to the recent revision of EAV nonstructural protein nomenclature (61), the names of some antisera have been adapted to correspond to the number of the replicase cleavage product which they recognize (56). Mouse monoclonal antibodies (MAbs) were used to visualize the localization of EAV ORF5-encoded glycoprotein GL (MAb 93B [24]) and the cellular enzyme PDI (MAb 1D3 [60]). An anti-histidine tag MAb (MAb 13/45/31 [66]) was used to detect hexahistidine-tagged expression products. As before (56), an antibromodeoxyuridine rat MAb (BU1/75 [ICR1]; Harlan Sera-Lab Ltd., Loughborough, England) was used to visualize BrUTP-labeled viral RNA. Biotinylated wheat germ agglutinin (WGA) and concanavalin A (ConA) were obtained from Sigma Chemical Co. (St. Louis, Mo.). The lectins were detected by using a mouse MAb to biotin (Sigma), followed by a rabbit anti-mouse immunoglobulin G antibody and a protein A-gold conjugate.
Immunofluorescence microscopy.
Cells were grown on coverslips, infected with EAV or vSinEAV(261-1677)His particles, and paraformaldehyde fixed as described by van der Meer et al. (56). Indirect immunofluorescence assays were carried out as described previously (56), and samples were examined by using an Olympus fluorescence microscope.
EM.
For conventional Epon embedding and sectioning, BHK-21 or RK-13 cells infected with EAV or recombinant Sindbis virus particles were fixed for 1 h in 1% glutaraldehyde in 200 mM cacodylate buffer (pH 7.4), washed repeatedly in aqua destillata, and incubated for 1 h in cacodylate buffer containing 1% OsO4 and 1.5% K3Fe(CN)6. Following two subsequent 30-min incubations in 1% tannic acid and 1.5% magnesium uranyl acetate, the samples were dehydrated by using ethanol and embedded in Epon. Ultrathin sections were prepared and stained with lead citrate.
For cryoimmuno-EM, infected BHK-21 or RK-13 cells were fixed by using 4% paraformaldehyde and 0.1% glutaraldehyde in 200 mM HEPES, pH 7.4. Cells were scraped from the dish, pelleted, and incubated in 2.3 M sucrose for 1 h at room temperature. Subsequently, cell pellets were mounted on silver pins and then flash frozen and stored in liquid nitrogen. The specimens were sectioned with a Reichert Ultracut S ultramicrotome with a Reichert FCS cryoattachment using a Drukker International diamond knife. Immunocytochemical labeling of thawed cryosections was performed essentially as described by Griffiths et al. (26). EM specimens were examined in a JEOL 1200EX or a Philips CM100 transmission electron microscope.
RESULTS
EAV nonstructural proteins and RNA synthesis localize to DMVs.
On the basis of immunofluorescence microscopy studies (56, 59), we have previously concluded that both EAV nonstructural proteins and viral RNA synthesis are associated with membranes which localize mainly to the perinuclear region of infected cells. To extend our analysis to the ultrastructural level, we employed cryoimmuno-EM of EAV-infected BHK-21 and RK-13 cells at various stages of infection. The results obtained with both cell lines were very similar.
A first cryo-EM analysis revealed the presence of unusual vesicular structures with a double membrane (Fig. (Fig.2)2) which were concentrated mainly in the perinuclear region of EAV-infected cells. The facts that the number of these double-membrane vesicles (DMVs) increased during the course of infection and that they were not found in uninfected cells (data not shown) indicated that the DMVs are induced by arterivirus infection, as previously concluded by others (8, 36, 52, 62, 64). Interestingly, immunogold labeling experiments revealed that the DMVs in EAV-infected cells could be labeled by using our EAV replicase antisera (see Materials and Methods). Not very abundant but highly specific labeling was obtained with rabbit antisera recognizing the replicase cleavage products nsp2 (Fig. (Fig.2a),2a), nsp3 (Fig. (Fig.2b),2b), nsp4, nsp7-8, nsp9, and nsp10 (data not shown).
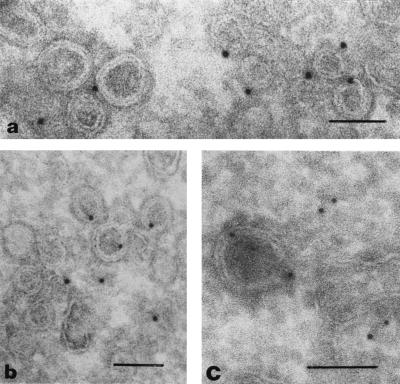
EAV replicase subunits and EAV RNA synthesis localize to DMV structures. (a and b) Cryosections of EAV-infected RK-13 cells (8 h p.i.) were labeled with rabbit anti-nsp2 (a) or anti-nsp3 (b) serum, followed by 10-nm protein A-gold. Specific but not very abundant labeling of the DMVs was observed. (c) Cryosection of EAV-infected BHK-21 cell (7.5 h p.i.) in which viral RNA synthesis had been labeled by using BrUTP in the presence of dactinomycin (see Materials and Methods). BrUTP-labeled RNA was detected by using an anti-BrUTP rat MAb, followed by 5-nm protein A-gold. Labeling of both DMVs and cytoplasm was observed. Bars, 100 nm.
