Abstract
Free full text

Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development
Abstract
To date, the in vivo importance of chromatin assembly factors during development in vertebrates is unknown. Chromatin assembly factor 1 (CAF-1) represents the best biochemically characterized factor promoting chromatin assembly during DNA replication or repair in human cell-free systems. Here, we identify a Xenopus homologue of the largest subunit of CAF-1 (p150). Novel dimerization properties are found conserved in both Xenopus and human p150. A region of 36 amino acids required for p150 dimerization was identified. Deletion of this domain abolishes the ability of p150 to promote chromatin assembly in vitro. A dominant-negative interference based on these dimerization properties occurs both in vitro and in vivo. In the embryo, nuclear organization was severely affected and cell cycle progression was impaired during the rapid early cleaving stages of Xenopus development. We propose that the rapid proliferation at early developmental stages necessitates the unique properties of an assembly factor that can ensure a tight coupling between DNA replication or repair and chromatin assembly.
Introduction
Chromatin assembly is fundamental during DNA replication and repair to ensure the maintenance of a functional eukaryotic genome (Wolffe, 1998; Adams and Kamakaka, 1999; Kornberg and Lorch, 1999; Kaufman and Almouzni, 2000; Verreault, 2000). After passage of the replication fork or repair of DNA lesions, formation of the basic unit of chromatin, the nucleosome (Workman and Kingston, 1998), requires first deposition of histones onto DNA to form a core particle. In vitro, using purified histones and DNA, this loading event can be stimulated by several histone chaperones (Ito et al., 1997; Philpott et al., 2000).
In human cells, a prime candidate to ensure the coordination between DNA synthesis and chromatin assembly is chromatin assembly factor 1 (CAF-1), the unique histone chaperone that preferentially facilitates in vitro chromatin assembly on newly synthesized DNA (Smith and Stillman, 1989, 1991; Kaufman et al., 1995; Gaillard et al., 1996). This three-subunit complex (p150, p60 and p48) co-purifies with newly synthesized isoforms of histone H3 and H4 (Verreault et al., 1996). The two largest subunits of this complex concentrate at replication foci during S phase (Krude, 1995; Martini et al., 1998; Shibahara and Stillman, 1999; Taddei et al., 1999) and at nucleotide excision repair (NER) sites outside of S phase (Martini et al., 1998). Based on these properties of CAF-1 at the crossroads of various DNA metabolic pathways (Ridgway and Almouzni, 2000; Verreault, 2000), one would expect that a deficiency in its function would have a profound effect in vivo.
Surprisingly, deletion of the genes encoding the three orthologous subunits in Saccharomyces cerevisae is not lethal and results in an increased sensitivity to UV irradiation and defects in transcriptional silencing in heterochromatic loci (Enomoto et al., 1997; Kaufman et al., 1997; Monson et al., 1997; Enomoto and Berman, 1998; Game and Kaufman, 1999). Based on these data, it is possible that in S.cerevisiae, CAF-1 is required predominantly for specialized functions associated with DNA repair of UV lesions and for maintenance of heterochromatic regions. The replication-associated function of CAF-1 may be fulfilled by redundant factors, and thus its importance in vivo remains unclear. Remarkably, none of the chromatin assembly factors identified to date in S.cerevisiae has proved essential for nucleosome assembly or viability in this organism (Verreault, 2000).
Key issues are thus raised concerning chromatin assembly factors and, more specifically, histone deposition factors and their exact function in vivo in different organisms and in various cell cycle contexts. Xenopus, a vertebrate with a defined development highly exploited for cell cycle studies and amenable to powerful biochemical strategies, appeared as an ideal system to gain insights into these questions (Kirschner et al., 1985; Dawid and Sargent, 1988). We thus cloned and identified a functional Xenopus p150 (xp150) CAF-1. Novel conserved dimerization properties of this subunit were discovered and their importance for CAF-1 function in vitro was assessed. A domain of 36 amino acids not present in other known proteins to date, critical for p150 dimerization, was found. This permitted the design of a dominant-negative strategy to assess the specific role of p150 CAF-1 in vitro and in vivo under conditions ensuring maximum specificity. This study demonstrates a critical role for the largest subunit of CAF-1 during Xenopus early embryonic development.
