Abstract
Free full text

Chromatin Assembly Factor I Mutants Defective for PCNA Binding Require Asf1/Hir Proteins for Silencing
Abstract
Chromatin assembly factor I (CAF-I) is a conserved histone H3/H4 deposition complex. Saccharomyces cerevisiae mutants lacking CAF-I subunit genes (CAC1 to CAC3) display reduced heterochromatic gene silencing. In a screen for silencing-impaired cac1 alleles, we isolated a mutation that reduced binding to the Cac3p subunit and another that impaired binding to the DNA replication protein PCNA. Surprisingly, mutations in Cac1p that abolished PCNA binding resulted in very minor telomeric silencing defects but caused silencing to be largely dependent on Hir proteins and Asf1p, which together comprise an alternative silencing pathway. Consistent with these phenotypes, mutant CAF-I complexes defective for PCNA binding displayed reduced nucleosome assembly activity in vitro but were stimulated by Asf1p-histone complexes. Furthermore, these mutant CAF-I complexes displayed a reduced preference for depositing histones onto newly replicated DNA. We also observed a weak interaction between Asf1p and Cac2p in vitro, and we hypothesize that this interaction underlies the functional synergy between these histone deposition proteins.
Chromatin assembly factor I (CAF-I) is a heterotrimeric protein complex that delivers histones H3 and H4 to DNA ( 43, 54) during DNA replication or DNA repair ( 6, 7, 16). Both the biochemical activity and primary structure of CAF-I have been evolutionarily conserved from budding yeast to humans ( 18 19 20, 33, 51). The human CAF-I subunit genes for p150, p60, and p48 are homologous to the yeast CAC1, CAC2, and CAC3 genes, respectively. The specificity of CAF-I for replicated DNA molecules is imparted by specific recognition of the DNA polymerase processivity factor PCNA by the large subunit of CAF-I in both yeast and human cells ( 37, 61).
In yeast, CAC gene deletions result in reduced position-dependent gene silencing at all three silenced loci: telomeres, the silent mating loci, and ribosomal DNA ( 4, 5, 19, 26, 41). Because mutations in histone termini ( 21, 24; reviewed in reference 10) and histone-modifying enzymes (reviewed in references 8 and 38) affect the magnitude of position-dependent gene silencing, this chromatin-mediated phenomenon is analogous to heterochromatic silencing in multicellular organisms. In mammalian cells, CAF-I interacts directly with heterochromatin protein 1 (HP1) and localizes to heterochromatin in late S phase ( 29, 47). Furthermore, overexpression of a fragment of the p150 CAF-I subunit interferes with chromatin-mediated transcriptional silencing in mouse cells ( 48). Together, these data suggest a conserved role for CAF-I in heterochromatin formation.
The yeast ASF1 gene is a member of a second histone deposition pathway important for chromatin assembly that largely overlaps the role of CAF-I in silencing. ASF1 was identified in two independent genetic screens for high-dosage disrupters of position-dependent gene silencing ( 23, 40). The Drosophila homologue of Asf1p was purified in a complex with acetylated forms of histones H3 and H4 as a factor that stimulates the nucleosome assembly activity of CAF-I in vitro ( 51). Yeast Asf1p also stimulates the activity of CAF-I in vitro ( 36), demonstrating that this activity is conserved. asf1Δ mutants display minor defects in heterochromatic gene silencing ( 23, 40, 51). However, cacΔ asf1Δ double mutants display synergistic reductions in heterochromatic gene silencing, consistent with the in vitro synergy data ( 51).
HIR genes, first identified as transcriptional repressors ( 31, 58), are part of the ASF1-mediated silencing pathway ( 36). Like cacΔ asf1Δ cells, cacΔ hirΔ double mutant cells display synergistic reduction of position-dependent gene silencing at both telomeres and the silent mating loci, exhibit slow growth after germination, and display increased sensitivity to methyl methanesulfonate ( 17, 32). These phenotypes occur regardless of which of the three cacΔ or four hirΔ gene deletions are combined ( 17, 32), suggesting that CAF-I and the Hir proteins function as independent complexes. Hir1p and Hir2p proteins interact with Asf1p in vitro and in cell extracts ( 36); in vivo interactions between Hir1p and Asf1p have been observed in two-hybrid experiments ( 46). Also, some mutant PCNA proteins block the contribution of ASF1 and HIR genes to telomeric silencing ( 36). Thus, silencing by both the CAF-I and Asf1p/Hir pathways is a PCNA-dependent process.
To better understand the contribution of CAF-I to heterochromatin formation, we conducted a genetic screen for silencing-impaired alleles of the CAC1 gene. Analysis of one allele resulted in discovery of an amino acid required for interaction between the Cac1p and Cac3p proteins. Another allele, termed cac1-13, encoded a single amino acid change in a consensus PCNA-binding motif and impaired binding to PCNA by Cac1p. Mutations in this motif caused mild telomeric silencing defects in otherwise wild-type cells, but caused dramatic silencing defects in the absence of ASF1 or the HIR1 genes. We also discovered a weak interaction between Asf1p and the Cac2p subunit of CAF-I in vitro. We propose that Asf1p/Hir proteins functionally assist CAF-I via this direct interaction and that this assistance is critical for nucleosome formation in vivo when the Cac1p-PCNA interaction is impaired.
MATERIALS AND METHODS
Plasmids.
For T7 promoter-driven Cac2p expression, a NotI fragment with a triple hemagglutinin tag ( 50) was cloned into a NotI site generated at the 3′ end of CAC2. A 1.4-kb NdeI fragment of this plasmid was inserted into pET15b (Novagen) to create pPK83. For T7 promoter-driven Cac1p expression, the 5′ end of the CAC1 gene was amplified by PCR to add a 2× polyomavirus epitope (PY) tag ( 35) to the N terminus using the oligonucleotides 5′-CCGGATCCGAAAATGGAATATATGCCAATGGGAATGGAATACATGCCTATGGAAATGGAGCAACATCTCTC-3′ and 5′-TAAAAGCTTGTCTTCAGC-3′. A 2.3-kb BamHI fragment containing the PY2-CAC1 gene was cloned into BamHI-digested pRSET-B (Invitrogen) to generate pPK122. For T7 promoter-driven Cac3p expression, the BamHI-SalI fragment from pPK115 ( 36) was cloned into BamHI-XhoI-digested pRSET-B to create pPK123.
For expressing glutathione S-transferase (GST) fusion proteins, fragments of the CAC1 gene were cloned into pGEX-KG ( 11); details of constructs used are available upon request. All GST fusion-expressing plasmids were sequenced to ensure that no errors were introduced during subcloning or PCR.
Yeast strains and medium.
All strains used in this study are isogenic with the W303 genetic background ( 49), with the genotype leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100. The URA3-VIIL telomere and cac1Δ, hir1Δ, and asf1Δ gene deletions in W303 were described previously ( 17, 19, 36). The ADE2-VR telomere was integrated into the W303 background using plasmid pHR10-6 ( 39). Standard procedures were used for genetic crosses and tetrad analysis, and standard yeast medium was used for scoring genetic marker segregation ( 15). SD is synthetic dextrose medium; 5-fluoro-orotic acid (FOA) was added to SD medium at a concentration of 1 mg/ml; medium with low adenine contained 5 mg/liter.