Next, viral RNA synthesis in EAV-infected cells was metabolically labeled by using BrUTP in the presence of dactinomycin as described previously (56). By using a BrUTP-specific MAb, the localization of de novo-generated EAV RNA was analyzed. A substantial part of the label was found to be associated with the DMVs (Fig. (Fig.2c),2c), although staining of the cytoplasm was also observed. The latter observation was not surprising, since a part of the viral transcripts, e.g., the subgenomic mRNAs encoding the viral envelope and nucleocapsid proteins, can be expected to be released from the replication-transcription complex to be translated elsewhere in the cytoplasm.
Taken together, our first cryoimmuno-EM data strongly suggested that the EAV replication complex is associated with the unusual double-membrane structures which are induced upon infection. To characterize the generation of the DMVs in more detail, we used conventional Epon embedding and sectioning of EAV-infected cells, focusing in particular on the earlier stages of infection, during which general cytopathic effects are minimal and virus assembly cannot yet be detected.
EAV-induced DMVs appear to be derived from the ER.
To study the intracellular origin and ultrastructure of EAV-induced DMVs, we examined single and serial sections of Epon-embedded BHK-21 and RK-13 cells at 4, 8, and 12 h p.i. For comparison, an immunofluorescence analysis showing the development of EAV replicase staining at the light microscopic level is shown in Fig. Fig.3.3. BHK-21 cells were double labeled for EAV nsp2 and the cellular marker PDI, which is a resident luminal protein of the ER and IC. The first signal was a punctate labeling of the perinuclear region which was observed around 4 h p.i. Subsequently, dense replicase staining in a large area around the nucleus developed, which substantially overlapped with the region of the cell that stained for PDI.
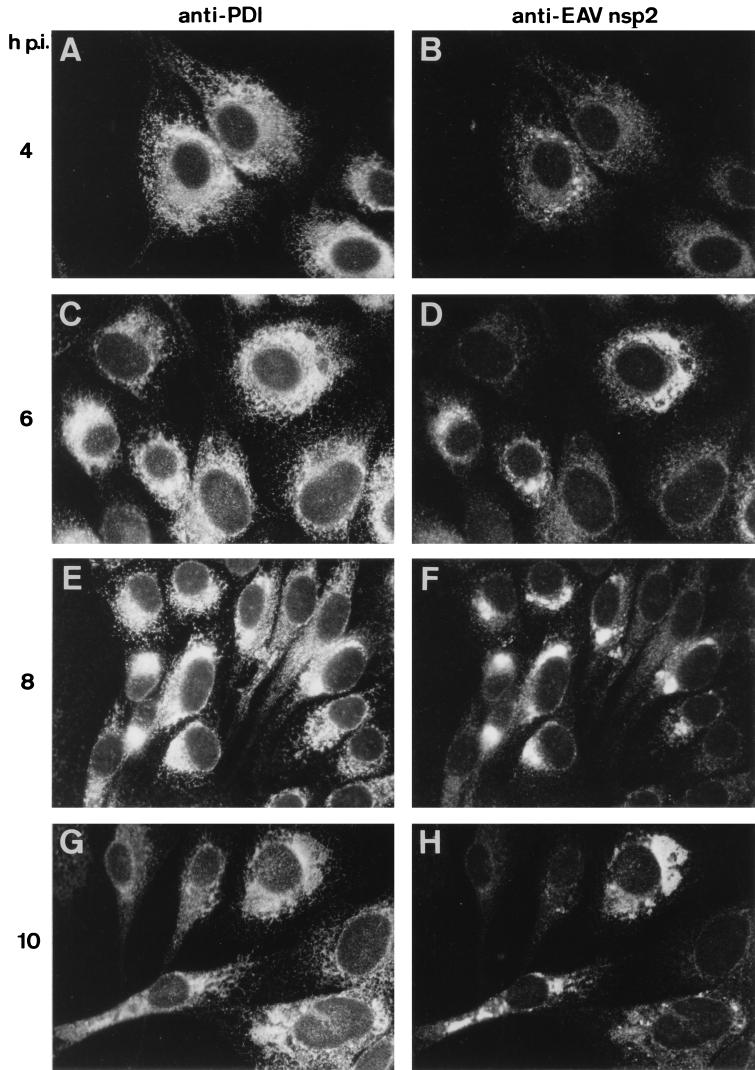
Immunofluorescence double labeling showing the intracellular distribution of the ER-IC marker PDI (A, C, E, and G) and the EAV replicase (B, D, F, and H) in EAV-infected BHK-21 cells. Cells were fixed at 4 (A and B), 6 (C and D), 8 (E and F), or 10 (G and H) h p.i. Subsequently, cells were processed for indirect double-immunofluorescence analysis using rabbit anti-nsp2 serum (48) and a mouse anti-PDI MAb (60). Photographs were generated by using the same exposure times for recording and printing.
