Abstract
Background
Nasopharyngeal qualitative reverse-transcription polymerase chain reaction (RT-PCR) is the gold standard for diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, but it is not practical or sufficient in every clinical scenario due to its inability to distinguish active from resolved infection. Alternative or adjunct testing may be needed to guide isolation precautions and treatment in patients admitted to the hospital.Methods
We performed a single-center, retrospective analysis of residual clinical specimens and medical record data to examine blood plasma nucleocapsid antigen as a candidate biomarker of active SARS-CoV-2. Adult patients admitted to the hospital or presenting to the emergency department with SARS-CoV-2 ribonucleic acid (RNA) detected by RT-PCR from a nasopharyngeal swab specimen were included. Both nasopharyngeal swab and a paired whole blood sample were required to be available for analysis.Results
Fifty-four patients were included. Eight patients had positive nasopharyngeal swab virus cultures, 7 of whom (87.5%) had concurrent antigenemia. Nineteen (79.2%) of 24 patients with detectable subgenomic RNA and 20 (80.0%) of 25 patients with N2 RT-PCR cycle threshold ≤ 33 had antigenemia.Conclusions
Most individuals with active SARS-CoV-2 infection are likely to have concurrent antigenemia, but there may be some individuals with active infection in whom antigenemia is not detectable. The potential for high sensitivity and convenience of a blood test prompts interest in further investigation as a screening tool to reduce reliance on nasopharyngeal swab sampling and as an adjunct diagnostic test to aid in clinical decision making during the period after acute coronavirus disease 2019.Free full text

Investigation of Blood Plasma Viral Nucleocapsid Antigen as a Marker of Active Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant Infection
Abstract
Background
Nasopharyngeal qualitative reverse-transcription polymerase chain reaction (RT-PCR) is the gold standard for diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, but it is not practical or sufficient in every clinical scenario due to its inability to distinguish active from resolved infection. Alternative or adjunct testing may be needed to guide isolation precautions and treatment in patients admitted to the hospital.
Methods
We performed a single-center, retrospective analysis of residual clinical specimens and medical record data to examine blood plasma nucleocapsid antigen as a candidate biomarker of active SARS-CoV-2. Adult patients admitted to the hospital or presenting to the emergency department with SARS-CoV-2 ribonucleic acid (RNA) detected by RT-PCR from a nasopharyngeal swab specimen were included. Both nasopharyngeal swab and a paired whole blood sample were required to be available for analysis.
Results
Fifty-four patients were included. Eight patients had positive nasopharyngeal swab virus cultures, 7 of whom (87.5%) had concurrent antigenemia. Nineteen (79.2%) of 24 patients with detectable subgenomic RNA and 20 (80.0%) of 25 patients with N2 RT-PCR cycle threshold ≤ 33 had antigenemia.
Conclusions
Most individuals with active SARS-CoV-2 infection are likely to have concurrent antigenemia, but there may be some individuals with active infection in whom antigenemia is not detectable. The potential for high sensitivity and convenience of a blood test prompts interest in further investigation as a screening tool to reduce reliance on nasopharyngeal swab sampling and as an adjunct diagnostic test to aid in clinical decision making during the period after acute coronavirus disease 2019.
Nasopharyngeal swab (NPS) qualitative reverse-transcription polymerase chain reaction (RT-PCR) is the gold standard for diagnosis of coronavirus disease 2019 (COVID-19), but it is not practical or sufficient in every clinical scenario. Nasopharyngeal swab RT-PCR frequently detects ribonucleic acid (RNA) from nonviable severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and is unable to distinguish active from resolved infection, resulting in diagnostic confusion when history of symptoms or prior infection is unclear and after acute COVID-19 in immunocompromised hosts where prolonged viral replication can occur [1, 2]. Ribonucleic acid detection beyond the usual duration of active infection is particularly confusing in the inpatient setting when RT-PCR is used for screening to determine need for isolation precautions, and when a patient presents with a respiratory syndrome that could be explained by another infectious or noninfectious process. The NPSs are also expensive, subject to sampling variability [3], and collection is uncomfortable for patients.