Yeast strains used in this study include PKY620 (MATa cac1Δ::hisG ADE2-VR), PKY557 (MATa cacΔ1::LEU2 ADE2-VR URA3-VIIL), PKY618 (MATα cac1Δ::hisG hir1Δ::HIS3 ADE2-VR URA3-VIIL), PKY619 (MATα cac1Δ::hisG ADE2-VR URA3-VIIL), PKY950 (MATα cac1Δ::LEU2 asf1Δ::HIS3,URA3-VIIL), and PKY2095 [MATα cac1Δ::hisG hir1Δ::HIS3 ade3Δ (hht1-hhf1)-1216/pPK107 (URA3 ADE3 CAC1 ARS/CEN)]. The (hht1-hhf1)-1216 allele was obtained in a genetic screen and causes viability of hir1Δ strains to be dependent on the CAC1 gene.
The CAC1-FLAG allele described previously ( 36) was shown to fully complement the telomeric silencing and UV sensitivity phenotypes of a cac1Δ strain (data not shown). Furthermore, we observed that the CAC1-FLAG allele restores wild-type levels of telomeric silencing to a cac1Δ hir1Δ strain, which is a more sensitive assay for CAC1 function ( 17).
Telomeric silencing assays using the URA3-VIIL reporter ( 9) were performed as previously described ( 36).
Mutagenesis of CAC1 and recovery of silencing-defective alleles.
In vitro PCR-based mutagenesis of CAC1 was performed as described ( 27); details of oligonucleotides used and PCR conditions are available upon request. Yeast strain PKY2095 was transformed simultaneously with mutagenized PCR products and XhoI- and BamHI-digested pPK150 (CAC1 TRP1 ARS/CEN) ( 36); colonies were grown on medium lacking tryptophan to select for recombination of the mutant alleles into the plasmid backbone. A total of 236 colonies were either replica-plated or patched to −Trp +FOA plates to cure PKY2095 of the wild-type CAC1 URA3 plasmid pPK107. Because (hht1-hhf1)-1216 cells require CAC1 for viability, transformants capable of growth on −Trp +FOA contained nonnull cac1 alleles. Trp+, FOA-resistant transformants were then mated to PKY620, which contains an ADE2-VR-marked telomere and wild-type HIR1 and HHT1-HHF1 genes but lacks the CAC1 gene. The resulting diploids therefore contained a colony color marker to assay telomeric silencing, had both chromosomal copies of CAC1 deleted, contained a mutated cac1 allele on the plasmid, had one functional copy of the HIR1 gene, and had three out of four wild-type histone H3/H4 loci.
Diploid cells were streaked onto −Trp +low adenine plates to select for the plasmid and test for telomeric silencing defects. Candidate cac1 alleles were isolated from strains that appeared mostly white or sectored, indicating a silencing defect. The mutations responsible for silencing defects were mapped by subcloning and sequenced on both strands. All mutant proteins accumulated to levels similar to the wild-type protein (data not shown). Also, addition of the C-terminal FLAG tag did not alter the silencing phenotypes of any of the alleles (data not shown). Additional alleles generated by site-directed mutagenesis all maintained viability of strain PKY2095 in the absence of a wild-type CAC1 gene (data not shown).
Protein purification.
Overproduction and purification of yeast PCNA in Escherichia coli were performed as described ( 1). CAF-I, CafI-13, CafI-20, and the Cac1/3 complex were purified from infected Sf9 cells, GST-Asf1p and Asf1p were purified from E. coli, and the Asf1p + H3/H4 complex was formed and purified as described ( 36). Other GST fusion proteins were prepared essentially as described ( 18) (details are available on request), except that the glutathione-agarose resin was washed with buffer containing 300 mM NaCl prior to elution.
Biochemical experiments.
For GST fusion coprecipitation experiments with Cac1p, Cac2p, and Cac3p, 2.5 μg of GST or GST-Cac1p or 10 μl of GST-Asf1p were bound to 10 μl of glutathione agarose (Sigma) and incubated with 5 μl of 35S-labeled in vitro-translated Cac1p, Cac2p, or Cac3p (T7 single tube protein transcription/translation; Novagen) in 200 μl of buffer A with 25 mM NaCl and 50 μg of ethidium bromide per ml. Reactions were rotated at 4°C for 1 h, followed by three 1-ml washes in buffer A-500 mM NaCl (GST-Cac1p experiments and GST-Asf1p + Cac2p) or buffer A-25 mM NaCl (GST-Asf1p + Cac3p) or buffer A-250 mM NaCl (GST-Asf1p + Cac1p). Bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography. For GST fusion coprecipitation experiments with PCNA, 4 μg of GST-Cac1p was incubated with 35 ng of PCNA in 100 μl of buffer A-100 mM NaCl-50 μg/ml ethidium bromide. Reactions were rotated at 4°C for 1 h followed by three washes of 1 ml of buffer A-500 mM NaCl. Bound proteins were eluted with SDS sample buffer and separated by SDS-PAGE followed by Western analysis.
To coimmunoprecipitate CAF-I and Asf1p, 100 ng of affinity-purified CAF-I or Cac1/3 was incubated with 60 ng of Asf1p ± H3/H4 for 10 min at 30°C in a 30-μl reaction in buffer A-50 mM NaCl followed by addition of 10 μl of 50% anti-PY antibody ( 35), cross-linked to protein A-Sepharose ( 13) and 10 μl of 50% G50-Sepharose. This mixture was rotated at 4°C for 1 h, followed by three washes of 1 ml of buffer A-100 mM NaCl. Proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting.
Glycerol gradient sedimentation was performed by incubating 200 ng of CAF-I, 350 ng of Asf1p + H3/H4, and/or 1,000 ng of Asf1p in 30 μl of buffer A-50 mM NaCl at 30°C for 10 min, followed by addition of 70 μl of buffer A-50 mM NaCl. Each protein mixture was overlaid onto a 5-ml 10 to 40% glycerol gradient and centrifuged at 200,000 × g at 4°C for 24 h. The gradients were separated into 380-μl fractions, and 5 μl was separated by SDS-PAGE followed by immunoblot analysis. Protein standards were chymotrypsinogen A (25 kDa), chicken egg albumin (45 kDa), bovine serum albumin (68 kDa), aldolase (158 kDa), and catalase (240 kDa).
Nucleosome assembly assays and micrococcal nuclease (MNase) digestions were performed as described ( 19, 36), except that the human cells used to make the replication extracts were HeLa and not 293 cells.
RESULTS
Isolation of silencing-defective cac1 alleles.
To isolate silencing-defective cac1 alleles, we made use of our discovery that CAC1 is essential for viability in yeast strains that lack the HIR1 gene and contain a mutation in the HHT1-HHF1locus, one of the two histone H3- and H4-encoding loci (M. D. Adams and P. D. Kaufman, unpublished results) (Fig. (Fig.1A).1A). To ensure that our screen would yield nonnull cac1 alleles, we first selected cac1 alleles that maintained the viability of a haploid cac1Δ hir1Δ hht1-hhf1 strain (Fig. (Fig.1A).1A). Viable cells were then mated to cac1Δ ADE2-VR cells (Fig. (Fig.1B).1B). In the resulting diploid cells, the only source of CAC1 was the mutagenized gene. The diploids also contained one wild-type copy of the HIR1 and HHT1-HHF1 genes, so any silencing defects detected by the telomeric ADE2-VR reporter resulted solely from the mutant cac1 allele. In the secondary screen, mutant cac1 alleles that reduced silencing of the telomeric ADE2 gene were then isolated based on colony color (Fig. (Fig.1B;1B; see Materials and Methods). This screen resulted in isolation of six silencing-defective CAC1 alleles (termed cac1-11 through cac1-16), and we present data here concerning a subset of these.