Results
Cloning and characterization of the Xenopus p150 CAF-1 homologue
A yeast two-hybrid screen was carried out using as bait a portion (C-terminus) of the largest subunit of human CAF-1 (hp150 CAF-1) and, as prey, a Xenopus oocyte cDNA library (Iouzalen et al., 1998). We did not retrieve the Xenopus p60 homologue in this screen. This may be due to a weak interaction with hp150, a low representation of p60 cDNA or the presence of the restriction site used to construct the Xenopus library within the xp60 cDNA. Unexpectedly, this screen enabled us to obtain the full-length sequence of a putative homologue of p150 CAF-1 in Xenopus. This result was not expected based on the available information concerning the CAF-1 network of interactions (Ridgway and Almouzni, 2000). Sequence comparison with the mouse p150 (DDBJ/EMBL/GenBank accession No. AJ132771) and hp150 sequences (Kaufman et al., 1995) showed a similar domain organization (Figure 1A and B). Blocks of conserved residues were found mainly in the C-terminal third of the proteins as well as in the large clusters of charged residues called KER and ED domains (Figure 1A and B), all regions critical for the chromatin assembly function of CAF-1 in vitro (Kaufman et al., 1995). In contrast, the N-terminal portion displayed weaker homology (Figure 1B). The sequence conservation in these domains suggested that our clone was the Xenopus homologue of p150 CAF-1 and hence it was named xp150.

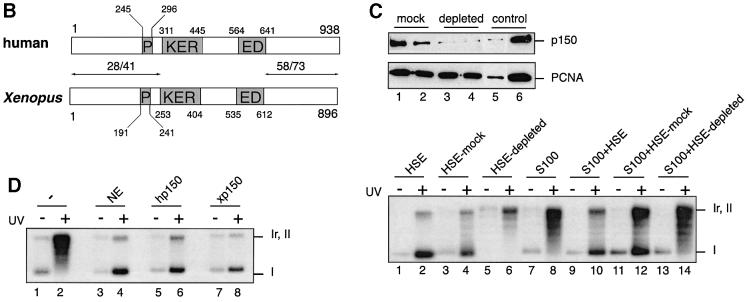
Fig. 1. A functional homologue of p150 CAF-1 in Xenopus. (A) Sequence alignment between human (hp150), mouse (mp150) and Xenopus p150 (xp150) CAF-1 obtained using ClustalW and Boxshade programs (BCM and ISREC web sites). The amino acid identity is black boxed and similarity is shown by grey boxes. The position of the KER and ED boxes (Kaufman et al., 1995) is indicated on the side. (B) Comparative schematic representation of the domain organization of human and Xenopus p150. The percentage similarity/identity in the N- and C-terminal ends is indicated above the arrows delineating areas of comparison. Residues delimiting domains are indicated for each species. P, PEST domain; KER, KER domain; ED, ED domain (Kaufman et al., 1995). (C) Depletion of xp150 impairs chromatin assembly coupled to DNA repair. Top: western blot analysis of a Xenopus egg extract (HSE) depleted of xp150. Anti-xp150 antibody (serum 566, 1/1000) and anti-PCNA antibody (PC10, DAKO) were used for detection. Lane 1, HSE depleted with control IgG; lane 2, HSE depleted with pre-immune serum; lane 3, HSE depleted with affinity-purified anti-xp150 antibody; lane 4, HSE depleted with anti-xp150 serum; lane 5, HSE diluted 1/10; lane 6, undiluted HSE equivalent to the depleted extract. Bottom: analysis of chromatin assembly by supercoiling on control and UV-irradiated DNA. The pBscript plasmid mock treated (–) or UV irradiated (+) (500 J/m2) (Gaillard et al., 1996) was incubated for 3 h at 23°C in HSE, mock-depleted HSE or HSE depleted with anti-xp150 antibody. Alternatively, the DNA was incubated for 3 h at 37°C in S100 human cytosolic extract (Smith and Stillman, 1989) or S100 extract complemented with HSE treated as indicated. [α-32P]dCTP was added to all samples to follow DNA repair synthesis. The migration of relaxed/nicked (Ir,II) and supercoiled DNA (I) is indicated. (D) Xenopus p150 complements S100 extracts for chromatin assembly. As in (A), DNA is incubated for 3 h at 37°C in S100 extract (–), S100 extract complemented with HeLa nuclear extract (NE) (Martini et al., 1998), or in vitro translated hp150 or xp150 proteins (TnT reticulocyte lysate system, Promega).
