Abstract
Free full text

Effects of Nonstructural Proteins NS1 and NS2 of Human Respiratory Syncytial Virus on Interferon Regulatory Factor 3, NF-κB, and Proinflammatory Cytokines
Abstract
Human respiratory syncytial virus (HRSV) is the leading cause of serious pediatric acute respiratory tract infections, and a better understanding is needed of the host response to HRSV and its attenuated vaccine derivatives. It has been shown previously that HRSV nonstructural proteins 1 and 2 (NS1 and NS2) inhibit the induction of alpha/beta interferon (IFN-α/β) in A549 cells and human macrophages. Two principal transcription factors for the early IFN-β and -α1 response are interferon regulatory factor 3 (IRF-3) and nuclear factor κB (NF-κB). At early times postinfection, wild-type HRSV and the NS1/NS2 deletion mutants were very similar in the ability to activate IRF-3. However, once NS1 and NS2 were expressed significantly, they acted cooperatively to suppress activation and nuclear translocation of IRF-3. Since these viruses differed greatly in the induction of IFN-α/β, NF-κB activation was evaluated in Vero cells, which lack the structural genes for IFN-α/β and would preclude confounding effects of IFN-α/β. This showed that deletion of the NS2 gene sharply reduced the ability of HRSV to induce activation of NF-κB. Since recombinant HRSVs from which the NS1 or NS2 genes have been deleted are being developed as vaccine candidates, we investigated whether the changes in activation of host transcription factors and increased IFN-α/β production had an effect on the epithelial production of proinflammatory factors. Viruses lacking NS1 and/or NS2 stimulated modestly lower production of RANTES (Regulated on Activation Normal T-cell Expressed and Secreted), interleukin 8, and tumor necrosis factor alpha compared to wild-type recombinant RSV, supporting their use as attenuated vaccine candidates.
Human respiratory syncytial virus (HRSV) is the most common cause of viral bronchiolitis and pneumonia in infants and children worldwide. HRSV is an enveloped, nonsegmented, negative-strand RNA virus that belongs to the genus Pneumovirus of the family Paramyxoviridae (7). Pneumoviruses express two putative nonstructural proteins, NS1 and NS2, from separate mRNAs encoded by the first two genes in the viral gene order. Recombinant HRSVs in which the NS1 and/or NS2 genes have been deleted (ΔNS1, ΔNS2, and ΔNS1/2) exhibit reduced replication in cultured cells that are competent to produce alpha/beta interferon (IFN-α/β), as well as in mice, monkeys, and chimpanzees (15, 16, 33, 34, 38). HRSVs lacking the NS1 and NS2 genes induced high levels of IFN-α/β in a human pulmonary epithelial cell line (A549) and in monocyte-derived macrophages (31). The NS1 and NS2 proteins of bovine RSV (BRSV) also had been previously shown to suppress the induction of IFN-α/β and have been shown to suppress the IFN-mediated antiviral state (5, 6, 27, 28, 37).
IFN-α/β are secreted by most eukaryotic cells in order to establish a first line of defense against viral infection. In human cells, the IFN response commences with the production of IFN-β and -α1, which then induce approximately a dozen other IFN-αs and additional amounts of IFN-β and -α1 in an autocrine and paracrine manner. IFN-β is induced at the level of transcription initiation by a number of regulatory factors. The key inducers of IFN-β are interferon regulatory factor 3 (IRF-3) and nuclear factor κB (NF-κB). Activator protein 1 (AP-1), a heterodimer of ATF-2 and c-jun, is a universal transcription factor that also plays a role in the induction of IFN-β. These three transcription factors can be activated by double-stranded RNA (dsRNA), among other inducers. Once activated, they form a transcriptional enhancer complex, called an enhanceosome, and bind to the IFN-β promoter (13). The IFN-β promoter is composed of an overlapping set of positive regulatory domains (PRDs) I to IV. IRF-3 binds to PRDs I and III, while NF-κB binds to PRD II and AP-1 to PRD IV. The IFN-α1 promoter contains virus response elements that resemble PRDs I and III of the IFN-β promoter (1).
IRF-3 is expressed constitutively in the cytoplasm and is activated by phosphorylation on serine and threonine residues in the C-terminal region mediated by kinases that are activated in response to viral dsRNA and viral nucleocapsid protein or nucleocapsid-like structures (9, 29, 35). This results in the formation of IRF-3 dimers that associate with the coactivators p300 and CBP and are translocated to the nucleus, where they bind to the IFN-β promoter (39). Activated IRF-3 also can directly up-regulate transcription of additional genes, such as the proinflammatory chemokine RANTES (regulated on activation normal T-cell expressed and secreted) (22). A number of viruses have been shown to encode proteins that interfere with activation of IRF-3, including Bunyamwera (18), influenza A (32), and Ebola (2) viruses. Recently, the NS1 and NS2 proteins of BRSV were implicated in inhibiting phosphorylation of IRF-3 (6). Here, we investigate the effects of the individual proteins of HRSV on IRF-3 translocation and show that the effect is time dependent.
In addition to its role in inducing IFN-α/β, NF-κB plays a role in the expression of a wide range of genes involved in inflammation and innate and adaptive immunity (20, 36). Inactive NF-κB is sequestered in the cytoplasm by association with the IκB regulatory proteins. In response to various inducers, such as dsRNA, cytokines, lipopolysaccharides, and IFN-α/β, a cascade of kinases is activated, resulting in the phosphorylation of IκBα and release of NF-κB, which is then translocated to the nucleus. The IκB proteins repress or sustain NF-κB function by controlling its trafficking between the cytoplasm and nucleus (20). The main activated form of NF-κB is a heterodimer of p50 and p65, which is persistently activated during infection of cultured cells with HRSV (3, 8, 12). Here, we have examined the activation of NF-κB in Vero cells, which lack the structural genes for IFN-α/β, in order to avoid the complication of autocrine effects of IFN-α/β. Under these conditions, the HRSV NS2 protein appeared to play the major role in stimulating activation of NF-κB. AP-1 was not significantly activated by infection with either wild-type (wt) HRSV or the NS1/NS2 mutants.