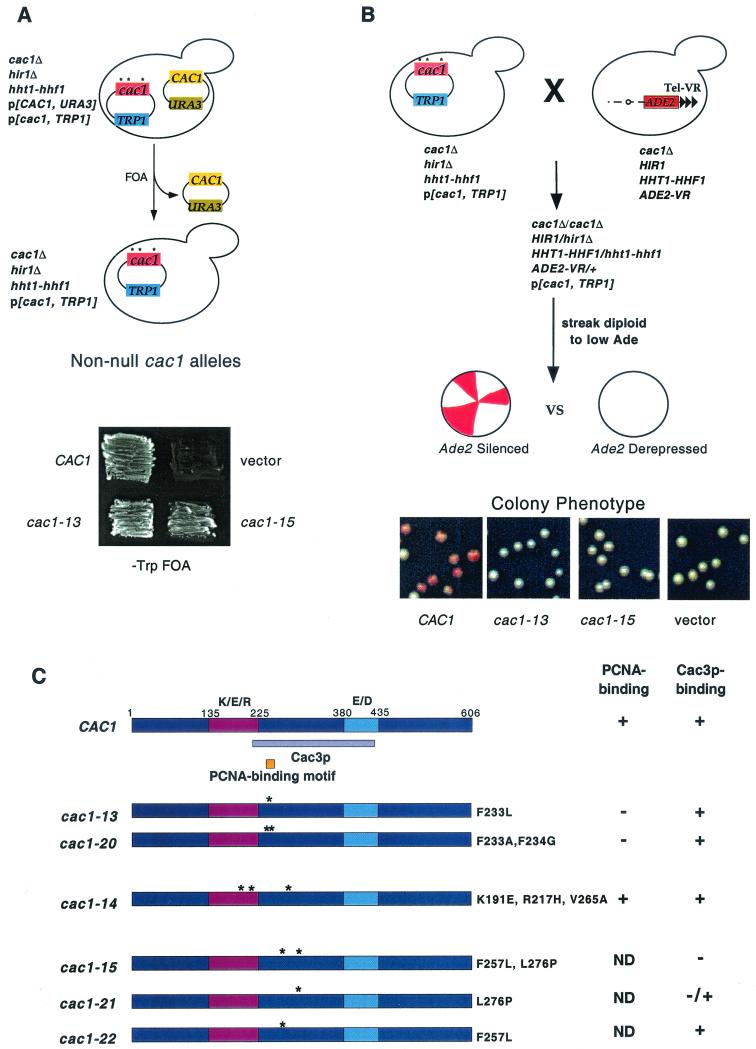
Isolation of silencing-impaired cac1 alleles. (A) Isolation of nonnull cac1 alleles (see text and Materials and Methods for details). Strain PKY2095 [cac1Δ hir1Δ (hht1-hhf1)-1216/pPK107 (URA3-CAC1)] was transformed with plasmids bearing the TRP1 gene and the indicated cac1 alleles or a vector control. Transformants were patched onto SC −Trp +FOA medium to select against plasmid pPK107 and incubated for 4 days at 30°C. Growth indicates the ability of the indicated cac1 allele to maintain viability of the hir1Δ (hht1-hhf1)-1216 cells. (B) Identification of cac1 alleles with telomeric silencing defects. Cells containing the ADE2 gene at telomere VR and the indicated cac1 alleles were plated on −Trp + low adenine plates and grown for 6 days at 30°C followed by 1 day at 4°C. (C) Map of the Cac3p (violet) and PCNA (orange) binding sites on Cac1p and locations of cac1 allele mutations. The highly charged K/E/R-rich and E/D-rich regions of the protein are indicated in purple and blue, respectively. Protein binding data from Fig. Fig.22 are summarized on the right. ND, not determined.
Mutation of the Cac3p-binding site of Cac1p.
The three alleles cac1-13, cac1-14, and cac1-15 each contained mutations within the region of Cac1p that we identify here as the Cac3p binding site (Fig. (Fig.1C).1C). To map the Cac3p binding site on Cac1p, we tested various GST-Cac1p protein fusions for the ability to coprecipitate in vitro-translated Cac3p. We observed that GST fused to Cac1p amino acids 87 to 429 would efficiently precipitate Cac3p and that Cac1p amino acids 215 to 429 were sufficient for this interaction (Fig. (Fig.2A,2A, lanes 2 and 7). In contrast, GST fused to Cac1p residues 87 to 306 or 427 to 606 or unfused GST did not bind Cac3p (Fig. (Fig.2A,2A, lanes 6, 8, and 9). We concluded that Cac1p residues 215 to 429 contain the Cac3 binding site (Fig. (Fig.1C1C).
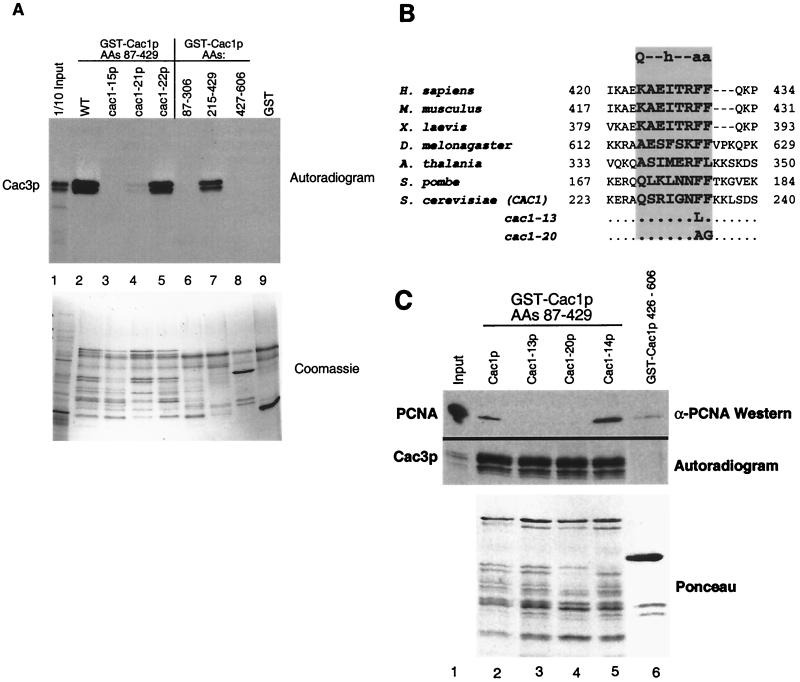
Protein binding defects of Cac1p mutants. (A) Cac1-15p does not bind Cac3p in vitro. GST fusion proteins (amino acids 87 to 429 of Cac1p, Cac1-15p, Cac1-21p, or Cac1-22p, amino acids 87 to 306, 215 to 429, or 427 to 606 of Cac1p, or an unfused GST control) were incubated with in vitro-translated Cac3p and washed extensively in the presence of 500 mM NaCl. Bound proteins were eluted with SDS sample buffer, separated by SDS-12.5% PAGE, and visualized by autoradiography. The input (lane 1) contains 1/10 of the amount of translation product input into the binding assays. The upper panel labeled autoradiogram shows in vitro-translated Cac3p, and the lower panel labeled Coomassie shows the input GST fusion proteins for the same experiment. The GST-Cac1p proteins were susceptible to proteolysis, resulting in the pattern of bands shown in the lower panel. (B) Evolutionary conservation of the consensus PCNA-binding motif in CAC1 homologues from human (GenBank accession number XM009408), mouse (NM013733), Xenopus laevis (AF222339), Drosophila melanogaster (AF367177), Arabidopsis thalania (AB027229), Schizosaccharomyces pombe (AL034463), and S. cerevisiae (NP015343). The consensus PCNA-binding motif is shown at the top of the shaded rectangle. Mutations in the cac-13 and cac1-20 alleles are indicated. Residues comprising the PCNA-binding motif are in bold. (C) Cac1-13p and Cac1-20p are defective in PCNA binding. GST fusion proteins (amino acids 87 to 429 of Cac1p, Cac1-13p, Cac1-14p, or Cac1-20p [lanes 2 to 5] or amino acids 427 to 606 of Cac1p [lane 6]) were incubated with purified recombinant yeast PCNA or in vitro-translated Cac3p. Samples were washed extensively in the presence of 500 mM NaCl, and bound proteins were eluted with SDS sample buffer and separated by SDS-15% PAGE. Cac3p was visualized by autoradiography, and PCNA was visualized by immunoblotting. The lower panel labeled Ponceau shows the input GST fusion protein for the PCNA coprecipitation experiment. For Cac3p, the input lane contains 1/30 of the amount of translation product input into the binding assays; for PCNA, the input lane contains 1/10 of the amount input (lane 1).