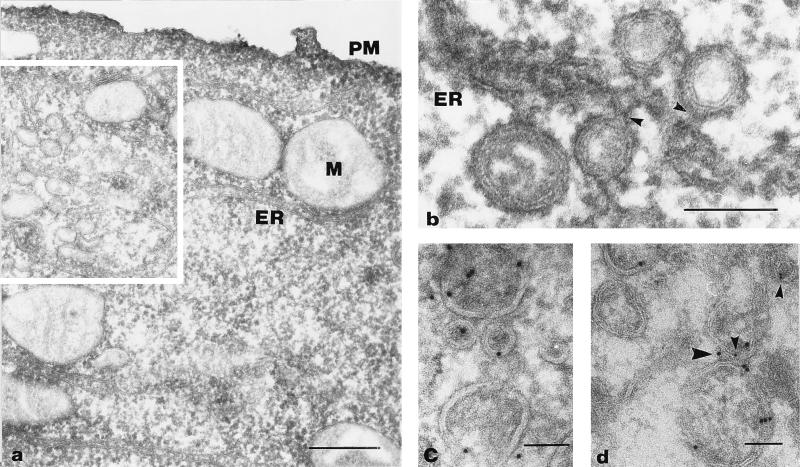
Our EM analysis (Fig. (Fig.4)4) revealed that at 4 and 8 h p.i., most of the cytoplasm did not show obvious signs of virus infection. DMVs surrounded by ER membranes were found mainly in the perinuclear region. The region where DMV formation was taking place also contained closely apposed ER membranes. The DMVs were approximately 80 nm in diameter, and in general, the inner and outer DMV membranes were tightly apposed. The electron density of the interior of most DMVs was similar to that of the cytoplasm. However, in some DMVs, a more electron-lucent interior was seen. Interestingly, a gap in the double membrane could be observed in some of the DMVs (Fig. (Fig.4b),4b), showing that the inner and outer DMV membranes had not (or not yet) been sealed. In other DMVs, the outer membrane was continuous with the membrane of a cellular compartment, most likely the ER (Fig. (Fig.4b).4b). However, many DMVs appeared to be closed and to have fully separated inner and outer membranes (Fig. (Fig.4c).4c). Figure Figure4d4d shows an example of a DMV which seemed to be forming from ER membranes. The outer membrane is clearly continuous with the ER, whereas the inner membrane appears to be fully separated. This electron micrograph suggests that, prior to DMV formation, the ER membranes become closely associated and form an electron-dense neck-like structure. Thus, our collective morphological observations strongly suggested that the EAV-induced DMVs originate from ER membranes.
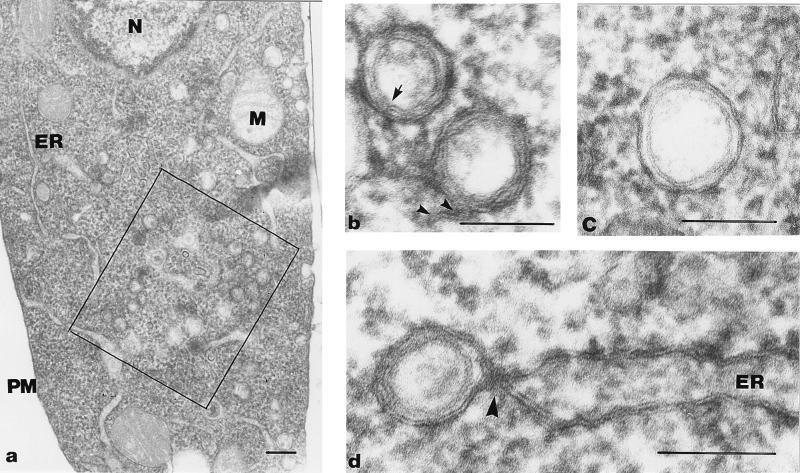
Formation of DMVs from paired ER membranes. Epon sections of EAV-infected BHK-21 cells at 4 (b and d) or 8 (a and c) h p.i. (a) Overview of an EAV-infected cell. The square indicates a region containing many DMVs surrounded by ER membranes. (b and c) DMVs at higher magnification. In panel b, the arrow indicates a region of the DMV where the inner and outer membranes are continuous, thereby creating a connection between the DMV interior and the cytoplasm. The two arrowheads show that the outer membrane of the other DMV is continuous with an intracellular membrane. (c) Image of a typical DMV with apparently sealed inner and outer membranes. (d) Possible intermediate in DMV formation. The DMV appears to arise by protrusion of paired ER membranes. An electron-dense, neck-like connection with the ER is clearly visible (arrowhead). The inner membrane of the DMV is separated, while the outer membrane is continuous with that of the ER. N, nucleus; M, mitochondrion; PM, plasma membrane. Bars, 1 μm (a) or 100 nm (b, c, and d).
During the course of infection, the ER membranes in infected cells appeared more irregular in shape and an increasing part became modified into DMVs or DMV-like structures. At 8 h p.i., the DMVs were less uniform in size than at 4 h p.i., but they were still spherical and concentrated mainly in the region surrounding the nucleus. At this stage of infection, part of the Golgi stacks were somewhat dilated. At the latest time point studied (12 h p.i.), the cells showed severe cytopathic effects and well-developed Golgi stacks were rare. Most ER membranes were closely apposed, and DMVs were spread throughout the cell. Furthermore, the size and shape of the DMVs were less uniform and their internal electron density was variable: whereas some DMVs contained an electron-dense matrix, others appeared to be partially or completely empty.
To analyze in more detail the DMV distribution in the cell and the relationship with the ER membranes, we prepared serial sections (thickness, approximately 50 nm) of BHK-21 cells at different stages of EAV infection (data not shown). These experiments confirmed the concentration of DMVs in the perinuclear region, in particular during the early stages of infection. The vesicular nature of the DMVs appeared to be corroborated by the fact that the same DMV was generally seen in only two (or sometimes three) consecutive sections, which rules out the presence of, e.g., long, tubular structures with a double membrane. DMVs with a gap in the outer membrane, which could be explained by a connection with the ER as shown in Fig. Fig.4d,4d, were rare. These observations strongly suggest that the DMVs are not permanently connected with ER membranes and that the images in which a neck-like connection with the ER was seen are likely to represent an intermediate stage of DMV formation.
nsp2 to nsp7 localize to the perinuclear region in the absence of EAV replication.