In addition to IFN-α/β, IRF-3 and NF-κB bind to the transcription promoters of numerous proinflammatory cytokines. As a measure of the overall impact of NS1 and NS2 on the epithelial cell response to HRSV, we measured the production of three factors: the CC chemokine RANTES, the CXC chemokine interleukin-8 (IL-8), and the cytokine tumor necrosis factor alpha (TNF-α). Understanding the role of the NS proteins in altering the cellular response to infection is important in understanding the basis of attenuation of live RSV vaccines in which these proteins have been ablated.
MATERIALS AND METHODS
Cells and viruses.
Vero and A549 cells were maintained in Opti-MEM supplemented with 4% fetal calf serum. The recombinant RSVs (rRSVs) ΔNS1, ΔNS2, and ΔNS1/2 were constructed in previous work and are derivatives of the wt A2 strain (wt rRSV) (31, 33, 34, 38). All of the viruses were grown in Vero cells, which lack the structural genes for IFN-α/β, and purified by centrifugation and banding in discontinuous 30 to 60% (wt/vol) sucrose gradients. The identity (presence of the desired mutation) and purity (absence of cross-contamination) of the specific virus stocks were confirmed by reverse transcription-PCR.
Infections.
Vero and A549 cells were infected at an input multiplicity of infection (MOI) of 3 PFU per cell for all experiments except for those using luciferase reporter plasmids, where the MOI was 1. Cells were mock infected or infected with wt rRSV, ΔNS1, ΔNS2, ΔNS1/2, or UV-inactivated wt rRSV in serum-free Opti-MEM for 2 h. The cells were then washed with serum-free Opti-MEM, which was replaced with Opti-MEM/2% fetal bovine serum/antibiotics, and incubated at 37°C.
Transfections and reporter assays.
As a positive control for IRF-3, NF-κB, and AP-1 activation, cells were transfected with 12 μg per 106 cells of polypoly(I:C) (Amersham Biosciences) using Lipofectamine (Invitrogen) in serum-free Opti-MEM for 2 h, after which the polypoly(I:C) was replaced with medium containing 2% fetal bovine serum and antibiotics.
IRF-3 activity was assayed using p55CIBLuc, in which the luciferase gene is under the control of the PRD I element of the IFN-β promoter (a generous gift from Takashi Fujita, Kyoto, Japan) (39). AP-1 and NF-κB activities were assayed using pAP-1-Luc and pNF-κB-Luc reporter plasmids, which contain promoter elements responsive to activity of the respective factors (Stratagene Pathdetect cis-reporting systems). As a positive control for luciferase activation, the IRF-3, NF-κB, and AP-1 reporter plasmids were cotransfected with the expression vector of a known activator, MEK kinase (pFC-MEKK; Stratagene; not shown). Subconfluent A549 and Vero cells grown in 12-well plates were transfected with 0.5 μg of vector using FuGENE 6 (Roche) in serum-free Opti-MEM. Ten hours after transfection with the reporter plasmids, cells were either mock infected or infected with wt rRSV, ΔNS1, ΔNS2, or ΔNS1/2 at an MOI of 1 as described above. At 8 or 14 h postinfection (p.i.), the cells were lysed directly in the wells with cell lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100) and clarified by centrifugation. Luciferase activity was measured with a commercial assay buffer (Promega) and a luminometer (Turner TD-20e).
Immunofluorescence.
Vero or A549 cells were grown on circular coverslips and were mock infected or infected as described above. At the indicated times p.i., the coverslips were washed with phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde in PBS (Electron Microscopy Sciences). The cells were blocked with PBS/5% bovine serum albumin (BSA), permeabilized with 1% Triton X-100 in PBS/3% BSA, and incubated for 1.5 h at 32°C with one of the following primary antibodies: rabbit anti-IRF-3 (Santa Cruz), diluted 1:500 in PBS/3% BSA; mouse anti-NF-κB (p65; Santa Cruz), diluted 1:200 in PBS/3% BSA; or rabbit antiserum against the C terminus of NS2, which cross-reacts with NS1 due to partial sequence identity in the peptide region, diluted 1:1,500 in PBS/3% BSA. After primary-antibody incubation, the cells were washed once with 1% Triton X-100 in PBS/3% BSA and twice with PBS. Secondary antibodies (anti-rabbit antibodies tagged with AF488 for IRF-3, anti-mouse-AF488 for NF-κB, and anti-rabbit-AF594 for NS1/NS2; Molecular Probes) were diluted 1:1,000 in PBS/3% BSA, and the cells were incubated for 1.5 h at 32°C. The cells were washed with 1% Triton X-100 in PBS/3% BSA and then twice with PBS and stained with 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) in PBS. The percentage of cells in which NF-κB or IRF-3 were brightly stained in the nucleus, or in which NS1 and/or NS2 was brightly stained, was calculated using 10 fields of view from two independent experiments, with DAPI-stained nuclei representing the total number of cells per field of view.
Preparation of subcellular extracts.
Nuclear and cytosolic fractions of A549 cells and Vero cells were prepared using methods modified from those of Tian et al. (36). The cells were washed in PBS and lysed by the addition of 3 cell volumes of buffer A, which contained 50 mM HEPES (pH 7.4), 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mg/ml of phenylmethylsulfonyl fluoride, 1 μg/ml of pepstatin A, 1 μg/ml of leupeptin, 10 μg/ml of soybean trypsin inhibitor, 10 μg/ml of aprotinin, and 0.5% IGEPAL CA-630 ([octylphenoxy] polyethoxyethanol). After 20 min on ice, the nuclei were pelleted by centrifugation at 6,000 × g for 20 min at 4°C. The supernatant constituted the cytosolic fraction. The nuclear pellet was washed in buffer A to remove traces of the cytosol. Proteins in the nuclei were extracted by incubation in buffer C, which contained 10% glycerol, 50 mM HEPES (pH 7.4), 400 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mg/ml of phenylmethylsulfonyl fluoride, 1 μg/ml of pepstatin A, 1 μg/ml of leupeptin, 10 μg/ml of soybean trypsin inhibitor, and 10 μg/ml of aprotinin. After 30 min on ice with frequent vortexing, the nuclear protein extract was clarified by centrifugation at 12,000 × g for 30 min at 4°C. The protein content was determined with a Pierce bicinchoninic acid protein assay kit using BSA as a standard: from a given number of cells, the cytosolic fraction contained ~4-fold more protein than the nuclear fraction. The efficacy of nuclear/cytosol separation was confirmed in each experiment by Western blot analysis using specific markers described below.