Virus culture assays of NPSs are used to detect viral replication in investigational studies, but they are likely unable to identify all cases of viable virus, impractical for clinical use, and require biosafety level-3 facilities [4]. Nasopharyngeal swab subgenomic RNA (sgRNA), a byproduct of coronavirus replication, is an alternative, although it likely persists beyond active infection [5, 6]. Because neither culture-based nor sgRNA assays are in widespread clinical use, qualitative RT-PCR cycle threshold (Ct) values are sought to clarify disease status in patients with detectable RNA; however, their use is not approved by the US Food and Drug Administration nor are they standardized across laboratories and platforms [4, 7].
The presence of SARS-CoV-2 nucleocapsid protein in blood plasma, or antigenemia, is highly prevalent in individuals with acute COVID-19 and has been observed in individuals with prolonged viral replication [8–10]. Observations of high prevalence of nucleocapsid protein antigenemia during acute COVID-19 has compelled interest in its potential as a biomarker of active infection, but this has not been directly investigated in the Omicron era. We performed a pilot study investigating antigenemia as a marker of active infection in patients presenting to the hospital through comparison with biomarkers in paired RT-PCR-positive NPSs.
METHODS
Patient Selection
A convenience sampling of adult patients evaluated and treated at Emory University Hospital in Atlanta, Georgia between January 4 and February 16, 2022 were included. Patients were screened once weekly from a list of individuals who had tested positive for SARS-CoV-2 RNA by RT-PCR from an NPS on the Cepheid GeneXpert platform the day before selection. Specimens were retrieved from any patient with a complete blood count (CBC) collected within 12 hours of the NPS according to the collection timestamp in the electronic medical record. Patients were excluded if the residual NPS or the residual whole blood specimen were unable to be retrieved from the clinical laboratory. Use of residual specimens and review of corresponding medical records were approved by the Emory University Institutional Review Board (Number STUDY00000510) before data collection. Written informed consent by the patients was not required.
Specimen Retrieval and Storage
Whole blood specimens were collected in lavender-top ethylenediaminetetraacetic acid (EDTA)-coated venipuncture tubes for CBC as part of routine clinical care. After CBC measurements on an automated hematology analyzer, specimens were stored at 4°C in the clinical laboratory and retrieved within 72 hours. Upon retrieval, whole blood specimens were centrifuged at 2000 ×g for 15 minutes. Blood plasma supernatant was removed and stored in aliquots at −80°C until assays were performed. All assays were performed after exactly one thaw.
Nasopharyngeal swab specimens were collected as a part of routine clinical care using a standard nasopharyngeal sampling swab and stored in ~2 mL universal transport media (see Supplementary Methods). Residual media after removal of a portion of the specimen for the Cepheid GeneXpert assay was aliquoted and stored at −80°C. All assays were performed after exactly 1 thaw.
Clinical Data
Clinical data were abstracted directly from the electronic medical record. Presence of COVID-compatible symptoms at time of testing, date of symptom onset, and reason for hospital admission were determined independently by 2 authors and discrepancies were reconciled by both authors. Patients were considered to have COVID-compatible symptoms if the presence of common symptoms listed by the Centers for Disease Control and Prevention ([CDC] fever or chills, cough, shortness of breath or difficulty breathing, fatigue, muscle or body aches, headache, new loss of taste or smell, sore throat, congestion or runny nose, nausea or vomiting, or diarrhea) were documented [11]. Abstracted data were stored in Microsoft Excel and imported into MATLAB (Mathworks, Inc.) for analysis. Additional details are provided in Supplementary Methods. Patient-level study data are included in Supplementary Table 2.
Virus Culture Assay
Infectivity was determined by inoculating of Vero E6-TMPRSS2-TA2-ACE2 cells (NR-54970; BEI Resources, Manassas, VA) with the NPSs after ultraspin and resuspension of virus. Viral production dynamics were monitored by measuring SARS-CoV-2 RNA levels in the cell culture supernatant longitudinally over 6 to 9 days (see Supplementary Methods).
Ribonucleic Acid Extraction and Molecular Testing
Ribonucleic acid was extracted from NPSs using a KingFisher Apex instrument and the MagMAX Viral RNA Isolation Kit (both from Thermo Fisher Scientific, Waltham, MA). Eluates were tested in a duplex assay for the N2 target in the SARS-CoV-2 genome and RNase P as an internal control (the N2RP assay) and a separate real-time RT-PCR for detection of SARS-CoV-2 N-gene sgRNA as previously described [12, 13]. Presumptive SARS-CoV-2 variants were detected in a single reaction of the Spike single nucleotide polymorphism (SNP) assay containing 5 probes to detect sequences that confer the following amino acids: K417 (detects ancestral sequence), 452R, 478K, 484K, and 501Y as previously described, except that all 5 probes were combined into a single reaction rather than performing 2 separate reactions [13, 14]. Variant calls were made by comparing Spike SNP assay results to the expected profile of known variants.