We then tested the effects of the mutations identified in the genetic screen on Cac3p binding by generating GST fusion proteins of amino acids 87 to 429 from each cac1 mutant. GST-Cac1-13p and GST-Cac1-14p bound to Cac3p with wild-type affinity (Fig. (Fig.2C).2C). In contrast, GST-Cac1-15p did not coprecipitate Cac3p (Fig. (Fig.2A,2A, lane 3). Cac1-15p contains two amino acid changes, F257L and L276P, and both reside within the Cac3p binding region. To determine which of these substitutions impaired Cac3p binding, we separated them, creating the cac1-21allele, encoding the single L276P substitution, and the cac1-22 allele, encoding the F257L change. GST-Cac1-21p only weakly associated with Cac3p; in contrast, GST-Cac1-22p bound Cac3p with wild-type efficiency (Fig. (Fig.2A,2A, lanes 4 and 5). We therefore concluded that the L276P substitution alone greatly impaired Cac3p binding. Thus, isolation of cac1-15 allowed us to identify a single amino acid important for interaction between Cac1p and Cac3p.
Mutation of the PCNA-binding site of Cac1p.
The cac1-13 allele encodes a single amino acid change at amino acid 233, converting a phenylalanine to leucine (Fig. (Fig.1C).1C). This mutation falls within a seven-amino-acid region, residues 227 to 234 (QSRIGNFF), which matches a characterized PCNA-binding motif [Qxx(I/L/V)xx(F/Y)(F/Y)] (Fig. (Fig.2B)2B) ( 55). The structure of a peptide containing this motif bound to human PCNA shows that the aromatic residues in this motif are buried in a hydrophobic pocket on the interdomain loop of PCNA ( 12). Furthermore, mutation of either of these aromatic residues to alanine in the cell cycle regulator p21 disrupts PCNA binding ( 30). We note that the aromatic residues of this motif are conserved in Cac1p homologs (Fig. (Fig.2B),2B), although the initial glutamine is not found in the genes from multicellular species (see Discussion).
Both human p150 and yeast Cac1p bind PCNA ( 25, 61), but the site of interaction on CAF-I had not been determined previously. To determine whether the conserved aromatic residues in Cac1p contribute to PCNA binding, we tested the interaction between mutant GST-Cac1p proteins and purified recombinant yeast PCNA. Consistent with previous experiments ( 61), GST-Cac1p amino acids 87 to 429 coprecipitated purified recombinant yeast PCNA (Fig. (Fig.2C,2C, lane 2). Notably, Cac1-13p was much less efficient at coprecipitating PCNA than was wild-type Cac1p. In contrast GST-Cac1-14p, which has mutations just outside of the PCNA-binding motif (amino acids 191, 217, and 265; see Fig. Fig.1C),1C), bound PCNA with wild-type affinity. GST-Cac1p and GST-Cac1-13p bound to Cac3p with equal efficiency, suggesting that Cac1-13p was specifically defective in PCNA binding (Fig. (Fig.2C,2C, autoradiogram). To more dramatically alter the PCNA-binding motif, we also directly changed both aromatic residues to create the cac1-20 allele (F233A, F234G; Fig. Fig.1C1C and and2B).2B). We observed that Cac1-20p also displayed reduced binding to PCNA but retained normal affinity for Cac3p (Fig. (Fig.2C,2C, lane 4). Therefore, Cac1-13p and Cac1-20p were specifically defective in PCNA binding, demonstrating that alterations of the canonical PCNA-binding motif disrupt the Cac1p-PCNA interaction.
Telomeric silencing phenotypes.
To ensure that the telomeric silencing defects observed in the genetic screen were not specific to the ADE2 gene at telomere VR, we also tested the cac1 alleles for telomeric silencing using the URA3 gene placed adjacent to telomere VIIL (Fig. (Fig.3A).3A). The drug FOA is toxic to cells expressing the URA3 gene ( 2); therefore, growth in the presence of FOA indicated that the URA3 gene was silenced. The URA3-VIIL assay also facilitated more quantitative differentiation of the silencing defects caused by the cac1 alleles, which could not be distinguished using ADE2-VR (compare Fig. Fig.1B1B and and3A).3A). For example, using URA3-VIIL, we observed that very mild reductions in telomeric silencing resulted from all alleles tested except cac1-15, which caused a strong defect (Fig. (Fig.3A).3A). Notably, although the amino acid changes in both the cac1-15 (F257L, L276P) and cac1-21 (L276P) alleles blocked Cac3p binding (Fig. (Fig.2A),2A), only cac1-15 caused a strong loss of silencing. Each mutant Cac1p protein accumulated to wild-type levels (data not shown); thus, differential protein stability was not responsible for the different silencing phenotypes. Therefore, we concluded that the strong silencing defect caused by cac1-15 required both the F257L and L276P amino acid changes.
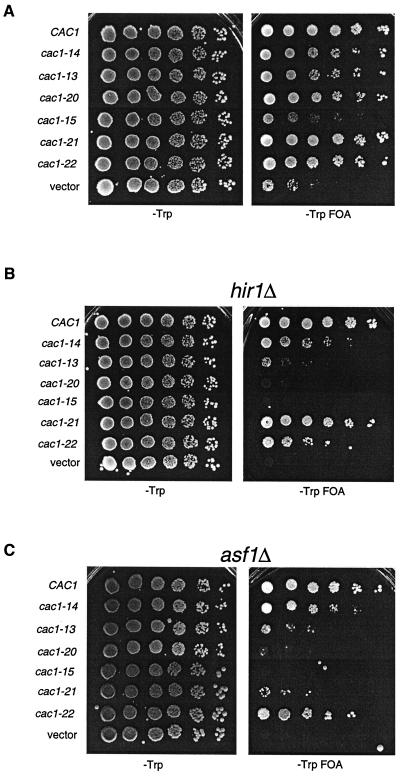
Telomeric silencing defects of cac1 alleles. Tenfold serial dilutions of cells containing the URA3-VIIL telomere and the indicated cac1 alleles were plated onto SC −Trp or SC −Trp +FOA plates and grown for 5 days at 30°C. Strains used were (A) PKY619 (cac1Δ), (B) PKY618 (cac1Δ hir1Δ), and (C) PKY950 (cac1Δ asf1Δ).
cac1 alleles sensitive to loss of ASF1 and HIR1 genes.