To prove a functional identity with the largest subunit of CAF-1, we used in vitro chromatin assembly assays that had served for the initial characterization of CAF-1 activity (Smith and Stilman, 1989, 1991; Kaufman et al., 1995; Gaillard et al., 1996; Verreault et al., 1996). Based on these assays, a CAF-1-like activity was found in Xenopus egg extracts (Gaillard et al., 1996; Kamakaka et al., 1996). These extracts were depleted using specific antibodies raised against a recombinant portion of xp150 (Figure 1C, top). More than 90% of the endogenous xp150 protein was removed without significant loss of other factors such as proliferating cell nuclear antigen (PCNA) or histones (Figure 1C, top and data not shown). The xp150-depleted extract was then tested for its ability to support chromatin assembly on UV-damaged plasmid DNA using a standard supercoiling assay (Figure 1C, bottom). The appearance of form I corresponding to supercoiled DNA in the control experiments indicated that minichromosomes were formed efficiently on the labelled DNA (Figure 1C, bottom, lanes 2 and 4). In contrast, this form was not accumulated in the xp150-depleted extract (Figure 1C, bottom, lane 6). Since migration of these same samples on a chloroquine gel did not show any increase in nicked DNA (not shown), these data are consistent with a loss of chromatin assembly activity in the xp150-depleted extracts.
The loss of CAF-1 activity in the depleted extracts was then tested in complementation assays using human S100 extract that lacks the largest subunit of the CAF-1 complex, p150 (Kaufman et al., 1995). Consistent with a p150 CAF-1 depletion, we found that the depleted Xenopus egg extracts were unable to complement the S100 extract (Figure 1C, bottom, compare lanes 12 and 14). Finally, using in vitro translated xp150, we could complement the S100 extract as usually obtained using a nuclear extract containing the endogenous hp150 or when using in vitro translated hp150 (Figure 1D, compare lanes 4, 6 and 8). This reaction was dependent on the presence of human p60 (hp60), since an S100 extract depleted in hp60 would not be complemented by the simple addition of xp150 (not shown). We thus concluded that we had identified the functional homologue of the p150 subunit of CAF-1 in Xenopus.
Evolutionarily conserved self-association properties of p150 CAF-1 proteins and their requirement for nucleosome assembly activity in vitro
The interaction of the conserved C-terminal third (Cter) of hp150 with its Xenopus counterpart in the two-hybrid screen raised two questions: (i) whether the interaction is direct and (ii) whether homodimerization or multimerization of p150 Cter can occur within the same species.
We first tested the interspecies interaction by co-expres sion in bacteria of histidine-tagged hp150 derivatives together with a glutathione S-transferase (GST)-tagged xp150 corresponding to the portion initially retrieved in our two-hybrid screen (amino acids 540–896) (Figure 2A, left). The GST–xp150 derivative could bind specifically to the hp150(620–938) peptide, but not to hp150(678–938), a peptide that is 58 amino acids shorter (Figure 2A, left, lanes 2, 3 and 5, 6). This interaction was stable up to 1 M salt extraction. Within the same species, the expression in bacteria of a combination of His6 and GST hp150 derivatives (Figure 2A, middle) demonstrated self-interaction properties again requiring this stretch of 58 amino acids (Figure 2A, middle, lanes 2, 3 and 5, 6). These interactions were confirmed by a yeast two-hybrid assay (data not shown). We also tested the homo-interaction in Xenopus. The full-length His6-tagged xp150 can interact with the GST–xp150(540–986) derivative but not with GST alone. This interaction is also stable up to 1 M salt extraction (Figure 2A, right, lanes 2, 3 and 5, 6). Further more, since aggregation of proteins could occur in GST pull-down assays, sedimentation analyses were performed with in vitro translated 35S-labelled (not shown) and bacterially expressed human and Xenopus full-length p150 proteins. When physiological salt conditions were used in the gradients, the majority of the bacterially expressed human and Xenopus full-length p150 proteins sedimented at a similar position to the 258 kDa molecular weight standard (Figure 2B, WT hp150 and WT xp150, centred around fraction 17). Remarkably, an internal deletion restricted to 36 amino acids in the critical region of hp150, outside of the ED domain (residues 642–678), gave rise to a protein sedimenting as a monomeric 150 kDa form when run in parallel under the same conditions [Figure 2B Δ(642–678), fraction 13]. When gradients were performed in the presence of 2 M NaCl to disrupt p150– p150 interactions, the majority of wild-type hp150, hp150Δ(642–678) mutant and xp150 behaved as monomeric 150 kDa proteins (Figure 2B).