We have recently reported that a number of ORF1a-encoded replicase subunits (in particular, nsp2, nsp3, and nsp5) which contain hydrophobic domains behave as integral membrane proteins in biochemical studies (56). Thus, we wanted to investigate whether these viral proteins are directly involved in DMV generation in EAV-infected cells. To this end, we used previously described alphavirus-based expression vector pSinEAV(261-1677)His (61), which allows the expression of a hexahistidine-tagged version of EAV nsp2-7 in the absence of EAV replication. The pSinRep expression system (7) is based on the infectious cDNA clone of the unrelated alphavirus Sindbis virus (SIN) and allows high-level cytoplasmic expression of heterologous proteins from the SIN subgenomic mRNA promoter. Furthermore, it is possible to package SIN RNA replicons into recombinant SIN particles which can be used for high-MOI infections of BHK-21 cells. Vector pSinEAV(261-1677)His was previously used to express, purify, and sequence replicase cleavage products encoded by the 3′-terminal part of ORF1a (61). The processing of the EAV nsp2-7His polyprotein expressed from this vector was similar to that in EAV-infected cells, indicating that the internally located EAV nsp2 and nsp4 protease domains were functional (61).
Obviously, the use of another positive-stranded RNA virus as an expression vector for EAV replicase proteins was a potential complication. However, the alphavirus replication complex is associated with modified endosomes and/or lysosomes (23) and not—as in the case of EAV—with ER membranes. Thus, we assumed that SIN replication would not necessarily interfere with the functions of the EAV replicative proteins. This assumption was supported by double-infection experiments with wild-type EAV and SIN in BHK-21 cells (data not shown). The two viruses, when present in the same cell, did affect each other’s replication and/or genome expression to a certain extent, as concluded from the somewhat reduced intensity of the staining obtained with various antibodies. However, nonstructural and structural proteins of both EAV and SIN were readily detected in double-label immunofluorescence studies, and the subcellular localization of these proteins was not affected by the presence and replication of the other virus. Finally, we used pSinRep/GFP, which expresses the green fluorescent protein, and pSinRep/GFP-derived recombinant virus particles as a control in all experiments to rule out the possibility that the observed changes (see below) resulted from SIN replication instead of EAV nsp2-7 expression.
BHK-21 cells were infected with vSinEAV(261-1677)His or vSinRep/GFP particles at an MOI of 5 to 10. At 6 and 9 h p.i. (for SIN), cells were fixed and processed for immunofluorescence assays (Fig. (Fig.5).5). The subcellular localization of the EAV nsp2 to nsp7 expression products strikingly resembled that in EAV-infected cells (compare, e.g., Fig. Fig.5A5A with Fig. Fig.3D,3D, F, and H) and substantially overlapped with the area of the cells which stained for PDI (Fig. (Fig.5B).5B). Antisera recognizing nsp2, nsp3, nsp4, and nsp7 all gave identical results. The presence of the hexahistidine tag at the C terminus of nsp7 allowed us to perform double-label experiments using a rabbit antiserum recognizing the N terminus of nsp2 and an anti-His tag mouse MAb. The results confirmed that the cleavage products carrying these two epitopes, which are at the extreme N and C termini of the nsp2-7His expression product, respectively, colocalized completely (Fig. (Fig.5C5C and D).
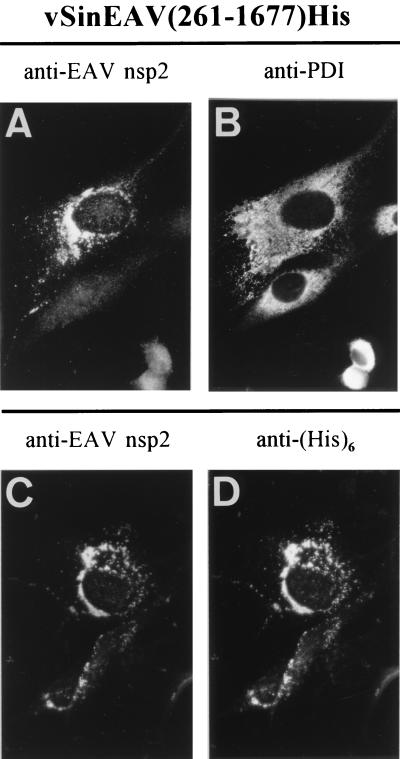
Immunofluorescence analysis of vSinEAV(261-1677)His-infected BHK-21 cells at 7 h p.i. (A and B) Double staining for EAV nsp2 (A) and the cellular enzyme PDI (B), a resident protein of the ER-IC. Note the striking resemblance to the nsp2 staining obtained with regular EAV-infected cells (Fig. (Fig.3).3). (C and D) Double staining for EAV nsp2 (C) and the hexahistidine tag (D), which was attached to the nsp7 C terminus of the vSinEAV(261-1677)His expression product (61).
Expression of EAV nsp2 to nsp7 induces DMVs.
