| PMC full text: | Published online 2023 Jun 8. doi: 10.1038/s41591-023-02327-2
|
Fig. 6
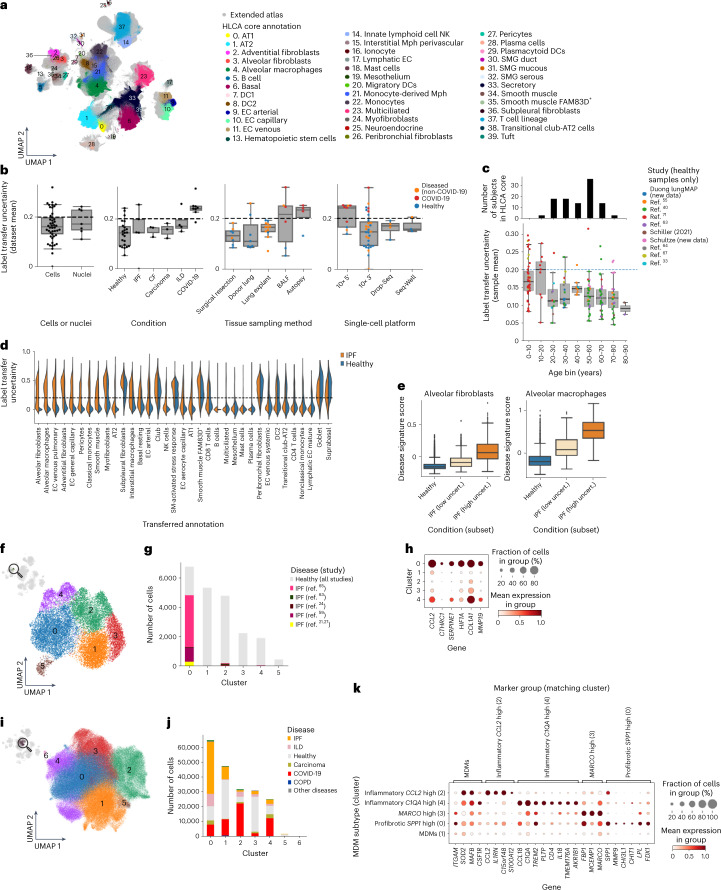
a, UMAP of the extended HLCA colored by coarse annotation (HLCA core) or in gray (cells mapped to the core). b, Uncertainty of label transfer from the HLCA core to newly mapped datasets, categorized by several experimental or biological features. Categories with fewer than two instances are not shown. The numbers of datasets per category were as follows: 30 cells, 7 nuclei, 23 healthy, 5 IPF, 3 CF, 3 carcinoma, 4 ILD, 8 surgical resection, 7 donor lung, 12 lung explant, 6 bronchoalveolar lavage fluid, 4 autopsy, 9 10x 5′, 31 10x 3′, 4 Drop-Seq and 3 Seq-Well. c, Bottom, mean label transfer uncertainty per mapped healthy lung sample in the HLCA extension, grouped into age bins and colored by study. The numbers of mapped samples per age bin were as follows: 43 for 0–10 years, 33 for 10–20
years, 33 for 10–20 years, 31 for 20–30
years, 31 for 20–30 years, 23 for 30–40
years, 23 for 30–40 years, 19 for 40–50
years, 19 for 40–50 years, 12 for 50–60
years, 12 for 50–60 years, 9 for 60–70
years, 9 for 60–70 years, 8 for 70–80
years, 8 for 70–80 years and 2 for 80–90
years and 2 for 80–90 years. Top, bar plot showing the number of donors per age group in the HLCA core. d, Violin plot of label transfer uncertainty per transferred cell type label for a single mapped IPF dataset64, split into cells from healthy donors (blue) and donors with IPF (orange). e, Uncertainty-based disease signature scores among alveolar fibroblasts and alveolar macrophages, split into cells from control donors (n
years. Top, bar plot showing the number of donors per age group in the HLCA core. d, Violin plot of label transfer uncertainty per transferred cell type label for a single mapped IPF dataset64, split into cells from healthy donors (blue) and donors with IPF (orange). e, Uncertainty-based disease signature scores among alveolar fibroblasts and alveolar macrophages, split into cells from control donors (n =
= 10,453 and 1,812, respectively), and low-uncertainty cells (n
10,453 and 1,812, respectively), and low-uncertainty cells (n =
= 1,419 and 200, respectively) and high-uncertainty cells (n
1,419 and 200, respectively) and high-uncertainty cells (n =
= 1,172 and 162, respectively) from donors with IPF. f, UMAP embedding of alveolar fibroblasts (labeled with manual annotation (core) or label transfer (five IPF datasets)) colored by Leiden cluster. g, Composition of the clusters shown in f by study, with cells from control samples colored in gray. h, Expression of marker genes for IPF-enriched cluster 0 per alveolar fibroblast cluster. Cluster 5 was excluded as 96% of its cells were from a single donor. i, UMAP of all MDMs in the HLCA, colored by Leiden cluster. j, Composition of the MDM clusters from i by disease. k, Expression of cluster marker genes among all MDM clusters excluding donor-specific clusters 5 and 6. For h and k, mean counts were normalized such that the highest group mean was set to 1 for each gene. For b, c and e, the boxes show the median and interquartile range. Data points more than 1.5 times the interquartile range outside the low and high quartile are considered outliers. Whiskers extend to the furthest nonoutlier point. BALF, bronchoalveolar lavage fluid; CF, cystic fibrosis; Drop-Seq, droplet sequencing; ILD, interstitial lung disease; Mph, macrophages; SM, smooth muscle; uncert., uncertainty.
1,172 and 162, respectively) from donors with IPF. f, UMAP embedding of alveolar fibroblasts (labeled with manual annotation (core) or label transfer (five IPF datasets)) colored by Leiden cluster. g, Composition of the clusters shown in f by study, with cells from control samples colored in gray. h, Expression of marker genes for IPF-enriched cluster 0 per alveolar fibroblast cluster. Cluster 5 was excluded as 96% of its cells were from a single donor. i, UMAP of all MDMs in the HLCA, colored by Leiden cluster. j, Composition of the MDM clusters from i by disease. k, Expression of cluster marker genes among all MDM clusters excluding donor-specific clusters 5 and 6. For h and k, mean counts were normalized such that the highest group mean was set to 1 for each gene. For b, c and e, the boxes show the median and interquartile range. Data points more than 1.5 times the interquartile range outside the low and high quartile are considered outliers. Whiskers extend to the furthest nonoutlier point. BALF, bronchoalveolar lavage fluid; CF, cystic fibrosis; Drop-Seq, droplet sequencing; ILD, interstitial lung disease; Mph, macrophages; SM, smooth muscle; uncert., uncertainty.