Nucleocapsid Assays
Viral nucleocapsid protein was measured on the Quanterix Simoa platform using the Simoa SARS CoV-2 N Protein Advantage Kit (Billerica, MA) [15]. Blood plasma and NPS specimens were assayed undiluted and at 10×, 100×, and 1000× dilutions as needed to produce a result in the calibrated range of the assay (approximately 0.1 to 800 pg/mL).
Antinucleocapsid Antibody Titers
Detection of SARS CoV-2 nucleocapsid-binding antibodies was performed as previously described [16]. Additional details are provided in Supplementary Methods.
Antinucleocapsid Blocking Experiment
To determine whether antinucleocapsid antibody was producing a blocking effect in the NPS culture-positive/antigenemia-negative specimen pair (specimen no. 28, antinucleocapsid immunoglobulin [Ig]G titer = 1:1009), a study was performed to compare it with 3 additional plasma specimens without detectable antigenemia but with the following antinucleocapsid IgG titers: 1:361 768 (specimen no. 17), 1:332 705 (specimen no. 39), and undetectable (specimen no. 51). Ten microliters of plasma was added to 90 µL of phosphate-buffered saline containing dilutions of recombinant nucleocapsid protein. Each 100-µL plasma/nucleocapsid dilution was then measured on the Quanterix platform as previously described.
Statistical Testing
Data were analyzed and figures were created in MATLAB. To calculate correlation coefficients (MATLAB function corrcoef) and perform linear regression (MATLAB function fit), undetectable nucleocapsid antigen was assigned a value of 0.1 pg/mL and regression was performed with the log-transformed values. Negative RT-PCR results were assigned a Ct value of 50. Two-sided Wilcoxon rank-sum tests (MATLAB function ranksum) were used to determine P values when comparing distributions of 2 subsets of the data. Data visualized with boxplots are depicted as the median (center), interquartile range (box), and range (whiskers).
RESULTS
Our study included 54 patients with available NPS in transport media positive for SARS-CoV-2 RNA (Cepheid GeneXpert RT-PCR system) and paired peripheral whole blood in EDTA, 30 of whom had antigenemia above the clinical cutoff of 3.0 pg/mL (Table 1). Mean time between collection of paired specimens was 1.5 hours (interquartile range [IQR], 0.1–2.0) (Supplementary Figure 1). Twenty-nine (53.7%) patients were female and median age was 54.5 years (IQR, 46–68). Forty-two (77.8%) encounters involved full hospital admission and the remaining were evaluated and treated in the emergency department. Forty-two (77.8%) patients had COVID-19-compatible symptoms at the time of collection for median of 7 days (range, 0–62) (Figure 1A). Thirty patients (55.6%) had no record of prior positive SARS-CoV-2 testing, whereas the remaining patients tested positive by any method a median 14 days before collection (range, 2–48). The Omicron 417variant/484 K/501Y genotype was detected by RT-PCR in 39 (72.2%) NPSs (Supplementary Figure 2A) [13, 14]. The remaining specimens had high RT-PCR Ct values suggesting insufficient viral RNA in the specimen to detect these polymorphisms. The Omicron variant was overwhelmingly dominant in circulation during the study period (Supplementary Figure 2B) [17].