The striking similarity between the subcellular localization of the EAV replication complex in infected cells and that of the pSinEAV(261-1677)His-derived expression products prompted us to extend our analysis to the EM level. BHK-21 cells were infected with vSinEAV(261-1677)His, fixed at 6 or 9 h p.i., and processed for both Epon embedding and cryosectioning. The Epon sections revealed the presence of closely apposed ER membranes and large numbers of DMVs at 6 h p.i. (Fig. (Fig.6a).6a). The DMVs were very similar to those observed upon EAV infection (compare, e.g., Fig. Fig.4).4). The remaining part of the cells seemed unaffected by the vSinEAV(261-1677)His infection. Three hours later, even more ER membranes were closely apposed and DMVs were dispersed throughout the cytoplasm. In general, the vSinEAV(261-1677)His-induced DMVs were more variable in size (up to 120 nm) and shape than those observed in EAV-infected cells. Figure Figure6b6b demonstrates the proximity of the DMVs to the closely apposed ER membranes in the perinuclear region. A connection between the DMV outer membrane and the ER can be observed, suggesting again that the DMV membranes originate from the ER. Closely apposed ER membranes and DMVs were not observed in control cells infected with vSinRep/GFP recombinant virus particles.
Formation of DMVs from ER membranes in vSinEAV(261-1677)His-infected BHK-21 cells. Epon sections (a and b) and cryosections (c and d) of cells fixed at 6 or 9 h p.i. were used. (a) Overview of an infected cell with the square indicating a region which shows abundant DMV formation and closely apposed ER membranes. (b) DMV formation from paired ER membranes. Note the electron-dense neck-like structures (small arrowheads) and the fact that the outer membrane of these DMVs is continuous with the ER membrane, whereas the inner membrane appears to be separated. In both models (see Discussion and Fig. Fig.8),8), the DMV structure would wrap around a part of the cytoplasm, while the narrow lumen between the two closely apposed DMV membranes would be equivalent to the ER lumen. (c) Cryoimmunolabeling of DMVs using anti-EAV nsp3 serum. (d) Double labeling for EAV nsp3 (large arrowhead, 10-nm gold) and PDI (small arrowheads, 5-nm gold), demonstrating that DMVs and closely apposed ER membranes contain both nsp3 and PDI. PM, plasma membrane; M, mitochondrion. Bars, 200 (a) or 100 (b, c, and d) nm.
Finally, we employed cryoimmuno-EM to prove the association of the EAV nonstructural expression products with DMVs. Figure Figure6c6c shows that both DMVs and closely apposed ER membranes were labeled for EAV nsp3. Similar observations were made for nsp2 and nsp4 (data not shown). The sections of vSinRep/GFP-infected cells were not labeled with any of the EAV replicase antisera (data not shown). Taken together, these data leave no doubt that DMV formation can be specifically induced by the expression of EAV nsp2-7, even in the absence of EAV RNA synthesis or ORF1b-encoded proteins. We postulate that these same replicase subunits are also responsible for the induction of the DMV structures which have been observed in cells infected with different arteriviruses.
DMV formation does not involve membranes of the Golgi complex.
An extended cryoimmuno-EM analysis of EAV- and vSinEAV(261-1677)His-infected BHK-21 cells was carried out to study DMV formation in more detail. The anti-PDI MAb was again used as an ER-IC marker, and a MAb specific for EAV glycoprotein GL (24, 56) was used as a marker for the Golgi complex. Furthermore, we used the lectins ConA and WGA to identify intracellular membrane compartments. Oligosaccharides on glycoproteins and glycolipids undergo sequential modifications during their transport from the ER through the Golgi complex to the trans-Golgi network, which are reflected in different specificities for lectins (25). ConA, which is specific for mannose, labels the ER (including the nuclear envelope), the entire Golgi complex, and the plasma membrane. WGA, on the other hand, binds to terminal N-acetylglucosamine and sialic acid and consequently labels only oligosaccharides which have been exposed to sugar transferases in the Golgi complex (37).
Figure Figure6d6d demonstrates the colocalization of PDI and nsp3 on vSinEAV(261-1677)His-induced DMVs. In single-labeling experiments with these antibodies, we observed one or two gold particles per DMV. DMVs containing both markers (PDI and nsp3) were less abundant than DMVs containing one of the markers. In EAV-infected cells, the ER membranes and the neck-like part of the presumed intermediates of DMV formation were labeled for ConA (Fig. (Fig.7a),7a), as were some of the DMVs to a limited extent (Fig. (Fig.7b).7b). At 12 h p.i., normal Golgi stacks (Fig. (Fig.7c)7c) were rare, and large cytoplasmic vacuoles were observed, which were extensively labeled for EAV GL (Fig. (Fig.7d).7d). DMVs (Fig. (Fig.7e),7e), ER membranes, and the neck-like structures were not labeled for WGA, whereas Golgi stacks and vacuoles (Fig. (Fig.7f)7f) and the plasma membrane were labeled abundantly. These observations suggested that these cytoplasmic vacuoles are dilated Golgi stacks which result from the effects of EAV infection. The DMVs were not labeled for the EAV GL protein, which, together with the results obtained with WGA, makes it unlikely that the DMV membranes are (partially) derived from the Golgi complex.