Western blot analysis.
For analysis of nuclear and cytoplasmic fractions, 90 μg of protein from each sample was heated at 95°C for 2 min in commercially obtained lithium dodecyl sulfate sample buffer (Invitrogen) that contained 1% (wt/vol) sodium dodecyl sulfate (SDS) supplemented with 100 mM dithiothreitol. The proteins were separated by electrophoresis on 12% polyacrylamide gels in the presence of SDS (12% SDS-polyacrylamide gel electrophoresis ) using commercially obtained precast gels that employed a Bis-Tris buffer system (Invitrogen), and transferred to polyvinylidene diflouride membranes in transfer buffer (NuPAGE; Invitrogen). The membranes were blocked in PBS containing 5% (wt/vol) dry skim milk for 1 h and immunoblotted with antibodies to either (i) USF2, as a marker for the nuclear fraction (Santa Cruz); (ii) α-tubulin, as a marker for the cytoplasmic fraction (Santa Cruz); (iii) RSV NS1/NS2 (polyclonal antibody described above); or (iv) NF-κB p65 (Santa Cruz). Bound antibodies were visualized by incubation with horseradish peroxidase-coupled goat anti-rabbit or anti-mouse immunoglobulin G antibodies and chemiluminescence (Pierce).
For analysis of RSV protein expression, infected cell pellets were heated in lithium dodecyl sulfate sample buffer (Invitrogen) as described above and centrifuged through Qiashredders (QIAGEN). Approximately 3.75 × 105 cell equivalents of each sample were separated by SDS-PAGE on commercially obtained 4 to 12% gradient gels in a Bis-Tris buffer system (Invitrogen), transferred to polyvinylidene diflouride membranes, and blocked as described above. The membranes were incubated with rabbit polyclonal antisera to RSV that had been purified by banding at the interface of a 30% to 60% (wt/vol) sucrose gradient. Bound antibodies were visualized as described above.
ELISA.
The secretion of IL-8, RANTES, and TNF-α from A549 cells was quantified by antigen capture enzyme-linked immunosorbent assay (ELISA; Biosource). Cell culture supernatants collected 16 h and 20 h p.i. were tested according to the manufacturers directions using controls supplied by the manufacturer as standards.
RESULTS
Kinetics of IRF-3 activation and deactivation. Indirect immunofluorescence was used to monitor activation and translocation of the transcription factor IRF-3 to the nuclei of A549 cells during infection with wt rRSV, UV-inactivated wt rRSV, and the ΔNS1, ΔNS2, and ΔNS1/2 gene deletion mutants (Fig. 1A and B). As a positive control, cells were transfected in parallel with polypoly(I:C) dsRNA. This positive control induced rapid, efficient translocation of IRF-3 to the nucleus, with peak accumulation at 4 to 6 h posttransfection (Fig. (Fig.1A,1A, bottom). This was followed by the rapid loss of nuclear IRF-3, reflecting deactivation of IRF-3 and recycling to the cytoplasm, followed by degradation.

Nuclear localization of IRF-3 in A549 cells in response to infection with wt rRSV or treatment with synthetic polypoly(I:C). (A) A549 cells were mock infected or infected at an MOI of 3 PFU/cell with wt rRSV, ΔNS1, ΔNS2, ΔNS1/2, or the equivalent amount of UV-inactivated wt rRSV or were transfected with polypoly(I:C), as indicated. The cells were fixed at the indicated times postinfection, permeabilized, and analyzed by indirect immunofluorescence with polyclonal antibodies to IRF-3 (green) or a polyclonal antiserum that reacts with both NS1 and NS2 (red). This antiserum detected NS1 more efficiently than NS2 (see the text). (B) The percentage of cells with nuclei positive for IRF-3 or that were positive for NS1/NS2 at each time point and for each treatment was calculated based on multiple fields of view from two independent experiments. The indicated (*) value of IRF-3 for the ΔNS1, ΔNS2, or ΔNS1/2 virus was confirmed to be significantly different from that of wt RSV (P = 9.1 × 10−8, 1.3 × 10−7, or 1.6 × 10−8, respectively; Student's t test). The values of IRF-3 at 14 h versus 10 h for wt RSV also were confirmed to be significantly different (P = 1.1 × 10−8).