To further differentiate among the silencing-defective cac1 alleles, we sensitized the telomeric silencing assay by deletion of the ASF1 or HIR gene, both of which contribute to a silencing pathway that functionally overlaps CAF-I ( 17, 36, 51). As observed previously, deletion of either the HIR1 or ASF1 gene in the presence of a wild-type CAC1 gene had little effect on silencing. In contrast, combination of either asf1Δ or hir1Δ gene deletions with the cac1 alleles reduced the efficiency of telomeric silencing up to 100,000-fold, depending on the particular cac1 allele (Fig. (Fig.3B3B and and3C3C).
The hir1Δ and asf1Δ deletions had an especially dramatic effect on silencing in combination with the cac1 alleles that impair PCNA binding. In otherwise wild-type cells, the cac1-13 and cac1-20 alleles resulted in nearly wild-type telomeric silencing (Fig. (Fig.3A).3A). However, deletion of HIR1 or ASF1 reduced silencing in cac1-13 cells approximately 1,000-fold, and silencing in cac1-20 cells was reduced approximately 100,000-fold (Fig. 3B and C). In contrast, the cac1-14 allele, which did not impair PCNA or Cac3p binding (Fig. (Fig.2C),2C), displayed much milder silencing defects when combined with the hir1Δ or asf1Δ deletion. Similar results were obtained by combining these cac1 alleles with hir2Δ deletions (data not shown), consistent with genetic and biochemical evidence indicating that Hir proteins function as a complex ( 36, 44). We note that the stronger silencing defect in the cac1-20 cells is consistent with the more drastic alteration of the PCNA-binding site in this allele compared to cac1-13 (Fig. (Fig.2B).2B). These data indicated that mutation of the PCNA-binding motif of CAC1 caused telomeric silencing to be almost completely dependent on the ASF1 and the HIR genes.
The cac1-15 mutation caused a significant loss of silencing, similar to that observed in the absence of CAC1, and deletion of either HIR1 or ASF1 similarly exacerbated these phenotypes (Fig. (Fig.3).3). The nearly null phenotypes caused by cac1-15 were stronger than those of the separated point mutations cac1-21 and cac1-22. For example, deletion of either HIR1 or ASF1 did not strongly affect silencing in cac1-22 cells. However, the cac1-21 allele that blocked Cac3p binding (Fig. (Fig.2A)2A) had an intermediate phenotype. In this case, telomeric silencing in cac1-21 hir1Δ cells was nearly wild type, but silencing in cac1-21 asf1Δ cells was reduced almost 10,000-fold compared to CAC1 asf1Δ cells (Fig. (Fig.3C).3C). These data indicated that disruption of the Cac1p-Cac3p interaction led to a greater requirement for Asf1p than for Hir1p for telomeric silencing. Thus, the ASF1 and HIR genes have at least one separable function in telomeric silencing (see Discussion).
Stimulation of CAF-I complexes defective in PCNA binding by Asf1p/H3/H4.
CAF-I promotes DNA replication-dependent chromatin assembly in vitro ( 42), and the Asf1p/H3/H4 complex stimulates the chromatin assembly activity of CAF-I ( 36, 51). Our genetic data suggested that mutant CAF-I complexes defective in PCNA binding were particularly reliant on the Hir proteins and Asf1p. We therefore tested whether these mutant CAF-I complexes were capable of replication-coupled chromatin assembly and synergy with Asf1/H3/H4 complexes.
Wild-type yeast CAF-I, CafI-13 (Cac1-13p, Cac2p, and Cac3p) and CafI-20 (Cac1-20p, Cac2p, and Cac3p) were overproduced in insect cells and purified to homogeneity by ion exchange and epitope affinity chromatography using a C-terminal epitope on Cac1p (Fig. (Fig.4A).4A). Purified CAF-I complexes were tested for chromatin assembly activity in an assay that detects nucleosome-mediated supercoiling of newly replicated DNA templates ( 45). In this assay, wild-type or mutant CAF-I was added to an in vitro DNA replication reaction containing a plasmid with a simian virus 40 (SV40) origin of replication, the SV40 replication initiation protein T-antigen, [α-32P]dATP to label newly replicated DNA, and HeLa S100 extract which contains proteins required for DNA replication.
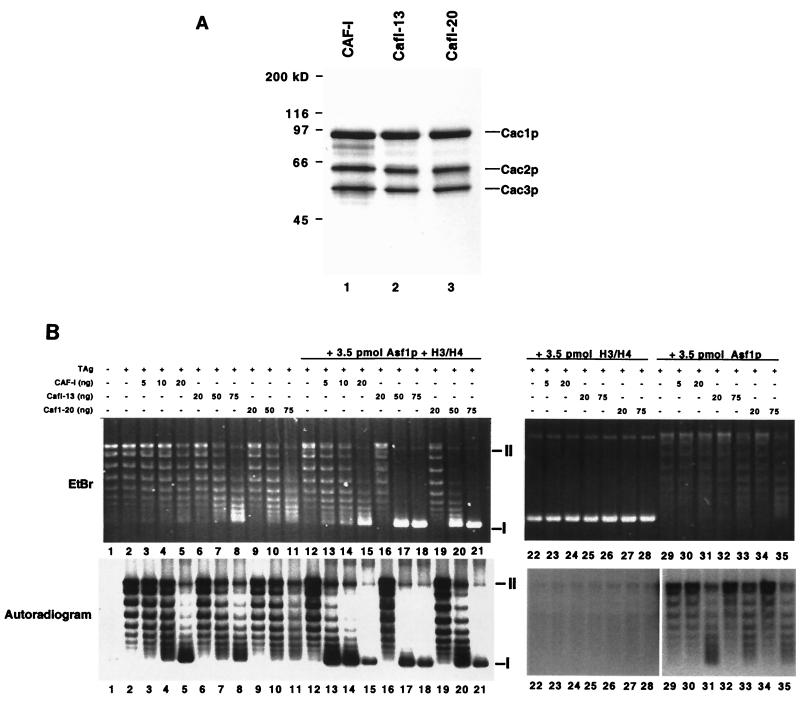
Chromatin assembly activity of CafI-20 complexes. (A) Purified recombinant CAF-I complexes. Purified CAF-I, CafI-13, and CafI-20 (2 μg) were separated by SDS-10% PAGE and stained with Coomassie blue. (B) Chromatin assembly activity. SV40 DNA replication reactions (50 μl) containing 100 ng of DNA (52 fmol of plasmid molecule) were performed with CAF-I complexes alone (lanes 1 to 11) or with the addition of 3.5 pmol of Asf1p/H3/H4 (lanes 12 to 21), H3/H4 alone (lanes 22 to 28), or Asf1p alone (lanes 29 to 35). Indicated amounts of purified CAF-I, CafI-13, or CafI-20 were added (10 ng of CAF-I complex = 59 fmol). The upper panel shows total DNA visualized by ethidium bromide (EtBr) staining; the lower panel shows replicated DNA visualized by autoradiography (autoradiogram). Migration of supercoiled form I DNA molecules is indicated by the symbol I at the right; migration of nicked form II molecules is indicated by the symbol II. Reactions lacking both the replication initiator T antigen and CAF-I or lacking only CAF-I are shown in lanes 1 and 2, respectively.
In the absence of CAF-I, the newly replicated DNA molecules visualized by autoradiography had a topoisomer distribution similar to that of the total DNA visualized by ethidium bromide staining (Fig. (Fig.4B,4B, lane 2). However, in the presence of sufficient CAF-I, most of the replicated DNA was recovered as form I supercoiled DNA, although most of the total DNA in the reactions was not supercoiled (Fig. (Fig.4B,4B, lane 5). Therefore, wild-type yeast CAF-I preferentially assembled nucleosomes onto newly replicated DNA molecules.