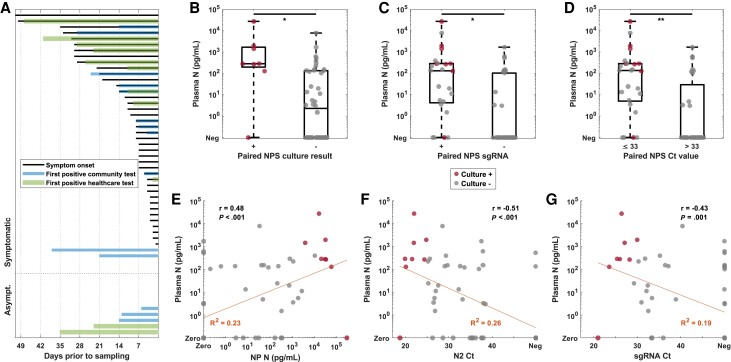
Nucleocapsid antigenemia is higher in individuals with nasopharyngeal swab (NPS) indicators of active infection and shows moderate correlation with respiratory biomarkers in hospitalized reverse-transcription polymerase chain reaction (RT-PCR)-positive individuals presenting with a wide range of symptoms and testing history. (A) Bar chart of symptom duration and time from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnosis by any test method reflect diversity of hospitalized patients with positive SAR-CoV-2 RT-PCR. (B) Distribution of plasma nucleocapsid protein in culture-positive and culture-negative individuals, (C) individuals with and without subgenomic ribonucleic acid (sgRNA), and (D) individuals with low cycle threshold (Ct) values demonstrate the association of higher antigen levels with NPS markers of active infection. (E–G) Plasma nucleocapsid level showed moderate correlation with NPS biomarkers. N, nucleocapsid; r, correlation coefficient with associated P value; R2, coefficient of determination for the linear model.
Table 1.
Demographic and Clinical Characteristics of Patients in the Study Stratified by Presence of Absence of Antigenemia Above the Clinic Threshold of 3.0 pg/mL
| Characteristics | Patients With Antigenemia (N = 30) | Patients Without Antigenemia (N = 24) |
|---|---|---|
| Female | 18 (60%) | 11 (45.8%) |
| Age, years; median (range) | 56 (21–79) | 54.5 (22–85) |
| Immunocompromised | 20 (66.7%) | 9 (37.5%) |
| SARS-CoV-2 Vaccine Documented | 11 (36.7%) | 10 (41.7%) |
 2 doses 2 doses | 7 (63.6%) | 5 (50%) |
 3 doses 3 doses | 4 (36.4%) | 3 (30%) |
 4 doses 4 doses | 0 (0%) | 2 (20%) |
| Prior positive COVID test (healthcare system) | 6 (20%) | 5 (20.8%) |
| Prior positive COVID test (reported) | 11 (36.7%) | 3 (12.5%) |
| Hours between blood and NP specimens; mean (range) | 0.2 (−5.4 to 11.2) | −0.1 (−6.7 to 4.2) |
| Asymptomatic | 4 (13.3%) | 6 (25%) |
| Any Reported COVID-19 Symptoms | 26 (86.7%) | 18 (75%) |
 Active at time of sampling Active at time of sampling | 24 (92.3%) | 18 (100%) |
| Days since COVID-19 symptom onset; median (range) | 8 (0–50) | 7 (0–62) |
| Encounter primarily for COVID-19 | 18 (60%) | 11 (45.8%) |
| Chest X-Ray Obtained | 25 (83.3%) | 24 (100%) |
 Abnormal Abnormal | 18 (72%) | 6 (25%) |
| WHO Ordinal Scale | ||
 4 4 | 22 (73.3%) | 15 (62.5%) |
 5 5 | 4 (13.3%) | 6 (25%) |
 6 6 | 3 (10%) | 0 (0%) |
 7 7 | 1 (3.3%) | 3 (12.5%) |
Abbreviations: COVID-19, coronavirus disease 2019; NP, nasopharyngeal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WHO, World Health Organization.
Increase in viral RNA levels of 1 log or greater in viral cell culture was observed with 8 (14.8%) of 54 NPSs, confirming presence of replication-competent virus (Supplementary Figure 3). Seven (87.5%) of eight culture-positive NPSs and 23 of 46 culture-negative NPSs had nucleocapsid protein greater than 3.0 pg/mL in the corresponding blood plasma specimen. Plasma nucleocapsid levels were higher in individuals with culture-positive NPSs (P = .003) (Figure 1B). Similarly, higher nucleocapsid levels were observed in individuals with detectable NPS sgRNA (P = .001) (Figure 1C) and Ct value ≤ 33 (P < .001) (Figure 1D). Although it was observed in high titer patient specimens, no antinucleocapsid blocking activity was observed in the blood sample from the antigenemia-negative, NPS culture-positive patient (Supplementary Figure 4).
Correlations between antigenemia and NPS biomarkers were moderate (Figure 1E–G, Supplementary Figure 5), but driven by patients without antigenemia (Supplementary Figure 6). Both antigenemia and culture-positive NPSs were found in subgroups stratified by either symptom duration or days since earliest positive test (Figure 2A and B). Antigenemia levels were higher in the subgroup with chest x-ray abnormalities (P = .008) (Figure 2C). Culture-positive NPSs were seen in patients with and without vaccination (Supplementary Figure 7), although vaccination records were limited.