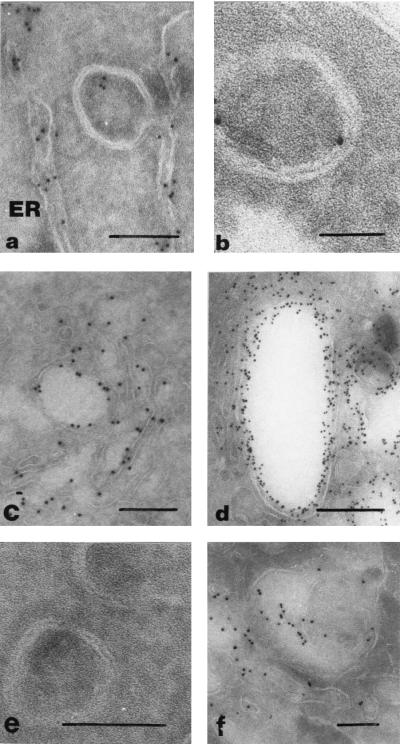
Cryoimmuno-EM analysis of EAV-infected BHK-21 cells (12 h p.i.) labeled by using ConA (a and b), an anti-EAV GL MAb (c and d), and WGA (e and f). (a) Possible intermediate in DMV formation. The ER lumen was extensively labeled by using ConA, whereas labeling was greatly reduced in the region where ER membranes are closely apposed. (b) DMV structure showing limited labeling with ConA. (c) Localization of the EAV GL protein in the Golgi apparatus. (d) Accumulation of the GL protein in vacuolar structures which are probably dilated Golgi stacks. (e) DMVs from WGA-labeled cryosections, showing the absence of labeling. (f) WGA labeling of (partially dilated) Golgi stacks in EAV-infected cells. Bars, 100 (a, e, and f), 50 (b), or 200 (c and d) nm.
DISCUSSION
Membrane association of the RNA synthesis of positive-stranded RNA viruses.
The RNA-synthesizing machinery of all of the eukaryotic positive-stranded RNA viruses studied to date is intimately associated with intracellular membranes (see, e.g., references 3 to 6, 9, 12, 13, 23, 27, 38, 41, 44, 55, 63, and 65 and references therein). The formation of a cytoplasmic complex consisting of viral RNA, viral replicative proteins, and host cell-derived membranes is one of the earliest steps in the viral life cycle. Furthermore, an increasing number of host cell proteins has been reported to be associated with positive-stranded RNA virus replication complexes (for a recent review, see reference 30). The reasons for the membrane association of viral RNA synthesis and the structure of the membrane-bound replication complexes are still poorly understood. The membranes may play a structural and/or functional role by offering a suitable microenvironment for viral RNA synthesis, or they may facilitate the recruitment of membrane-associated host cell proteins for the purpose of viral replication and transcription.
In many cases, positive-stranded RNA virus infection dramatically affects both the structure and function of the host cell membrane system. Different forms of membrane-bound vesicular structures and cytoplasmic vesicles, bound by either a single or a double membrane, are induced by members of different virus families. Alphaviruses (23, 27) and rubiviruses (31, 32) modify endosomes and lysosomes, the replication of bromoviruses and potyviruses is associated with the ER (38, 41), and the alfalfa mosaic virus replication complex is bound to chloroplast membranes (13). Paired ER membranes and vesicular structures are induced upon flavivirus infection and label for nonstructural proteins and double-stranded RNA (63). One of the most extensively studied positive-stranded RNA virus replication complexes is that of poliovirus, which is also associated with virus-induced vesicular structures (5, 12, 54). These structures were initially thought to be single-membrane vesicles, but by using more advanced (cryo)fixation protocols, Schlegel et al. (42) more recently demonstrated the presence of a double membrane. In particular, the poliovirus 2BC and/or 2C proteins are involved in the reorganization of intracellular membranes for the purpose of viral RNA synthesis and can also induce these changes upon expression in a heterologous system (1, 2, 11, 53). Although the ER was concluded to be a major source for poliovirus-induced membrane vesicles, they contained markers from throughout the secretory pathway (42), suggesting that the membranes of different compartments are used by the virus.
The data presented in this paper and a previous report (56) provide convincing evidence that arterivirus RNA synthesis also occurs in association with intracellular membranes. In this case, the double-membrane structures which are induced for this purpose appear to be specifically derived from the ER. Nidovirus RNA synthesis comprises both genome replication and discontinuous subgenomic mRNA transcription (for reviews, see references 17 and 45). Both genomic and subgenomic replicative intermediates are found in cells infected with arteriviruses (14) and coronaviruses (40, 44). Although the EAV replicase proteins appear to localize exclusively to the DMV structures, it remains to be determined whether the complexes involved in genomic and subgenomic RNA synthesis are, in fact, one and the same.
ORF1a-encoded subunits form a membrane-associated scaffold for the EAV replication complex.
The cryoimmuno-EM studies presented in this paper have clearly shown that most EAV replicase subunits localize to the paired membranes and DMVs which are induced upon EAV infection. The results obtained with the heterologous SIN expression vector prove that DMV formation is a distinct step in the arterivirus life cycle, which is directed by ORF1a-encoded replicase subunits and does not depend on other viral proteins and/or EAV-specific RNA synthesis. In single-immunolabeling experiments, many EAV replicase subunits (with the exception of nsp1) localized to the same region of infected cells (56, 59). However, their colocalization had not yet been proven formally because the fact that all of our replicase antisera had been raised in rabbits prevented double-labeling experiments. The presence of a His tag at the C terminus of the nsp2-7 expression product of pSinEAV(261-1677)His and the availability of a mouse MAb specific for this epitope (66) have allowed us to show that different replicase processing products do, indeed, colocalize perfectly (Fig. (Fig.5C5C and D). More importantly, the use of the SIN expression system will allow convenient and straightforward delineation of the domains within the 1,417-residue-long nsp2 to nsp7 region which are required for DMV formation. Conserved hydrophobic domains in nsp2, nsp3, and nsp5 have previously been implicated in membrane association (56). However, the mechanism by which these domains are inserted into the membrane is unclear, since they are part of a large polyprotein which does not contain an N-terminal signal sequence.