CAF-I functions stoichiometrically during nucleosome assembly ( 18, 22, 36, 42), so that addition of substoichiometric amounts of CAF-I results in incomplete supercoiling of DNA templates. Consistent with previous data ( 36), addition of 20 ng (120 fmol) of recombinant wild-type yeast CAF-I was required to promote efficient supercoiling of replicated templates in reactions containing 52 fmol of total plasmid molecules (Fig. (Fig.4B,4B, lane 5). In contrast, addition of 20 ng of either CafI-13 or CafI-20 did not result in supercoiling (Fig. (Fig.4B,4B, lanes 6 and 9). Addition of larger amounts (75 ng [0.44 pmol]) of CafI-13 or CafI-20 did result in increased DNA superhelicity (Fig. (Fig.6B,6B, lanes 8 and 11), but this effect was not specific to replicated DNA molecules, as the total DNA visualized by ethidium bromide displayed increased migration in these cases. These data indicate that mutation of the PCNA-binding site of Cac1p reduced the specific activity of CAF-I, and particularly interfered with the preference for newly replicated DNA molecules.
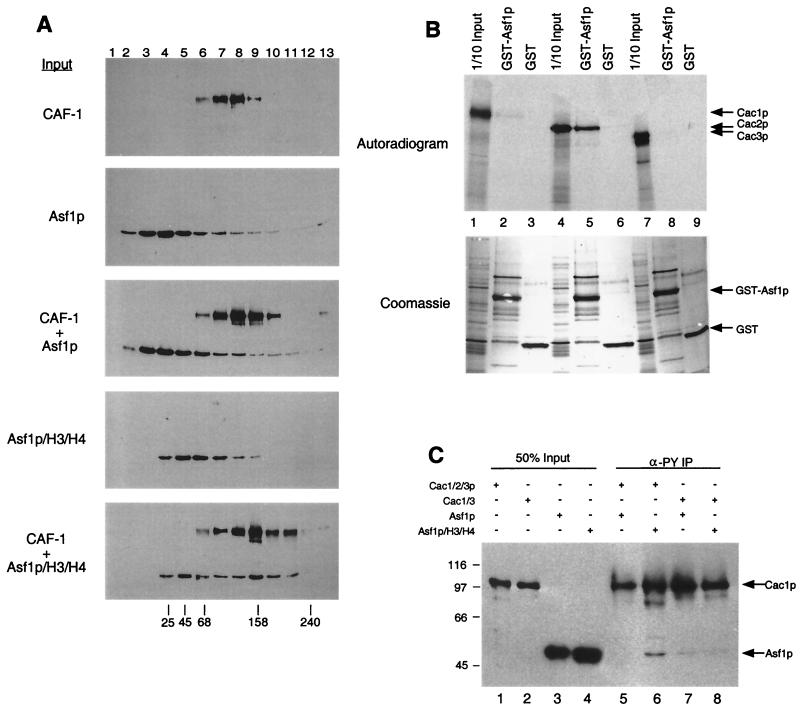
Direct physical interaction between Asf1p and CAF-I. (A) Glycerol gradient sedimentation of CAF-I and Asf1p. Purified recombinant CAF-I, Asf1p, or Asf1p complexed with histones H3 and H4 were sedimented on linear 10 to 40% glycerol gradients. Thirteen fractions were collected and separated by SDS-12.5% PAGE followed by immunoblotting with an anti-FLAG-M2 epitope antibody (Sigma). Cac1p and Asf1p were both tagged with the FLAG epitope. Sedimentation of molecular size standards is indicated at the bottom (in kilodaltons). (B) Asf1p binds Cac2p in vitro. GST-Asf1p was incubated with in vitro-translated Cac1p (lanes 1 to 3), Cac2p (lanes 4 to 6), or Cac3p (lanes 7 to 9). Samples were washed extensively (see text); bound proteins were eluted with SDS sample buffer, resolved by SDS-12.5% PAGE, and visualized by autoradiography. Input lanes represent 1/10 of the protein added to the reactions. (C) The Asf1p/H3/H4 complex coimmunoprecipitates with CAF-I. Purified recombinant CAF-I (Cac1/2/3p), Cac1/3p, Asf1p, or Asf1p/H3/H4 was mixed and immunoprecipitated with the anti-PY antibody (which recognizes PY-Cac3p). Samples were analyzed by SDS-12.5% PAGE followed by immunoblotting using anti-FLAG-M2 epitope antibody, which recognized Cac1p and Asf1p.
To confirm that the DNA supercoiling that we observed resulted from nucleosome formation, the reaction products were digested with MNase. MNase preferentially cleaves chromatin in the linker region between nucleosomes, resulting in a characteristic ladder of DNA products ( 34). In the absence of CAF-I, some mononucleosome- and subnucleosome-sized DNA fragments were protected from digestion, indicative of a background level of histone interaction with replicated DNA (Fig. (Fig.5A,5A, lanes 1 to 4). As observed previously ( 36), addition of 20 ng of wild-type yeast CAF-I resulted in formation of a spaced ladder of MNase-protected DNA species, indicative of formation of polynucleosomal arrays (Fig. (Fig.5A,5A, lanes 5 to 8). Furthermore, addition of 75 ng of either CafI-13 or CafI-20 also resulted in nucleosome formation (Fig. (Fig.5A,5A, lanes 9 to 16). These data demonstrated that the supercoiling of DNA by high levels of the mutant CAF-I complexes did indeed reflect nucleosome assembly.
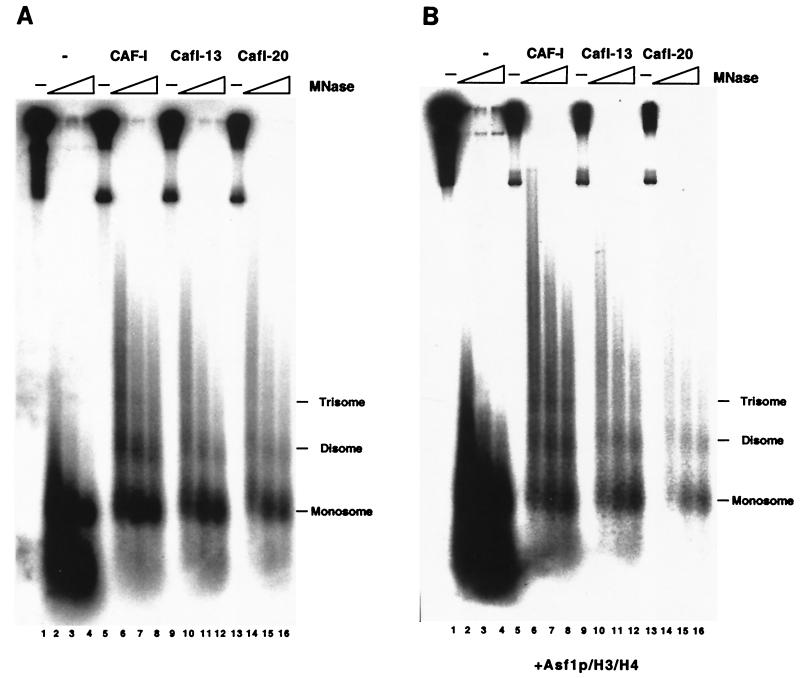
MNase digestion of DNA replication products. DNA replication reactions containing 20 ng of CAF-I, 75 ng of CafI-13, or 75 ng of CafI-20 (A) without or (B) with 3.5 pmol of Asf1p/H3/H4 were digested with 0.2 U of MNase (Sigma) for 0, 10, 20, and 30 min at 22°C. Recovered DNA was separated on a 2% agarose gel and visualized by autoradiography.