Infection with wt rRSV and the ΔNS1, ΔNS2, and ΔNS1/2 gene deletion mutants efficiently induced nuclear translocation of IRF-3 during the first 10 h p.i. (Fig. 1A and B). Translocation of IRF-3 was not observed when cells were exposed to UV-inactivated wt rRSV, indicating that RSV replication was necessary for IRF-3 activation in A549 cells (Fig. (Fig.1A).1A). In cells infected with wt rRSV, the average percentage of nuclei positive for IRF-3 reached a peak of ~26% at 10 h p.i., a value very similar to the peak of 35% observed with polypoly(I:C) (Fig. (Fig.1B).1B). One difference was that the peak of nuclear accumulation of IRF-3 in response to wt rRSV occurred somewhat later than in response to polypoly(I:C) (10 h versus 4 to 6 h), which presumably reflects a lag necessary for viral macromolecular synthesis to occur sufficient to activate IRF-3. The percentage of cells with intense nuclear IRF-3 staining then dropped dramatically in wt rRSV-infected cells to ~1% at 14 h p.i., mirroring the export and degradation of IRF-3 as observed following treatment with polypoly(I:C). In cells infected with the NS1/NS2 deletion mutants, the percentage of nuclei positive for IRF-3 reached a maximum of ~35% by 10 h (ΔNS1) or ~30% by 14 h (ΔNS2, ΔNS1/2) p.i. These values did not subsequently decline, indicating that IRF-3 activation continued in cells infected with the gene deletion mutants. Thus, the difference between wt rRSV and the NS1/NS2 gene deletion mutants was not in the ability to activate IRF-3, but rather in the duration of the response. The ability of NS1 and NS2 to individually affect the activation of IRF3 had not been investigated for HRSV or BRSV. Here, deletion of either NS1 or NS2 greatly alleviated the block to activation. This suggested that the cooperative effect of the two proteins in inhibiting IRF-3 activation is much greater than those of the individual proteins.
The expression of the NS1 and NS2 proteins in parallel cultures was visualized with a polyclonal antiserum that recognizes both proteins (Fig. (Fig.1A).1A). This showed that the expression of NS1 was readily detectable by 10 h p.i. (illustrated by the ΔNS2 mutant in Fig. Fig.1A),1A), whereas NS2 was detected less efficiently (illustrated by the ΔNS1 mutant). The antiserum used here detects NS1 much more efficiently than NS2, as we have observed in previous experiments (unpublished data) and as was confirmed in the present study by Western blot analysis (e.g., see Fig. Fig.3B).3B). In wt rRSV-infected cells, the percentage of cells in which expression of NS1 and NS2 was readily detected increased from ~8% to 27% from 10 to 14 h p.i. (Fig. (Fig.1B),1B), which coincided with the loss of IRF-3 from the nucleus. This suggests that the de novo synthesis of NS1 and NS2 in wt rRSV-infected cells reached a sufficient level by ~10 h p.i. to block further activation and translocation of IRF-3, so that the normal deactivation pathway led to a rapid depletion of nuclear IRF-3.
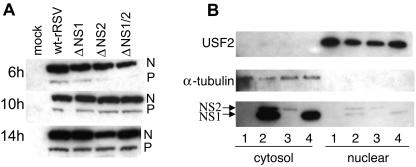
Viral protein expression in A549 cells infected with wt rRSV and the NS1/NS2 gene deletion mutants. (A) Wt rRSV and the NS1/NS2 deletion mutants display comparable levels of gene expression, as illustrated by the levels of N and P proteins. A549 cells were infected with the indicated viruses at an MOI of 3 PFU/cell and harvested 6 h, 10 h, or 14 h postinfection. Equivalent amounts of the cell lysates were subjected to SDS-PAGE and Western blot analysis with a polyclonal serum raised against purified RSV virions. The portion of the blot containing the N and P proteins is shown. (B) NS1 and NS2 are located predominantly in the cytosolic fraction of wt-rRSV-infected A549 cells. Cells were either mock infected (lanes 1) or infected with wt rRSV (lanes 2), ΔNS1 (lanes 3), or ΔNS2 (lanes 4) at an MOI of 3. The cells were harvested 20 h postinfection, and nuclear and cytosolic fractions were prepared and analyzed by Western blotting. The nuclear fraction is represented in fourfold-greater relative concentration than the cytosolic fraction. The efficacy of cell fractionation was confirmed using antibodies to USF2 as a marker for nuclear content and α-tubulin as a marker for cytosolic content. The localization of NS1 and NS2 was identified using a polyclonal antiserum that reacts with both proteins but with NS1 more efficiently than NS2.
The effects of NS1 and NS2 on the activation of IRF-3 were also investigated using a reporter plasmid containing the luciferase gene under the control of an IRF-3-responsive promoter. At 8 h p.i., there was significant IRF-3-responsive promoter activity in A549 cells infected with wt rRSV, ΔNS1, ΔNS2, and ΔNS1/2, with little difference between the various viruses (Fig. (Fig.2).2). However, there was no increase in IRF-3-responsive promoter activity between 8 h and 14 h p.i. in cells infected with wt rRSV. As luciferase activity is cumulative, this suggests that transcription from the IRF-3-responsive reporter plasmid had ceased. In contrast, cells infected with the NS1/NS2 gene deletion mutants exhibited a continued increase in IRF-3-responsive promoter activity between 8 h and 14 h p.i. This confirmed the results from the immunofluorescence study that wt rRSV and the NS1/NS2 gene deletion mutants each activated IRF-3 efficiently early in infection but that continued activation was observed only in cells infected by the NS1/NS2 gene deletion mutants.
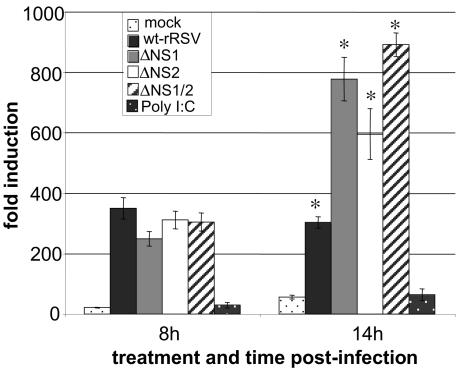
Activation of IRF-3 monitored using an IRF3-responsive reporter gene. A549 cells were transfected with p55CIBLuc, a reporter plasmid containing the luciferase gene under the control of the PRD I element of the IFN-β promoter. Ten hours posttransfection, the cells were mock infected or infected with the indicated viruses at an MOI of 1 PFU/cell. At 8 and 14 h postinfection, cells were harvested and assayed for luciferase activity. Relative light units are given as induction (n-fold) compared to cells that were transfected with the reporter plasmid but not mock infected or infected. The mean values and standard errors of three independent infections are shown. The indicated (*) value for the ΔNS1, ΔNS2, or ΔNS1/2 virus was confirmed to be significantly different from that of wt RSV (P = 1.6 × 10−5, 0.004, or 2.1 × 10−8, respectively; Student's t test).