It is tempting to speculate on a regulatory role for the ORF1a-encoded proteases in the membrane association of the arterivirus replicase. Two early and essential cleavages (at the nsp2/3 and nsp4/5 sites) occur immediately upstream of predicted membrane-spanning domains (56). Processing of the nsp2/3 junction (Fig. (Fig.1)1) occurs within 15 min after polyprotein synthesis (48) and liberates the N-terminal hydrophobic domain of nsp3. A very strong interaction between nsp2 and nsp3, or nsp3-containing processing intermediates, was described previously (48, 61). We recently obtained evidence that one of the probable functions of the nsp2-nsp3 interaction is the formation of a complex between nsp2 and nsp3-8, the C-terminal half of the ORF1a protein (Fig. (Fig.1).1). This complex appears to be required to allow nsp2 to act as a cofactor for cleavage of the nsp4/5 junction by the nsp4 SP (61). In the absence of the nsp2 cofactor, the nsp4 SP is able to cleave various other sites in the polyprotein, suggesting that the cofactor is not essential for the basic enzymatic activity of the SP. Thus, it appears that the formation of the nsp2-nsp3-8 complex is required for the correct presentation of the nsp4/5 site to the nsp4 SP. Processing of the latter site liberates the hydrophobic N terminus of nsp5, which seems to confer membrane association on many downstream processing intermediates (e.g., nsp5-7 and nsp5-8), some of which appear to extend to the C terminus of the ORF1b polyprotein (56). The elucidation of the interplay between polyprotein processing and membrane association, two of the main functions of the ORF1a protein, will be an important aspect of future studies on nidovirus replication complexes.
Consequences and mechanism of DMV formation.
The formation of closely paired membranes and DMVs in arterivirus-infected cells was described many years ago (8, 36, 52, 62, 64), but they were never implicated in viral RNA replication or transcription. In contrast to the generation of poliovirus-induced DMVs (42), EAV DMV formation appears to involve ER membranes only. This may be explained by a fundamental difference between the two viruses: whereas poliovirus is a nonenveloped virus, EAV acquires an envelope by budding into pre-Golgi intracellular compartments. Consequently, arteriviruses depend on a functional exocytotic pathway to be able to leave the host cell. In contrast, protein secretion in general is severely inhibited in poliovirus-infected cells (18, 19) and typical Golgi stacks are not found (11). Poliovirus RNA synthesis is blocked by brefeldin A, an inhibitor of ER-to-Golgi transport (28, 34). A preliminary study has shown that this drug—added either before or during EAV infection—blocks virus production but does not interfere with the formation or function of the replication complex (data not shown).
For porcine reproductive and respiratory syndrome virus, Weiland et al. (62) proposed that the DMVs originate from the ER by protrusion and detachment, a conclusion which seems to be confirmed by our observations for EAV (Fig. (Fig.44 and and6).6). Especially the formation of tightly paired membranes, which seems to precede DMV formation, argues against the only alternative model, a double-budding event (Fig. (Fig.8A).8A). As also proposed by Schlegel et al. for poliovirus (42), tightly paired membranes may form an initially horseshoe-shaped structure, which wraps around a part of the cytoplasm and could subsequently be sealed (Fig. (Fig.8B).8B). This process would bear similarity to the formation of autophagic vacuoles (12, 21, 42) and (to a certain extent) to the assembly of the intracellular mature form of vaccinia virus from crescent-shaped double IC membranes (39, 51). In poliovirus-infected cells, horseshoe-shaped (putative) intermediates of DMV formation were observed quite frequently (42). In the case of EAV, however, structures like the one shown in Fig. Fig.4d4d were rarely observed and can, in fact, be explained in the context of both models (depending on the plane of sectioning). Our analysis of serial sections suggested that most EAV DMVs are closed vesicles, but additional experiments are certainly required to prove this point. The presence of a permanent, narrow (neck-like) connection with the ER cannot be excluded.
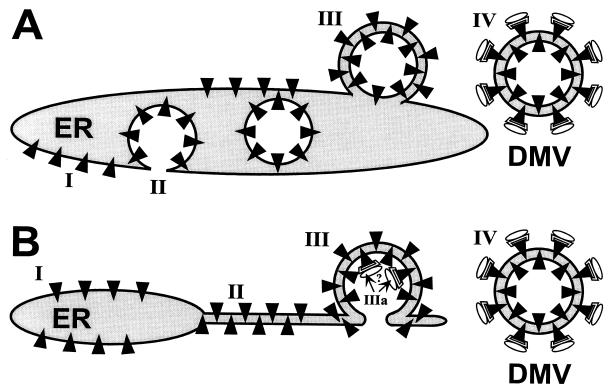
Two models of the formation of arterivirus DMVs from ER membranes. (A) Double-budding mechanism. According to this model, ORF1a-encoded replicase subunits (black triangles) associate with ER membranes (I) and induce the budding of a vesicular structure into the ER lumen (II). Subsequently, this vesicle undergoes a second budding event (III, resulting in an intermediate structure which could resemble the one shown in Fig. Fig.4d)4d) and finally detaches from the ER to give rise to a DMV (IV). Possibly, the ORF1b-encoded replicase subunits involved in RNA synthesis (represented by globular structures) associate with the cytoplasmic face of the DMV only after its formation. (B) Protrusion-and-detachment model. ORF1a-encoded replicase subunits associate with the ER (I) and induce the formation of tightly paired membranes (II). Subsequently, a vesicular double-membrane structure could be formed (III), resembling, e.g., the structures seen in Fig. Fig.6b.6b. If the double-membrane structure remained continuous with the ER, its interior would be connected with the cytoplasm and might function as a specific environment for viral RNA synthesis (IIIa). Alternatively, the structure may be sealed and form a DMV (IV), which could carry complexes involved in viral RNA synthesis at its surface, as also proposed by model A.