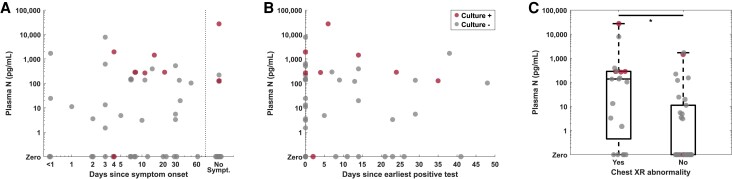
Antigenemia is found in the majority of culture-positive individuals throughout the range of symptom duration, suggesting it may be a useful adjunct test to distinguish residual nonviable ribonucleic acid from prolonged active infection in individuals beyond the typical period of acute infection. Antigenemia is found in the majority of individuals with positive viral culture throughout the range of (A) symptom duration or (B) time from earliest positive test, especially when nasopharyngeal swab (NPS) culture is positive beyond the expected 10-day period of acute coronavirus disease 2019. (C) Antigenemia levels are higher in individuals with chest x-ray (XR) abnormalities, suggesting it may reflect lower respiratory tract disease. Specimens from patients with positive NPS virus culture are depicted in red throughout. N, nucleocapsid.
Diagnostic performance metrics were evaluated for antigenemia and symptom duration with respect to 3 surrogate reference standards: NPS virus culture, sgRNA, and N2 Ct value ≤33. Antigenemia produced greater area under the receiver operating characteristic curve (Figure 3A–C) and greater positive percent agreement and negative percent agreement (NPA) (Figure 3D–E) compared with symptom duration, which were calculated using the clinical threshold of 3.0 pg/mL for positive antigenemia.
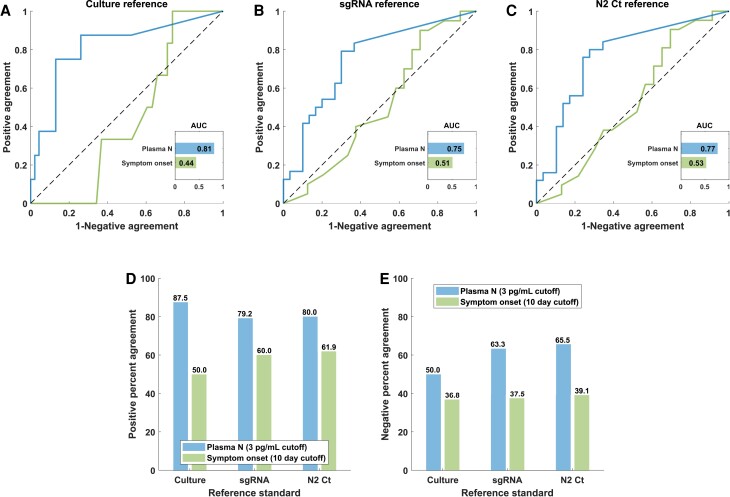
Nucleocapsid (N) antigenemia is superior to symptom duration as a diagnostic indicator of active infection. Receiver operating characteristic analysis (A–C), positive agreement (D), and negative agreement (E) with respect to nasopharyngeal swab virus culture, subgenomic ribonucleic acid (sgRNA), and N2 cycle threshold ([Ct] cutoff of ≤33). AUC, area under the receiver operating characteristic curve.
DISCUSSION
Our pilot study was designed to inform need for further investigation in the use of nucleocapsid antigenemia as a biomarker of active SARS-CoV-2 infection. In residual clinical specimens from 54 patients presenting to the hospital, we found that 87.5% of patients with culture-positive NPSs, 79.2% of patients with sgRNA detected in the NPS, and 80.0% of patients with NPS Ct ≤33 had concurrent antigenemia.
We performed analysis using 3 different reference standards because no perfect gold standard for active infection exists: negative NPS viral culture does not completely rule out active infection [4], whereas sgRNA likely persists in the nasopharynx beyond clearance of replicating virus [5, 6]. Our requirement for ultraspin to concentrate virus in NPSs and minimize the cytotoxic effects of transport media, degradational, and dilutional effects likely also limited our ability to detect all viable virus present in fresh specimens. Furthermore, cases have been reported in which active virus in the lower respiratory tract is evident but no viral RNA is present in the NPS, highlighting the potential for discordance between the nasopharynx and other sites of infection [8, 18, 19].