Fig. 2. Inter- and intraspecies p150 dimerization: identification of a critical domain and role in chromatin assembly. (A) p150 self-association by GST pull-down assay. Left: GST–xp150(540–896) was co-expressed in bacteria (Studier et al., 1990) containing hp150(620–938) or hp150(678–938) tagged with His6. Middle: His6-tagged hp150(620–938) was co-expressed in bacteria with either GST–hp150(620–938) or GST–hp150(678–938). Right: equal volumes of separate bacterial extracts containing the full-length His6-xp150 and GST–xp150(540–986) were mixed at 2 M NaCl; the salt concentration was lowered by sequential dilution to 350 mM NaCl, and glutathione–Sepharose resin was then added. Protein production and purification on glutathione–Sepharose resin were performed essentially as described (Moggs et al., 2000). Co-purification of His-tagged hp150 proteins was detected by western blot probed with anti-His6. The xp150 proteins were detected using the specific anti-xp150 serum 566. I, input; B, bound fraction; B*, 1 M NaCl-resistant bound fraction. The positions of His6-hp150(620–938) and His6-hp150(678–938) are indicated. Asterisks mark degradation products. (B) Glycerol gradient sedimentation of p150 proteins. Bacterially produced recombinant proteins wild-type hp150, hp150Δ(642–678) and wild-type xp150 were sedimented on a linear 10–40% glycerol gradient. Every second fraction of the gradient was run on SDS–PAGE. The proteins were detected by the specific anti-hp150 and anti-xp150 sera on western blots. The sedimentation positions of the molecular weight standards for each gradient are indicated above the gel: BSA (66 kDa) and catalase (258 kDa). Arrows mark the mean position of the proteins. The salt concentrations used in the gradients are as indicated. (C) Chromatin assembly analysis by supercoiling in the presence of hp150Δ(642–678). pBscript control (–) and UV-irradiated (500 J/m2) (+) were incubated as in Figure 1D with S100 extract (–), or S100 extract complemented by HeLa nuclear extract (NE) or in vitro translated hp150 wild-type (lanes 5–6), hp150Δ(642–678) and xp150 wild type. The positions of the relaxed/nicked (Ir,II) and supercoiled DNA (I) are indicated.
Therefore, the self-association of the p150 CAF-1 subunit involves primarily dimeric forms, both in human and in Xenopus, and requires a conserved stretch of 36 amino acids, outside of the ED domain, in the C-terminal third of the proteins. Remarkably, searching through data banks for this motif did not retrieve any other protein, indicating that it is unique to p150. Based on these data, we then tested the functional importance of this domain in p150 using the in vitro assay for chromatin assembly coupled to repair of UV-damaged DNA as above. In vitro translated wild-type human and xp150 as well as the hp150Δ(642–678) mutant were used to complement the S100 extracts. Both full-length human and Xenopus p150 promoted nucleosome assembly onto damaged DNA as efficiently as a nuclear extract providing a functional CAF-1 complex (Figure 2C, lanes 4, 6 and 10). In contrast, the hp150Δ(642–678) mutant that does not homodimerize (Figure 2B) failed to complement the S100 extract (Figure 2C, lane 8). Thus, deletion of the 36 amino acid motif that was found critical for p150 self-association also abrogates p150 competence for chromatin assembly in vitro.