We considered the possibility that the continued activation of IRF-3 in cells infected with the NS1/NS2 gene deletion mutants might somehow be due to altered kinetics of viral macromolecular synthesis. However, Western blot analysis of cell lysates harvested at various times following infection with wt rRSV or the gene deletion viruses showed that there was little difference between viruses in the level of accumulation of the N and P proteins, as markers of RNA replication and gene expression (Fig. (Fig.3A).3A). Furthermore, it was demonstrated in a previous study (31) that the intracellular accumulations of viral genomic RNA were similar for wt rRSV and the NS1/2 mutants.
The patterns of immunofluorescence of NS1 and NS2 in infected cells indicated that the two proteins accumulated significantly in the vicinity of the nucleus (Fig. (Fig.1A),1A), although the staining pattern was not clearly indicative of nuclear accumulation compared to that of IRF-3. If nuclear accumulation of NS1 or NS2 occurred, it raised the possibility that they might directly affect events in the IRF-3 activation-deactivation cycle in that compartment. However, fractionation of A549 cells infected with wt rRSV, ΔNS1, or ΔNS2 showed that in each case NS1 and NS2 accumulated mostly in the cytoplasm (Fig. (Fig.3B3B).
The NS2 protein is important for activating NF-κB.
NF-κB can be efficiently activated by a variety of inducers that include IFN-α/β, which strongly up-regulates the expression of the double-stranded-RNA-activated protein kinase PKR (41). Since wt rRSV and the NS1/NS2 gene deletion mutants vary considerably in their abilities to induce IFN-α/β (31), in particular with mutants lacking NS1 being capable of inducing a very high level, we controlled for this variable by investigating the activation of NF-κB in Vero cells, which lack the structural genes for IFN-α/β. Translocation of NF-κB to the nucleus was monitored by indirect immunofluorescence at various intervals p.i. using a mouse monoclonal antibody specific to p65 (Fig. 4A and B), which is the major form activated in response to HRSV (3). The inactive form of NF-κB, which is sequestered in a complex in the cytoplasm, could be detected as a diffuse staining pattern at each time point (Fig. (Fig.4A).4A). In cells infected with wt rRSV, ΔNS1, ΔNS2, or ΔNS1/2, nuclear accumulation of NF-κB was evident in each case by 6 h p.i. (Fig. (Fig.4A).4A). Unexpectedly, however, the level of nuclear accumulation of NF-κB was significantly lower in cells infected with ΔNS2 or ΔNS1/2, each of which lacks the NS2 gene. Thus, the presence of the NS2 gene, and presumably expression of its protein, appeared to be important in the activation of NF-κB. However, the effect was not absolute, since there was a residual level of activation by the ΔNS2 viruses. This background was dependent on viral macromolecular synthesis, as indicated by the lack of NF-κB activation by UV-inactivated virus.

Nuclear localization of NF-κB in Vero cells in response to infection with wt rRSV or the NS1/NS2 gene deletion mutants. (A) Vero cells were mock infected or infected at an MOI of 3 PFU/cell with either wt rRSV, ΔNS1, ΔNS2, ΔNS1/2, or the equivalent amount of UV-inactivated wt rRSV. The cells were fixed at the indicated times postinfection, permeabilized, and analyzed by immunofluorescence with a monoclonal antibody against NF-κB p65 (green) or a polyclonal antiserum against NS1 and NS2 (red) that reacted more strongly with NS1 than with NS2. (B) The percentage of cells with nuclei positive for NF-κB or that were positive for NS1/NS2 at each time point and for each treatment was calculated based on multiple fields of view from two independent experiments. The indicated value of NF-κB for the ΔNS2 or ΔNS1/2 virus was confirmed to be significantly different from that of wt RSV at 10 h (*, P = 5 × 10−6 and 5.7 × 10−6, respectively; Student's t test) and at 14 h (**, P = 3.3 × 10−6 and 2.9 × 10−6, respectively). In contrast, the value for the ΔNS1 mutant was not significantly different from that for wt RSV at each time point.
The effect of NS2 on the activation of NF-κB was also investigated using a reporter plasmid containing the luciferase gene under the control of an NF-κB-responsive promoter (Fig. (Fig.5).5). This showed that the magnitude of activation of the NF-κB-responsive promoter (measured in this example at 14 h p.i.) was greatest for cells infected with wt rRSV, was somewhat less for cells infected with ΔNS1, and was very low for cells that were mock infected or infected with ΔNS2 or ΔNS1/2.

NF-κB activation monitored with an NF-κB-responsive reporter gene. Vero cells were transfected with a luciferase construct driven by a promoter element responsive to NF-κB. Ten hours posttransfection, the cells were infected with the indicated viruses at an MOI of 1 PFU/cell. At 14 h postinfection, cells were harvested and assayed for luciferase activity. Relative light units are given as induction (n-fold). The results show the mean values and standard errors of three independent infections. The values for ΔNS2 and ΔNS1/2 were significantly different from that for wt RSV (*, P = 6.5 × 10−9 and 1.2 × 10−10, respectively) and ΔNS1 (**, P = 1.2 × 10−5 and 1.2 × 10−7, respectively).
We also prepared nuclear extracts from the infected Vero cells at various times and subjected them to Western blot analysis with an antibody against p65 as an independent method of monitoring NF-κB activation (Fig. (Fig.6A).6A). This confirmed that the nuclear accumulation of NF-κB p65 was substantially higher in cells infected with wt rRSV or ΔNS1 than in cells infected with ΔNS2 or ΔNS1/2 and confirmed that some activation occurred in the absence of NS2. Nuclear extracts were also analyzed using an antibody to USF2 to confirm the purity of the extracts (not shown).