Most existing literature characterize antigenemia as a biomarker for acute COVID-19, which can be defined by timing since test positivity and symptom onset [9, 10, 20–23]. These studies, taken together, exhibit heterogeneity and varying rigor in defining cases, and they are not designed to evaluate antigenemia as a biomarker of viral replication. A recent study published by Mathur et al [24], which compared antigenemia with anterior nares swab virus culture before emergence of the Omicron variant, showed results consistent with our findings.
Our study does not definitively determine the sensitivity of antigenemia for active infection due to a small sample size. The observation of antigenemia in 7 of 8 NPS culture-positive individuals suggests that it may be a sensitive marker—that replication-competent virus in the NPS is unlikely in the absence of antigenemia. The single discordant patient suggests there likely exists a phenotype in which antigenemia is absent during active infection. Our sample size is insufficient to characterize prevalence of this phenotype or to determine its relationship to symptom severity, vaccination status, and other patient characteristics. These warrant further investigation.
Our study suggests that specificity of antigenemia as a marker of active infection is poor. More than one third of individuals without positive NPS culture, NPS sgRNA, or NPS Ct value ≤33 still had antigenemia. This is consistent with observations of detectable antigenemia beyond 10–14 days from illness onset in prior literature [9]. However, it should be recognized that NPA measured in our study likely severely underestimates specificity of the test in a broader population, because our study population only includes those with viral RNA detectable in the nasopharynx. Many with negative culture, absent sgRNA, and high Ct values are likely recovered from recent infection and thus in a period in which a high false-positive antigenemia rate may be expected. Studies examining individuals without a history of SARS-CoV-2 infection and prepandemic samples report specificity near 100% [9, 10, 20–23].
Although Ct values and antigen levels derived from each NPS specimen (Supplementary Figure 5) were the best predictors of culture positivity, such intraspecimen consistency is expected—specimens harboring replication-competent virus are expected to contain the highest concentrations of viral protein and RNA. Our study does not assess the consistency of NPS biomarkers across variables that may affect their ability to be used as surrogates for active infection, such as healthcare worker performing sample collection, swab type, and storage conditions. Rather than demonstrate inferiority or superiority of antigenemia to NPS biomarkers, our study was designed to explore potential for utility of the blood biomarker to play a role in certain clinical scenarios in which it may be considered to replace or supplement an NPS.
The CDC guidance recommends testing all symptomatic individuals for SARS-CoV-2 by NPS RT-PCR but leaves screening for asymptomatic infection to the discretion of the institution [25]. Universal NPS screening is expensive (we estimate added cost of approximately US $16.00 per patient screened compared with antigenemia testing) (see Supplementary Methods), uncomfortable for patients, and exerts high demand on molecular platforms in the clinical laboratory, and the detection of nonviable RNA may lead to unnecessary isolation [26]. Adjudication of these cases is burdensome for infection control staff and often relies on timing from symptom onset or earliest positive testing, which are poor indicators of positive culture, presence of sgRNA, or Ct value ≤33 in our study (Figure 3). The Society for Healthcare Epidemiology has advised against universal asymptomatic screening in the current testing paradigm [26].
On the other hand, abandoning screening altogether may lead to missed isolation of infectious patients with subclinical SARS-CoV-2 infection and missed opportunities to treat early mild disease in patients admitted for reasons other than COVID-19. Almost all hospitalized patients undergo venipuncture for common assays (such as complete blood counts), and blood specimens can be secondarily tested for antigenemia with common immunoassay platforms. Antigenemia may be sufficiently sensitive yet low cost and low burden as an alternative screening approach (Figure 4A). On the other hand, it remains possible that such a strategy would still uncover high numbers of antigenemia-positive, NP RT-PCR-positive individuals in the uncertain period beyond 10–14 days from illness onset, producing a similar conundrum to RT-PCR-screening strategies. Further investigation of antigenemia in this context alongside real-world cost analysis are warranted to determine whether universal antigenemia screening might limit missed diagnoses of subclinical SARS-CoV-2 infection that present risk of nosocomial transmission or could benefit from antiviral therapy, while relieving the burden of a universal NPS testing strategy. Comparison of an antigenemia-based screening algorithm to rapid nasal antigen testing should also be investigated, because this is an approach that has been implemented by many institutions and can more easily be self-collected in outpatient settings where blood sampling is not available [27].