We then explored the biochemical synergy between CAF-I and Asf1p. As reported previously ( 36), Asf1p/H3/H4 stimulated wild-type CAF-I activity. In the absence of Asf1p/H3/H4, 20 ng (0.12 pmol) of CAF-I was required for supercoiling of replicated DNA. However, only 10 ng (59 fmol) of CAF-I was required in the presence of 3.5 pmol of Asf1p/H3/H4 (Fig. (Fig.4B,4B, lanes 5 and 14). Although Asf1p/H3/H4 was added in much higher concentrations than CAF-I, this amount of Asf1p/H3/H4 did not result in nucleosome formation in the absence of CAF-I (Fig. (Fig.4B,4B, lane 12). Additionally, 3.5 pmol of Asf1p in the absence of bound histones was unable to stimulate the activity of CAF-I, CafI-13, or CafI-20 to the same degree observed with equivalent amounts of the Asf1p/H3/H4 complex (Fig. (Fig.4B,4B, lanes 29 to 35). Furthermore, 3.5 pmol of histones H3 and H4 in the absence of Asf1p potently inhibited DNA replication and converted the DNA to a rapid-migrating form (Fig. (Fig.4B,4B, lanes 22 to 28), confirming previous experiments demonstrating the ability of Asf1p to prevent nonspecific electrostatic interactions between DNA and histones ( 28, 36).
Because telomeric silencing in cac1-13 and cac1-20 cells is largely dependent on the ASF1 gene (Fig. (Fig.3C),3C), we then tested whether Asf1p/H3/H4 complexes could stimulate nucleosome assembly by CafI-13 or CafI-20. Indeed, Asf1p/H3/H4 did stimulate DNA supercoiling by CafI-13 and CafI-20 (Fig. (Fig.4B,4B, lanes 16 to 21), and MNase digestion analysis confirmed that this reflected nucleosome assembly (Fig. (Fig.5B).5B). However, as observed in the absence of Asf1p/H3/H4, the mutant CAF-I complexes were required at two- to fourfold greater levels than was wild-type CAF-I to observe efficient nucleosome assembly (Fig. (Fig.4B,4B, lanes 16 to 21). Addition of Asf1p/H3/H4 to the mutant CAF-I complexes resulted in a similar distribution of total DNA and newly replicated DNA, supporting the hypothesis that the mutant complexes displayed a reduced preference for newly replicated DNA molecules. Similar results were observed when wild-type CAF-I was added in twofold excess of the amount required for nucleosome assembly (Fig. (Fig.4B,4B, lane 15). Together, these data suggest that when present in high levels, CAF-I was able to deposit histones in a manner independent of interaction with PCNA and that CafI-13 and CafI-20 could only function in this way. However, this replication-unlinked mode of activity was still stimulated by Asf1p/H3/H4.
Interaction between Asf1p and Cac2p.
Because of the biochemical and genetic synergies between CAF-I and Asf1p, we tested for direct interaction between these proteins. We did this in three types of experiments: cosedimentation, GST fusion protein coprecipitation, and coimmunoprecipitation. First, purified recombinant yeast CAF-I, Asf1p, and Asf1p complexed with H3/H4 were analyzed by glycerol gradient sedimentation. Yeast CAF-I sedimented with a peak in fractions 7 to 8 (150 kDa), while Asf1p sedimented with a peak in fractions 3 to 5 (30 kDa) (Fig. (Fig.6A).6A). Preincubation of CAF-I with Asf1p did not significantly alter the sedimentation of either protein. In contrast, Asf1p complexed with H3/H4 sedimented with a peak in fractions 5 to 6 (≈67 kDa). The Asf1p/H3/H4 complex sedimented more broadly when it was preincubated with CAF-I, with one peak similar to Asf1p/H3/H4 alone and a second more rapidly migrating peak in fractions 9 to 10 (≈160 kDa) (Fig. (Fig.6A,6A, CAF-I + Asf1p/H3/H4). These data implied that a fraction of the added Asf1p/H3/H4 complexes associated directly with CAF-I in solution.
To determine which subunit of CAF-I interacted with Asf1p, we performed coprecipitation experiments using GST-Asf1p and in vitro-translated Cac1p, Cac2p, and Cac3p (Fig. (Fig.6B).6B). Neither Cac1p nor Cac3p was coprecipitated by GST-Asf1p (Fig. (Fig.6B,6B, lanes 1 to 3 and 7 to 9). In contrast, Cac2p was efficiently coprecipitated by GST-Asf1p (Fig. (Fig.6B,6B, lanes 4 to 6); this interaction was stable to washing in the presence of 500 mM NaCl. Although Asf1p required the presence of histones H3 and H4 for association with CAF-I in solution (Fig. (Fig.6A),6A), histones were not added in these coprecipitation experiments. There are several possibilities that could explain this. The interaction between Asf1p and Cac2p could be modulated by other CAF-I subunits or by the presence of the GST moiety. Alternatively, other proteins in the reticulocyte extract used for the translation, including histones, could be involved. We note that GST-Asf1p also binds to Hir1p ( 36), the yeast protein that displays the highest homology to Cac2p ( 17).
We also analyzed the Asf1p-Cac2p interaction in coimmunoprecipitation experiments with purified recombinant proteins. CAF-I was incubated with Asf1p or Asf1p complexed with H3/H4, followed by immunoprecipitation of CAF-I with an anti-Py antibody that recognizes Py-tagged Cac3p. A fraction of the input Asf1p/H3/H4 complexes was coimmunoprecipitated with CAF-I, and this association was much less efficient in the absence of histones (Fig. (Fig.6C,6C, lanes 5 and 6), consistent with the sedimentation experiments (Fig. (Fig.6A).6A). Additionally, a purified recombinant Cac1/3p complex lacking Cac2p did not efficiently coimmunoprecipitate Asf1p regardless of added histones H3/H4 (Fig. (Fig.6C,6C, lanes 7 and 8). Together, these data are consistent with Cac2p’s serving as a binding site for the Asf1p/H3/H4 complex.
DISCUSSION
Cac3p binding site on Cac1p.
Our results indicate that multiple protein-protein interactions contribute to the function of the CAF-I protein complex. For example, our screen for silencing-defective cac1 alleles recovered an allele (cac1-15) that strongly reduced silencing and prevented binding to the Cac3p subunit of the CAF-I complex. Cac1-15p contained two amino acid changes, F257L and L276P. The L276P amino acid change alone, encoded by the cac1-21 allele, resulted in loss of Cac3p binding. However, telomeric silencing was largely unaffected by this mutation. Furthermore cac1-21 cells became greatly dependent on the ASF1 but not the HIR1 gene for telomeric silencing. Previous genetic analysis of the silencing roles of ASF1 and HIR1 genes at telomeres or the silent HML mating locus revealed no separation of function ( 36). We propose that this novel genetic separation of ASF1 and HIR gene function reflects a function for Asf1p that is not shared by Hir proteins. For example, Cac3p is a member of a family of histone H3/H4-binding proteins ( 53). Asf1p, another histone H3/H4-binding protein ( 3, 28, 36, 51), may directly or indirectly assist in recruiting histones to the CAF-I complex in the absence of an efficient Cac1p-Cac3p interaction.