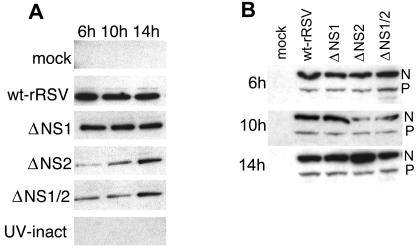
Western blot analysis of (A) the nuclear accumulation of NF-κB p65 and (B) the synthesis of RSV N and P proteins in Vero cells infected with wt rRSV or the NS1/NS2 gene deletion mutants. (A) Nuclear accumulation of NF-κB p65. Cells were mock infected or infected at an MOI of 3 PFU/cell with either wt rRSV, ΔNS1, ΔNS2, ΔNS1/2, or UV-inactivated wt rRSV. The nuclear fractions from replicate cultures were harvested at the indicated times, and equivalent amounts of protein were subjected to SDS-PAGE, followed by Western blotting with a monoclonal antibody against NF-κB p65. (B) Expression of RSV N and P proteins. Cells were infected with the indicated viruses and harvested 6 h, 10 h, and 14 h postinfection. Equal amounts of cell extract were subjected to SDS-PAGE and Western blot analysis of RSV N and P proteins using a polyclonal antiserum to purified virions.
In order to rule out the possibility that the low level of NF-κB activation in response to ΔNS2 and ΔNS1/2 might reflect reduced kinetics or magnitude of viral RNA replication and gene expression in Vero cells, Western blot analysis was used to measure the levels of accumulation of the N and P proteins as markers for viral RNA synthesis (Fig. (Fig.6B).6B). The accumulation of N protein at 10 h p.i. was somewhat lower for cells infected with ΔNS1 and ΔNS1/2, reflecting experimental variability. By 14 h p.i., the accumulations of both N and P proteins were similar for wt RSV and the gene deletion mutants.
Lack of activation of AP-1 by wt rRSV, ΔNS1, ΔNS2, or ΔNS1/2.
The activation of the transcription factor AP-1 in response to infection with wt rRSV and the NS1/NS2 gene deletion viruses was evaluated in Vero and A549 cells using a reporter plasmid under the control of an AP-1-responsive promoter (Fig. 7A and B, respectively). As a positive control, cells were cotransfected with a plasmid expressing the known activator MEK kinase, which activated AP-1 to a level 120 to 130 times higher than in mock-infected cells, confirming the efficiency of transfection. There was no significant increase in expression of the AP-1-dependent reporter plasmid in A549 or Vero cells infected with wt rRSV, ΔNS1, ΔNS2, or ΔNS1/2 compared to mock infection.

AP-1 activation monitored with an AP-1-responsive reporter gene was not detected within 14 h of infection with RSV. (A) Vero cells or (B) A549 cells were transfected with a luciferase reporter plasmid. Ten hours posttransfection, the cells were mock infected or infected with the indicated viruses at an MOI of 1 PFU/cell. At 14 h postinfection, cells were harvested and assayed for luciferase activity. Relative light units are given as induction (n-fold) compared to cells that were transfected with the reporter plasmid but were not infected or mock infected. Results show the mean values and standard errors of three independent infections.
Secretion of cytokines from A549 cells infected with the NS1/NS2 gene deletion mutants.
Activation of IRF-3 and NF-κB and the production of IFN-α/β led to changes in the expression of a broad array of cellular genes, including ones involved in inflammation and innate and adaptive immunity. The deletion of NS1 and/or NS2 had the potential for unpredictable effects on the epithelial cell response: for example, since IRF-3 can up-regulate transcription of the RANTES chemokine (22), vaccine candidates in which NS1 or NS2 have been deleted might have the potentially undesirable attribute of increased RANTES expression. We therefore investigated the expression of three proinflammatory factors by A549 cells in response to infection with wt rRSV and the NS1/NS2 gene deletion mutants: the chemokine RANTES, the chemokine IL-8, and the cytokine TNF-α. We chose these factors, as they have been suggested to contribute to disease in RSV-infected infants (17, 30). Our goal was not to attempt to dissect the signaling pathways or determine their relative contributions to the expression of each cytokine, but rather to determine the gross effect of the NS1 and NS2 gene deletions on the human epithelial cell response. ELISA of medium supernatants harvested at 16 and 20 h p.i. confirmed the secretion of IL-8, RANTES, and TNF-α in response to wt rRSV (Fig. 8A, B, and C), as has been previously observed (36, 42). This experiment showed that the secretion of IL-8, RANTES, and TNF-α in response to the gene deletion mutants was reduced compared to wt rRSV, providing an indication of safety for these vaccine candidates.