The largest subunit of human CAF-1 expressed in the early Xenopus embryo interacts with Xenopus p150 and arrests development
The discovery that a unique, conserved 36 amino acid stretch in p150 is required for homodimerization of the protein and for its chromatin assembly activity prompted us to consider the use of a dominant-negative strategy (Herskowitz, 1987) to study the requirements for CAF-1 in vivo. The Xenopus embryo and its rapid divisions at early stages (Kirschner et al., 1985) was an ideal in vivo system to test the importance of a factor potentially critical for coordinating chromatin assembly with DNA replication and repair. The RNA encoding the hp150 subunit was injected into one blastomere of a two-cell stage Xenopus embryo, and both the interaction with xp150 and its effect on development were examined. Immunoprecipitations using anti-xp150 or anti-hp150 antibodies were performed on extracts derived from embryos injected with hp150 RNA (Figure 3A). As shown in Figure 3B, when anti-xp150 antibodies were used in the immunoprecipitation all the detectable hp150 protein present in the embryo was found associated with xp150 (Figure 3B, compare lanes 2 and 3, hp150). Conversely, only a fraction of the xp150 present in the embryo was bound to hp150 (Figure 3B, lanes 4 and 5, xp150). This was a good indication that the amount of exogenously expressed protein was within the range of the endogenous counterpart and that hp150 was not overexpressed in our experiments. We also searched in these immunoprecipitates for proteins known to associate with hp150, such as PCNA (Shibahara and Stillman, 1999; Moggs et al., 2000) and acetylated histone H4 (Verreault et al., 1996). No detectable amounts of these factors were found associated with either human or Xenopus p150 proteins (Figure 3B, lanes 3 and 5, PCNA, AcK5-H4). We also verified whether other histone chaperones such as nucleoplasmin or N1/N2, previously shown to participate in nucleosome formation in Xenopus oocyte or egg extracts (Dilworth et al., 1987; Kleinschmidt et al., 1990), were bound to human or Xenopus p150. Under our experimental conditions, we did not find any significant co-precipitation of these proteins with either of the p150 proteins (Figure 3B, lanes 3 and 5, nucleoplasmin; and data not shown). Although we cannot formally exclude minor or transient interactions with other factors, the majority of the hp150 expressed in the early embryo was bound to the endogenous xp150.
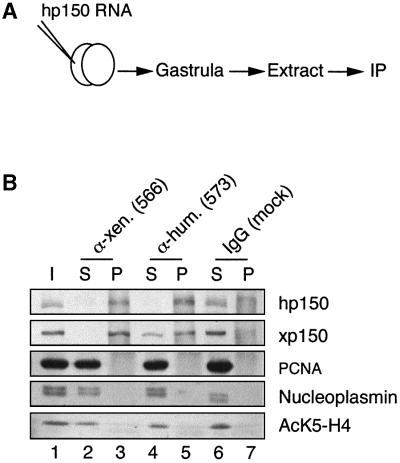
Fig. 3. Human p150 expressed in Xenopus embryos associates with Xenopus p150. (A) Scheme of the experiment: Xenopus embryos at the two-cell stage prepared as described in Vize et al. (1991) were injected into one blastomere using a nanoject injector (Drummond) with 1 ng of RNA encoding hp150 CAF-1. Embryos collected at the gastrula stage were processed for immunoprecipitation assay. (B) Immunoprecipitation of extracts from embryos injected with hp150 RNA using anti-xp150 antibody, anti-hp150 antibody or control antibody as indicated on top. Western blot of the different fractions: I, input; S, supernatant; P, pellet. hp150 and xp150 were detected by the polyclonal 573 and 566 antibodies, respectively; PCNA by the PC10 monoclonal antibody; nucleoplasmin by the monoclonal PAC35 antibody; and histone H4 acetylated at Lys5 by the polyclonal anti-H4AcK5 antibody.
We then analysed whether expression of hp150 in the early Xenopus embryo and its interspecies association with xp150 had any effect on development. The appearance of the protein was monitored by western blotting in parallel (Figure 4A, bottom). Control embryos, injected only with buffer or RNA encoding green fluorescent protein (GFP) as a cell lineage marker, developed normally (Figure 4A, buffer and GFP RNA). In the latter case, GFP fluorescence visible around the mid-blastula transition (MBT) was dispersed widely on one side of the embryo, but was virtually absent on the other (Figure 4A, GFP RNA), as previously reported (Zernicka-Goetz et al., 1996). In contrast, the injection of hp150 RNA severely affected embryo development, which did not reach the mid-blastula stage (Figure 4A, hp150 + GFP RNA). This arrest of Xenopus early development was obtained reproducibly when 1 ng of RNA encoding hp150 was injected (a usual amount for embryo injection) (Vize et al., 1991). However, amounts of RNA <100 pg were not effective, suggesting that a threshold in the expression of hp150 must be reached, probably to titrate the endogenous xp150 quantitatively in order to induce the developmental interference. Since these abnormal embryos were only detected when hp150 RNA was injected, we could exclude non-specific effects due to the microinjection itself or to exogenous RNA overexpression. Importantly, the timing of the effect, before the MBT, during a period of active division without significant transcription, together with the detection of large cells deriving from the injected side of the embryo (Figure 4A, see white arrowhead, Figure 4C) argues for a defect in cell cycle progression. For a closer examination of these aberrant large cells, embryos injected with buffer and hp150 + GFP RNAs were processed for cryosection at the early gastrula stage when GFP was detectable (Figure 4B). Strikingly, in the embryos co-injected with GFP and hp150 RNAs, 4′,6-diamidino-2-phenylindole (DAPI) staining was excluded from the area where GFP was detected (Figure 4B, top and bottom, hp150 + GFP RNA, white arrowheads) whereas a normal DAPI staining was observed in the embryos injected only with buffer. Immunofluorescence staining with the anti-hp150 monoclonal antibody (mAb1) confirmed that the distribution of hp150 overlapped with that of the GFP lineage marker (Figure 4B, bottom, GFP + hp150). We thus concluded that expression of hp150 was not compatible with the detection of normal nuclei at this developmental stage.