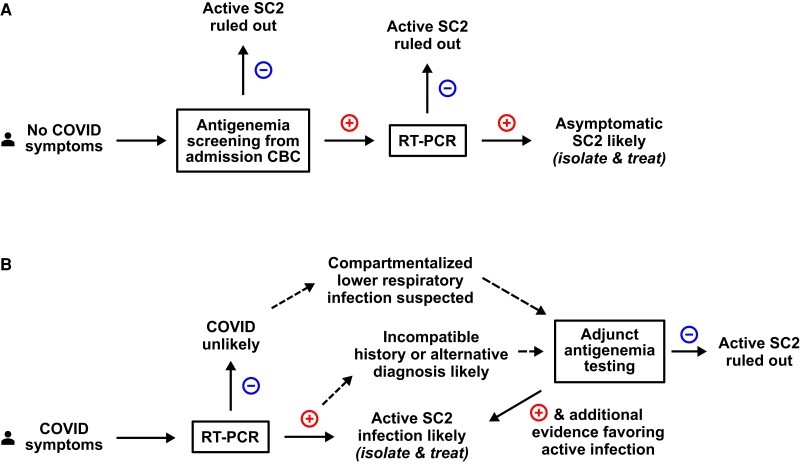
Antigenemia may add value to screening or diagnostic testing strategies in specific clinical scenarios: eg, (A) as a screening strategy for hospitalized patients without coronavirus disease (COVID) symptoms where only antigenemia-positive individuals receive nasopharyngeal swab (NPS) testing. This may allow for universal asymptomatic screening at lower costs than NPS screening while reducing unnecessary isolation of patients with nonviable ribonucleic acid and providing advantages over abandoning asymptomatic screening altogether. (B) Antigenemia may be a useful adjunct test in individuals with COVID-compatible symptoms but confusing clinical history, such as recent verified severe acute respiratory syndrome coronavirus 2 (SC2) infection in the preceding weeks but beyond 10 days from initial diagnosis. Absent antigenemia in this scenario should favor consideration of an alternative diagnosis, whereas presence of antigenemia should be interpreted in the context of the history timelines, reverse-transcription polymerase chain reaction (RT-PCR) cycle threshold value, deep respiratory tract sampling when indicated, and radiographic characteristics to aid the diagnostic impression. Each of these applications warrant further investigation. CBC, complete blood count.
Antigenemia testing should be considered as a method to clarify disease status in patients with respiratory symptoms or findings and who remain RT-PCR positive beyond the acute COVID-19 period or those in whom suspicion of active SARS-CoV-2 remains despite negative NPS RT-PCR. In this setting, antigenemia may aid diagnostic reasoning or influence use of SARS-CoV-2-directed antivirals or immunotherapies (Figure 4B). Our data suggest that in the immediate period (>10 days to a few weeks from symptom onset), detectable antigenemia may not sufficiently rule in active SARS-CoV-2, but absence of antigenemia in this time period favors diagnoses other than active SARS-CoV-2. Furthermore, in immunocompromised patients who may develop protracted SARS-CoV-2 infection, this biomarker may be even more sensitive and specific for a period of weeks to months after acute COVID-19. Although our study was not designed to specifically evaluate protracted infection, antigenemia in 3 individuals with culture-positive NPSs after more than 10 days in the subgroup with immune compromise (Supplementary Figure 8 and Table 1) adds to emerging literature supporting antigenemia as an adjunct diagnostic tool in these scenarios [8]. Finally, when considering the possibility of discordance between the nasopharynx and lower respiratory tract—for example, in an NPS RT-PCR-negative patient for whom clinical suspicion for active SARS-CoV-2 remains—antigenemia may be an important indicator. Association of higher levels of nucleocapsid antigenemia with chest x-ray abnormalities in our study adds to literature and suggests that viral replication in the lower respiratory tract may be associated with antigenemia [8, 28].
The dilemma presented by the lack of a perfect gold standard for active infection is that it may be impossible to fully characterize the utility of antigenemia with comparison to an NPS alone. Prospective studies are needed to examine the potential impact of antigenemia screening on outcomes such as nosocomial transmission, and studies to determine the utility of antigenemia as an adjunct diagnostic test should focus on immune-compromised populations with history of COVID-19 and prolonged pulmonary symptoms.