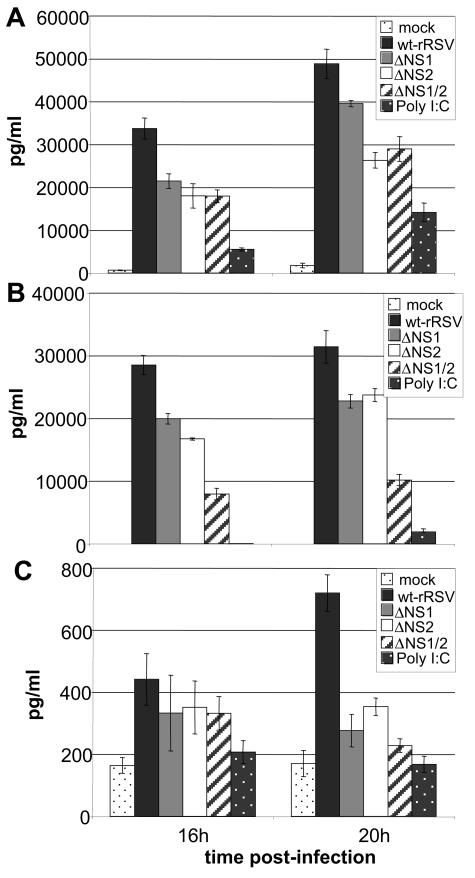
Secretion of (A) IL-8, (B) RANTES, and (C) TNF-α protein from A549 cells infected with wt rRSV, ΔNS1, ΔNS2, or ΔNS1/2; mock infected; or transfected with polypoly(I:C). Expression levels were measured with Human ELISA kits (BioSource) and calibrated using standards for each cytokine supplied with the kits. IL-8 secretion from cells infected with each of the deletion mutants was statistically lower than that from cells infected with wt RSV at both 16 h p.i. (P = 0.007 for ΔNS1 and ΔNS2 and 0.0006 for ΔNS1/2) and 20 h p.i. (P = 0.06 for ΔNS1 and 0.006 for ΔNS2 and ΔNS1/2). RANTES secretion from cells infected with each of the mutants was also statistically lower than secretion from cells infected with wt RSV at both 16 h p.i. (P = 0.007 for ΔNS1, 0.008 for ΔNS2, and 0.0003 for ΔNS1/2) and 20 h p.i. (P = 0.006 for ΔNS1, 0.007 for ΔNS2, and 0.0001 for ΔNS1/2). The secretion levels of TNF-α from cells infected with the mutants and wt RSV were statistically similar at 16 h p.i. (P = 0.50, 0.49, and 0.34 for ΔNS1, ΔNS2, ΔNS1/2, respectively, compared to wt RSV). However, at 20 h p.i., secretion was significantly lower from cells infected with the mutants (P = 0.003 for ΔNS1, 0.002 for ΔNS2, and 0.004 for ΔNS1/2).
DISCUSSION
Infection by viruses triggers the activation of host cell signaling pathways, including gene products that contribute to rapid intracellular defense against infection and influence innate and adaptive immune responses. Type 1 interferons (IFN-α/β) are part of this defense and establish an antiviral state within infected and neighboring cells. The IFN-α/β response in human cells first involves the production of IFN-β and -α1. Transcription of IFN-β and -α1 is mediated by the transcription factors IRF-3, NF-κB, and AP-1. Once secreted, IFN-β and -α1 bind to cell surface receptors in an autocrine and paracrine fashion. This induces the JAK/STAT signaling pathway, which produces the full complement of IFN-αs, additional IFN-β, and a wide array of interferon-stimulated genes, including ones that establish an intracellular antiviral state. IRF-3 and NF-κB also directly mediate the transcription of certain interferon-stimulated genes and other cellular genes with host defense functions. For example, IRF-3 can directly activate transcription of the RANTES chemokine (22), and NF-κB has been shown to mediate antiviral protection against HRSV in an epithelial cell line independently of IFN-α/β (4).
Viruses have evolved mechanisms to counteract the IFN-α/β response that may involve blocking initial induction, blocking JAK/STAT signaling, or inhibiting the antiviral state at some subsequent step (11, 13). We have shown previously (31) that the NS1 and NS2 proteins of HRSV inhibit the induction of IFN-α/β. This had also been shown for the NS1 and NS2 proteins of BRSV, the bovine counterpart of HRSV (5, 6, 28, 37). For each virus, the proteins exerted their inhibitory activities individually, as well as cooperatively, although the systems were not exactly parallel in that NS1 played the greater role for HRSV compared to NS2 for BRSV.
In the present study, we found that wt HRSV resembled the ΔNS1, ΔNS2, and ΔNS1/2 deletion mutants in the ability to activate IRF-3 during the first ~10 h of infection. Thereafter, the activation of IRF-3 was strongly suppressed in wt HRSV-infected cells, whereas a high level of activation continued in cells infected with each of the gene deletion mutants. In normal cells, IRF-3 is present constitutively and is shuttled continually between the cytoplasm and nucleus but is predominantly cytoplasmic. In response to an inducer, such as viral dsRNA, IRF-3 is phosphorylated and nuclear import is increased over nuclear export. However, without continual stimulation, the phosphorylated form is exported from the nucleus and degraded by the proteosome pathway in the cytoplasm (21). This cycle of IRF-3 activation and degradation was evident in the present study using polypoly(I:C), where a strong activation and subsequent reversal were observed (Fig. 1A and B). Our interpretation of the loss of nuclear IRF-3 in cells infected with wt rRSV is that the accumulation of NS1 and NS2 proteins reached sufficient levels by ~10 h p.i. to block further activation, whereas the nuclear export and degradation part of the cycle continued. Studies with BRSV showed that NS1 and NS2 interfere with IRF-3 activation by blocking its phosphorylation, although the roles of the individual proteins were not investigated (6). It is reasonable to anticipate that the same mechanism holds for HRSV. The present study indicates that deletion of either NS1 or NS2 provides substantial activation of IRF-3, indicating that the two proteins function much more efficiently together than either does individually.
The inhibition of IRF-3 activation is employed by many viruses to suppress the induction of IFN-α/β. The V protein of simian virus 5 (SV5) interferes with both IRF-3 and NF-κB activation, resulting in poor IFN-β induction in wt-infected cells (14, 26). However, unlike HRSV, wt SV5 does not activate IRF-3 even in the early stages of infection, prior to viral protein accumulation. HRSV also differs from SV5 in that NF-κB is activated in wt-HRSV-infected cells. This may explain why wt RSV is a better inducer of IFN-α/β than SV5 and certain other paramyxoviruses (40). The Ebola virus nonstructural protein VP35 and the NS3/4A protease of hepatitis C virus also inhibit IFN-α/β production by inhibiting IRF-3 phosphorylation (2, 10). The herpes simplex virus type 1 protein ICP0 blocks the activation of both IRF-3 and IRF-7 (23); however, the mechanism for this remains to be determined.