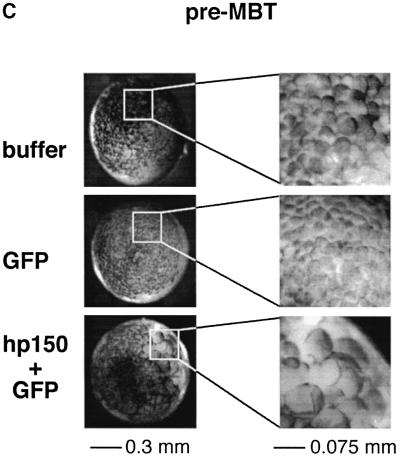
Fig. 4. Human p150 expressed in Xenopus embryos specifically perturbs early development. Xenopus embryos were injected with RNA as in Figure 3A. (A) Development of embryos injected with: buffer, 2 ng of RNA encoding GFP (GFP) or a mixture of RNA encoding GFP and hp150 CAF-1, 1 ng each (hp150 + GFP) was followed at 23°C. Time after fertilization and corresponding developmental stages (Nieuwkoop and Faber, 1994) are indicated. Images of the embryos recorded at the indicated times after fertilization are presented as merges of the visible and GFP channels. The white arrowhead indicates the area of the embryo where the defect is first visible (pre-MBT, top right corner). The scale bar is indicated. For each stage, expression of hp150 in embryos injected with buffer (–) or with hp150 + GFP RNA (+) was monitored by western blotting (bottom) using the polyclonal 573 antibody. (B) Embryos injected with buffer and hp150 + GFP RNA were selected at early gastrulation stages and processed for cryosection and immunodetection. The injected RNAs or buffer are indicated on the left. Top: whole embryo sections, stained with DAPI to visualize DNA, were observed in bright field (left column) and appropriate filters for DAPI (DNA, middle) and GFP (right) fluorescence as indicated. Bottom: embryo cryosections were processed for immunofluorescence, and hp150 was detected using the mAb1 monoclonal antibody. DNA (revealed by DAPI), GFP and hp150 (Texas red) fluorescence were recorded with appropriate filters as indicated on the top. White arrowheads indicate areas of the section where GFP and hp150 are expressed but where DAPI staining is absent. The scale bar is indicated at the bottom. (C) Same pre-MBT Xenopus embryos as in (A). For each case, a higher magnification of the injected area is presented on the right. Scale bars are indicated at the bottom.
The 36 residues of p150 required for dimerization are critical to generate the negative interference in the embryo
The next step was to determine whether the domain of hp150 crucial for the dimerization was critical to generate the effects observed in the in vivo interference assay. Various truncated forms of hp150 were expressed in the Xenopus embryo (Figure 5A). Equivalent expression levels were verified by western blotting (data not shown) to compare the effects on embryonic development. Injection of RNA corresponding to a form of hp150 lacking the ED or KER box produced developmental defects comparable to those observed for wild-type hp150 (Figure 5A, WT, ΔED; and data not shown). Since the regions of hp150 containing the ED and KER boxes have been proposed to mediate interactions with histones (Kaufman et al., 1995), a simple histone titration appeared unlikely to explain the observed phenotype. This is in agreement with the results of the immunoprecipitation shown in Figure 3. Importantly, development was normal when the injected RNA encoded a hp150 mutant lacking the conserved C-terminal third of the protein [Figure 5A, (1–649)]. We thus expressed this latter region of hp150 in the embryo and found it sufficient to generate the abnormal phenotype [Figure 5A, (620–938)]. We next tested whether p150 mutants that cannot dimerize in vitro have an effect on early Xenopus development. Injection of the RNA coding for a truncated form of hp150 lacking the critical domain for self-association [Figure 5A, (678– 938)], as well as full-length hp150 lacking this internal domain [Figure 5A, Δ(642–678)], did not impair embryo development. These results are consistent with the interspecies association of the largest subunit of CAF-1 to mediate the arrest of early embryo development.