PCNA-binding motifs.
The cac1-13 allele encodes a single point mutation (F233L) in one of the aromatic residues of the consensus PCNA-binding motif ( 55), and Cac1-13p displayed reduced affinity for PCNA in vitro. Additionally, mutation of the PCNA-binding motif of Cac1p crippled its DNA replication-linked nucleosome assembly activity in vitro and caused telomeric silencing to be dependent on the ASF1/HIR pathway in vivo. However, the silencing phenotypes of the PCNA-binding-defective cac1 alleles were nearly wild type in an otherwise wild-type strain. These data underscore the degree of functional overlap between CAF-I and the Asf1p/Hir silencing pathways in vivo. One model consistent with our data is that Asf1p/Hir proteins provide an alternative route for recruitment of Cac1-13p and Cac1-20p to DNA. We propose that the interaction between Asf1p and Cac2p underlies this functional synergy.
The aromatic residues in the PCNA-binding motif of Cac1p have been conserved in evolution. Although human p150 protein binds PCNA ( 25, 37), it lacks the glutamine residue that begins the defined consensus motif that is present in the budding and fission yeast genes (Fig. (Fig.4A)4A) ( 56). Mutational analysis of several PCNA-binding proteins has demonstrated that residues outside of the PCNA-binding motif are important for stable PCNA binding ( 30, 62), and variations of this consensus sequence exist ( 57). For example, the large subunit of DNA polymerase delta from several species contains a diverged PCNA-binding motif lacking the initial glutamine ( 60), demonstrating that complete conservation of the PCNA-binding motif is not absolutely required for PCNA binding. Additionally, the transcriptional coactivator p300 binds PCNA but lacks this consensus altogether, and novel PCNA-binding peptides have been identified in phage display experiments ( 14, 59). These latter results underscore the fact that not all PCNA-binding proteins will necessarily interact with the interdomain loop bound by the canonical PCNA-binding motif.
Almouzni and coworkers have demonstrated that human p150 binds to Xenopus PCNA in vitro, and this interaction was mapped using GST fusion proteins. Their data suggested that the first 244 amino acids of p150 were important for PCNA binding and that the first 31 amino acids are required for the interaction ( 25). However, amino acids 1 to 296 of human p150 are dispensable for replication-coupled chromatin assembly in vitro ( 18), and this region of the protein is the least well conserved with homologs from more distant organisms. In the absence of point mutation analyses, it is difficult to reconcile these data. One possibility is that PCNA binding by human p150 is complex, containing regions that bind to PCNA in addition to the degenerate PCNA-binding motif shown in Fig. Fig.4A.4A. Consistent with this possibility, the large subunit of DNA polymerase delta appears to contain both canonical and noncanonical PCNA-binding regions ( 59).
Role of Asf1p during DNA synthesis.
We demonstrated that the Asf1p/H3/H4 complex interacted with CAF-I by binding the Cac2p subunit. This interaction is conserved in evolution; the Drosophila homologs of these proteins (dASF1 protein and the p105 subunit of dCAF-I) have recently been shown to interact directly ( 52). However, the interaction of the Drosophila proteins is independent of histones, whereas we have observed that the presence of histones H3 and H4 is important for observing the interaction with recombinant protein complexes in solution (Fig. (Fig.6A6A).
We also observed that yeast Asf1p was not quantitatively associated with CAF-I in the sedimentation and coimmunoprecipitation experiments and that an excess of Asf1p/H3/H4 was required to stimulate CAF-I activity (Fig. (Fig.44 and and6).6). Therefore, the interaction between these yeast protein complexes is inefficient and/or transient. Additionally, other proteins or protein modifications may regulate the CAF-I-Asf1p interaction. For example, Asf1p in yeast forms a stable complex in the absence of histones with Rad53p, a kinase central to the DNA damage and intrA-S-phase checkpoints ( 3). When cells are arrested in S-phase with hydroxyurea or after DNA damage, Asf1p is released from Rad53p and associates with histones. This provides a mechanism by which the chromatin assembly activity of Asf1p is limited to periods of DNA synthesis. These data also suggest that sequestration of Asf1p by Rad53p would prohibit Asf1p from interacting with CAF-I in the absence of DNA synthesis.
The mechanism underlying the functional synergy between Asf1p and CAF-I remains unknown, and several possibilities exist. In vitro, CAF-I functions stoichiometrically to deposit histones onto DNA ( 18, 22, 36, 42). Stimulation of reduced amounts of CAF-I by Asf1p could result from more efficient reloading of histones onto CAF-I. Alternatively, Asf1p may assist in more efficient recruitment of CAF-I to replicating DNA. These models are not the only possibilities, nor are they mutually exclusive. Future biochemical experiments will be required to determine how the CAF-I reaction cycle and subunit interactions lead to chromatin assembly in vivo.
Acknowledgments
We thank Peter Burgers, Bruce Futcher, Daniel Gottschling, and Jasper Rine for providing plasmids and antibodies, Jim Kadonaga and Andrew Emili for communication of results prior to publication, Melissa Adams for strain generation, Jasper Rine, Ann Kirchmaier, anonymous reviewers, and members of the Kaufman lab for comments, and Jay Chuang for excellent technical assistance.
This work was supported by Department of Energy funds awarded to P.D.K. and administered through the Lawrence Berkeley National Laboratory and by National Institutes of Health grant 1 R01 GM55712 and National Science Foundation grant MCB-9982909.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.22.2.614-625.2002
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/22/2/614.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Structural Basis for the Interaction Between Yeast Chromatin Assembly Factor 1 and Proliferating Cell Nuclear Antigen.
J Mol Biol, 436(16):168695, 04 Jul 2024
Cited by: 0 articles | PMID: 38969056
Disordered regions and folded modules in CAF-1 promote histone deposition in Schizosaccharomyces pombe.
Elife, 12:RP91461, 20 Feb 2024
Cited by: 2 articles | PMID: 38376141 | PMCID: PMC10942606
Chromatin assembly factor-1 preserves genome stability in ctf4Δ cells by promoting sister chromatid cohesion.
Cell Stress, 7(9):69-89, 14 Aug 2023
Cited by: 1 article | PMID: 37662646 | PMCID: PMC10468696
Structural and functional studies of PCNA from African swine fever virus.
J Virol, 97(8):e0074823, 03 Aug 2023
Cited by: 4 articles | PMID: 37534905 | PMCID: PMC10506467
A novel single alpha-helix DNA-binding domain in CAF-1 promotes gene silencing and DNA damage survival through tetrasome-length DNA selectivity and spacer function.
Elife, 12:e83538, 11 Jul 2023
Cited by: 2 articles | PMID: 37432722 | PMCID: PMC10335832
Go to all (119) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (1 citation) ENA - AF222339
- (1 citation) ENA - AF367177
- (1 citation) ENA - AB027229
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing.
Curr Biol, 11(7):463-473, 01 Apr 2001
Cited by: 195 articles | PMID: 11412995
The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae.
Genes Dev, 15(23):3169-3182, 01 Dec 2001
Cited by: 108 articles | PMID: 11731480 | PMCID: PMC312838
Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I.
Mol Cell Biol, 18(8):4793-4806, 01 Aug 1998
Cited by: 125 articles | PMID: 9671489 | PMCID: PMC109065
Chromatin assembly: the kinetochore connection.
Curr Biol, 12(7):R256-8, 01 Apr 2002
Cited by: 10 articles | PMID: 11937044
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: 1 R01 GM55712
Grant ID: R01 GM055712