Assuming that DMV formation does, indeed, involve the protrusion of a horseshoe-shaped double-membrane structure from the ER, the prior association of EAV nonstructural proteins with this compartment can be expected. The formation of paired membranes may even result from direct interactions across the ER lumen between the luminal domains of transmembrane nonstructural proteins. Alternatively, the association of nonstructural proteins with the cytoplasmic face of the ER might induce DMV formation by forcing the membrane into a curved structure. This mechanism would resemble the action of, e.g., coatomer and clathrin during the formation of single-membrane vesicles with a function in cellular membrane trafficking (33, 43). If the arterivirus DMVs are not sealed and a connection between the DMV interior and the cytoplasm remains, the cavity of this structure can be envisioned to form a specific environment for viral RNA synthesis (Fig. (Fig.8B,8B, step IIIa). When DMVs are, indeed, closed vesicular structures, as our analysis of serial sections seems to suggest, both cytoplasm and nonstructural proteins associated with the DMV inner membrane would be trapped inside. As a result, catalytic replicase subunits associated with the inner membrane would be lost for, e.g., viral genome replication and transcription. Such problems would be circumvented if the RNA-synthesizing machinery assembled on the cytoplasmic face of the DMV outer membrane only after this structure had been sealed. In fact, this model may (partially) explain why ORF1a-encoded replicase subunits are overexpressed compared to ORF1b-encoded proteins. The expression of the latter, which probably have a more catalytic and less structural role in viral RNA synthesis, is downregulated by ribosomal frameshifting in all nidoviruses. Surprisingly, a recent study on the replication complex of the coronavirus mouse hepatitis virus has revealed major differences from our findings on EAV (55). Various mouse hepatitis virus replicase subunits and also viral RNA synthesis were concluded to localize to late endosomal structures. Thus, despite the common replicase organization and expression strategy of arteriviruses and coronaviruses, they may use essentially different mechanisms to form a membrane-associated scaffold for their replication complex.
ACKNOWLEDGMENTS
We gratefully acknowledge Leonie van Dinten, Hans van Tol, and Fred Wassenaar (Department of Virology, Leiden University Medical Center) and Tove Bakar, Espen Stang, and Andreas Brech (Department of Biology, Oslo University) for technical assistance and comments. We thank Jacomine Krijnse Locker and Gareth Griffiths (EMBL, Heidelberg, Germany) for helpful discussions and reviewing the manuscript. We are indebted to S. Fuller (EMBL) for the anti-PDI MAb, A. L. Glaser (Cornell University, Ithaca, N.Y.) for the anti-EAV GL MAb, H. Zentgraf (DKFZ, Heidelberg, Germany) for the anti-His tag antibody, R. E. Johnston and E. Beebe (University of North Carolina, Chapel Hill) for anti-Sindbis virus E2 MAb, R15, and C. M. Rice (Washington University, St. Louis, Mo.) for the SinRep/GFP vector.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.73.3.2016-2026.1999
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/73/3/2016.full.pdf
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/content/full/73/3/2016
Free to read at jvi.asm.org
http://jvi.asm.org/cgi/content/abstract/73/3/2016
Free after 4 months at jvi.asm.org
http://jvi.asm.org/cgi/reprint/73/3/2016
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.73.3.2016-2026.1999
Article citations
PRRSV non-structural protein 5 inhibits antiviral innate immunity by degrading multiple proteins of RLR signaling pathway through FAM134B-mediated ER-phagy.
J Virol, 98(10):e0081624, 12 Sep 2024
Cited by: 0 articles | PMID: 39264156
Mechanism, structural and functional insights into nidovirus-induced double-membrane vesicles.
Front Immunol, 15:1340332, 11 Jun 2024
Cited by: 0 articles | PMID: 38919631 | PMCID: PMC11196420
Review Free full text in Europe PMC
Host Subcellular Organelles: Targets of Viral Manipulation.
Int J Mol Sci, 25(3):1638, 29 Jan 2024
Cited by: 0 articles | PMID: 38338917 | PMCID: PMC10855258
Review Free full text in Europe PMC
Classification, replication, and transcription of Nidovirales.
Front Microbiol, 14:1291761, 24 Jan 2024
Cited by: 1 article | PMID: 38328580 | PMCID: PMC10847374
Review Free full text in Europe PMC
Viral regulation of organelle membrane contact sites.
PLoS Biol, 22(3):e3002529, 05 Mar 2024
Cited by: 0 articles | PMID: 38442090
Go to all (213) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex.
J Virol, 72(8):6689-6698, 01 Aug 1998
Cited by: 115 articles | PMID: 9658116 | PMCID: PMC109868
Adaptive Mutations in Replicase Transmembrane Subunits Can Counteract Inhibition of Equine Arteritis Virus RNA Synthesis by Cyclophilin Inhibitors.
J Virol, 93(18):e00490-19, 28 Aug 2019
Cited by: 2 articles | PMID: 31243130 | PMCID: PMC6714810
Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex.
J Gen Virol, 82(pt 5):985-994, 01 May 2001
Cited by: 131 articles | PMID: 11297673
Biogenesis and architecture of arterivirus replication organelles.
Virus Res, 220:70-90, 09 Apr 2016
Cited by: 55 articles | PMID: 27071852 | PMCID: PMC7111217
Review Free full text in Europe PMC