We also examined the activation of NF-κB in response to wt rRSV and the gene deletion mutants. However, because these viruses differed greatly in their abilities to induce IFN-α/β (31), and because IFN-α/β can augment NF-κB activation, these experiments were performed in Vero cells, which lack the structural genes for IFN-α/β. Unexpectedly, we found that activation of NF-κB was substantially lower in cells infected with ΔNS2 and ΔNS1/2, which each lack the NS2 gene. This effect could not be attributed to an obvious difference in viral gene expression. This strongly suggests that NS2 is involved in NF-κB activation in HRSV-infected cells. The fact that this effect was observed in the context of a permissive HRSV infection, rather than from an individual expressed cDNA, for example, lends authenticity to the observation. Previously, the F glycoprotein was shown to activate NF-κB in human monocytes through the TLR4 cell surface receptor and CD14 (19). In the present study, UV-inactivated virus did not stimulate NF-κB activation, suggesting that binding of the virus to the epithelial cell and fusion with the plasma membrane were not sufficient to significantly activate NF-κB. Expression of the HRSV P protein from a transfected plasmid also was reported to activate NF-κB (3). In the present study, a low level of NF-κB activation did occur in response to HRSV infection in the absence of NS2, and this presumably represents contributions from dsRNA and the F and P proteins, as well as whatever contribution might occur from oxidative stress or other potential inducers. Clearly, however, the NS2 protein appeared to play a major role.
In the present study we show that AP-1 is not significantly activated in response to RSV infection in either Vero or A549 cells. The limited responsiveness of AP-1 to viral infection has also been demonstrated for SV5 (26) and BRSV (6). The activation of AP-1 in HRSV-infected A549 cells has been shown to be sensitive to the redox state (24), and it might be that variability in the reported results represents differences in the conditions for cell culture. In any event, the constitutive level of AP-1 activation in our experiments was ample for a very robust IFN-α/β response (31).
There is presently no available licensed HRSV vaccine. However, live attenuated viruses containing deletion of the NS1 and/or NS2 genes, among other attenuating mutations, are under development and appear to be promising candidates (34, 38). One of the advantages of the NS1/NS2 deletion mutants as vaccine candidates is the promotion of IFN-α/β expression, which can enhance both innate and adaptive immune responses (31). However, a number of proinflammatory cytokines and chemokines contain NF-κB or IRF-3 binding sites in their promoters (17, 22) and can also be upregulated indirectly by IFN-α/β.
The recent use of cDNA and oligonucleotide microarrays has demonstrated that airway epithelial cells (A549 and primary small airway epithelial cells) infected with RSV show increased expression of a number of chemokines, including I-309, Exodus-1, TARC, RANTES, MCP-1, MDC, MIP-1α/β, IL-8, GRO α/β/γ, ENA-78, I-TAC, and fractalkine (25, 36, 42). It has also been demonstrated that human infants with RSV infection and bronchiolitis have increased secretion of IL-8, MCP-1, TNF-α, eotaxin, MIP-1α, and RANTES (17, 30). Vaccine candidate viruses that stimulate sustained IRF-3 activation, elevated NF-κB levels, and/or elevated IFN-α/β production have the potential to stimulate increased production of proinflammatory chemokines and cytokines that have been hypothesized to contribute to bronchiolitis and airway hyperreactivity. However, in the present study, the NS1/NS2 deletion mutants were found to be associated with reduced, rather than augmented, production of IL-8, RANTES, and TNF-α from A549 cells. This provides additional support for the development of viruses lacking NS1 and/or NS2 as safe live attenuated vaccines.
Acknowledgments
We thank T. Fujita, Tokyo Metropolitan Institute of Medical Science, for the kind gift of the luciferase reporter plasmid p55CIBLuc, and Siddhartha Mahanty, NIAID, for helpful discussion and for reviewing the manuscript.
REFERENCES
 and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [Abstract] [Google Scholar]
and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [Abstract] [Google Scholar]Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.79.9.5353-5362.2005
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/79/9/5353.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
[Vaccination against respiratory syncytial virus (RSV)-For the protection of infants and older adults].
Inn Med (Heidelb), 65(11):1066-1075, 25 Oct 2024
Cited by: 0 articles | PMID: 39453450
Review
Respiratory Syncytial Virus Vaccine Design Using Structure-Based Machine-Learning Models.
Viruses, 16(6):821, 22 May 2024
Cited by: 1 article | PMID: 38932114 | PMCID: PMC11209532
Mutations in the F protein of the live-attenuated respiratory syncytial virus vaccine candidate ΔNS2/Δ1313/I1314L increase the stability of infectivity and content of prefusion F protein.
PLoS One, 19(4):e0301773, 09 Apr 2024
Cited by: 0 articles | PMID: 38593167 | PMCID: PMC11003679
Clinical research on RSV prevention in children and pregnant women: progress and perspectives.
Front Immunol, 14:1329426, 24 Jan 2024
Cited by: 1 article | PMID: 38327765 | PMCID: PMC10847284
Review Free full text in Europe PMC
The implication of infection with respiratory syncytial virus in pediatric recurrent wheezing and asthma: knowledge expanded post-COVID-19 era.
Eur J Clin Microbiol Infect Dis, 43(3):403-416, 28 Dec 2023
Cited by: 1 article | PMID: 38153660
Review
Go to all (187) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness.
J Virol, 79(14):9315-9319, 01 Jul 2005
Cited by: 198 articles | PMID: 15994826 | PMCID: PMC1168759
Tumour necrosis factor-alpha and interferon-gamma synergistically activate the RANTES promoter through nuclear factor kappaB and interferon regulatory factor 1 (IRF-1) transcription factors.
Biochem J, 350 Pt 1:131-138, 01 Aug 2000
Cited by: 73 articles | PMID: 10926836 | PMCID: PMC1221234
The NS2 protein of human respiratory syncytial virus suppresses the cytotoxic T-cell response as a consequence of suppressing the type I interferon response.
J Virol, 80(12):5958-5967, 01 Jun 2006
Cited by: 31 articles | PMID: 16731934 | PMCID: PMC1472589
Respiratory syncytial virus mechanisms to interfere with type 1 interferons.
Curr Top Microbiol Immunol, 372:173-191, 01 Jan 2013
Cited by: 48 articles | PMID: 24362690
Review