CONCLUSIONS
Endemic SARS-CoV-2 mandates consideration of new diagnostic approaches. We conclude from a convenience sampling of NPS and blood specimens that nucleocapsid antigenemia measurements may hold predictive value toward discerning disease status. Further work is needed to fully characterize the performance of antigenemia in asymptomatic screening of hospitalized patients and the evaluation of individuals with possible protracted infection.
Acknowledgments
Financial support. This work was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health ([NIH] under Award Number U54EB027690) as part of the Rapid Acceleration of Diagnostics (RADx) initiative, NIH National Cancer Institute (Award Number U54CA260563), “Immune Regulation of COVID-19 Infection in Cancer and Autoimmunity” (to JDR), and NIH National Center for Advancing Translational Sciences (Award Number UL1TR002378), Georgia Clinical & Translational Science Alliance (Georgia CTSA). This work was also supported by funding from the Marcus Foundation (to JDR).
Contributor Information
Gregory L Damhorst, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA. The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA.
Nils Schoof, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
Phuong-Vi Nguyen, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA.
Hans Verkerke, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Eli Wilber, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA.
Kaleb McLendon, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
William O’Sick, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Tyler Baugh, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Suneethamma Cheedarla, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Narayanaiah Cheedarla, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Victoria Stittleburg, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA.
Eric C Fitts, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Margaret A Neja, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
Ahmed Babiker, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA. Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Anne Piantadosi, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA. Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
John D Roback, Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia, USA.
Jesse J Waggoner, Division of Infectious Diseases, Department of Medicine, Emory University, Atlanta, Georgia, USA. The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA. Department of Global Health, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Maud Mavigner, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
Wilbur A Lam, The Atlanta Center for Microsystems-Engineered Point-of-Care Technologies, Atlanta, Georgia, USA. Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA. Aflac Cancer and Blood Disorders Center at Children's Healthcare of Atlanta, Atlanta, Georgia, USA. Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
Articles from Open Forum Infectious Diseases are provided here courtesy of Oxford University Press
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/147300827
Article citations
Diagnostic Utility of SARS-CoV-2 Nucleocapsid Antigenemia: A Meta-analysis.
Open Forum Infect Dis, 11(10):ofae561, 02 Oct 2024
Cited by: 0 articles | PMID: 39431150 | PMCID: PMC11487748
SARS-CoV-2 RNA and Nucleocapsid Antigen Are Blood Biomarkers Associated With Severe Disease Outcomes That Improve in Response to Remdesivir.
J Infect Dis, 230(3):624-634, 01 Sep 2024
Cited by: 2 articles | PMID: 38657001 | PMCID: PMC11420797
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nucleocapsid Antigenemia Is a Marker of Acute SARS-CoV-2 Infection.
J Infect Dis, 226(9):1577-1587, 01 Nov 2022
Cited by: 10 articles | PMID: 35877413 | PMCID: PMC9384592
Comparison of a novel antigen detection test with reverse transcription polymerase chain reaction assay for laboratory diagnosis of SARS-CoV-2 infection.
Infection, 51(1):91-96, 05 May 2022
Cited by: 4 articles | PMID: 35513690 | PMCID: PMC9070611
Measurement of Severe Acute Respiratory Syndrome Coronavirus 2 Antigens in Plasma of Pediatric Patients With Acute Coronavirus Disease 2019 or Multisystem Inflammatory Syndrome in Children Using an Ultrasensitive and Quantitative Immunoassay.
Clin Infect Dis, 75(8):1351-1358, 01 Oct 2022
Cited by: 20 articles | PMID: 35213684 | PMCID: PMC8903440
Diagnostic Performance Assessment of Saliva RT-PCR and Nasopharyngeal Antigen for the Detection of SARS-CoV-2 in Peru.
Microbiol Spectr, 10(4):e0086122, 18 Jul 2022
Cited by: 2 articles | PMID: 35867471 | PMCID: PMC9430815
Funding
Funders who supported this work.
Marcus Foundation
NHLBI NIH HHS (1)
Grant ID: T32 HL069769
NIH National Cancer Institute (1)
Grant ID: U54CA260563
NIH National Center for Advancing Translational Sciences (1)
Grant ID: UL1TR002378
National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (1)
Grant ID: U54EB027690