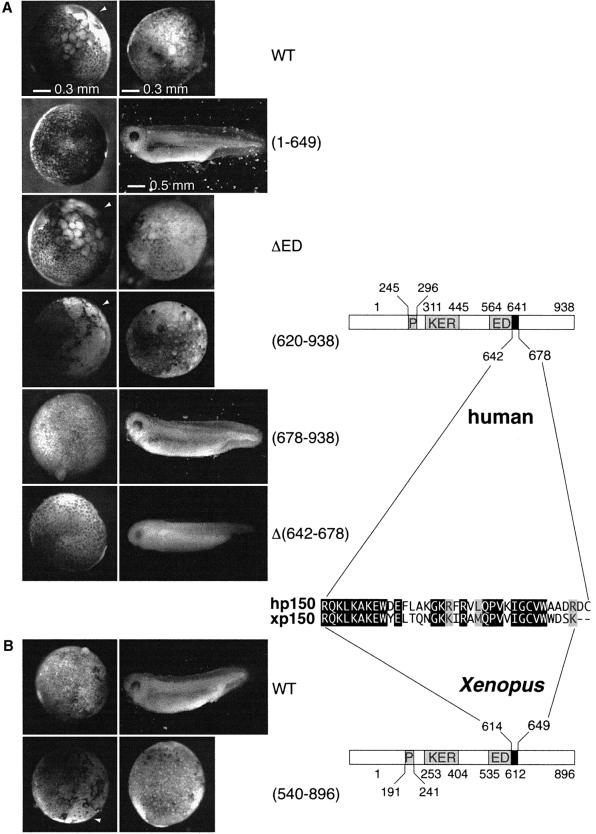
Fig. 5. The dimerization domain of p150 is required to perturb Xenopus early development. (A) Xenopus embryos were injected with buffer or RNA encoding the full-length hp150 (WT) and truncated forms of hp150 RNA: (1–649), ΔED, (620–938), (678–938) and Δ(642–678), as indicated on the side. Images were recorded under a dissecting microscope at stages 9 and 36 of development (as judged from control embryos). The white arrowheads point to the area of the embryo where the defect is first visible. Scale bars are indicated. (B) Injections were as in (A) but with RNA encoding the full-length xp150 (WT) or xp150(540–896). On the right, a schematic representation of hp150 and xp150 proteins and a sequence alignment of dimerization domain for each species are shown.
Consistent with a dominant-negative effect, injection of mRNA coding for the xp150 Cter (amino acids 540–896) also impaired embryo development [Figure 5B, (540– 896)] whereas embryos injected with full-length xp150 RNA were not perturbed (Figure 5B, WT). The latter result allowed us to exclude a toxic effect simply due to a dosage variation of the CAF-1 p150 subunit. In conclusion, the in vivo interference with early Xenopus development mediated by hp150 or by truncations of xp150 protein required the presence of 36 residues in the C-terminal third of p150 that are critical for its dimerization and are conserved between the human and the Xenopus protein (Figure 5, right).
Co-expression of hp60 rescues the developmental defects mediated by expression of hp150
Remarkably, injection of RNA encoding hp60, the mid-subunit of CAF-1, did not alter embryonic development when injected alone (Figure 6, hp60), but could counteract the effect caused by expression of full-length hp150 (Figure 6A). In the same way, co-injection of hp60 RNA counteracted the effect caused by expression of hp150(620–938), which contains the p150 dimerization domain and is able to interact with hp60 in vitro, but is unable to support CAF-1 activity (Kaufman, 1995) (Figure 6B). This effect also argues once again for an interference specifically related to CAF-1. Furthermore, the fact that hp60 counteracts the interference caused by hp150(620–938) (Figure 6B) indicates that hp60 is acting by titrating out hp150 expressed in the embryo rather than by forming a functional complex.

Fig. 6. Expression of hp60 counteracts the developmental perturbation induced by hp150 expression. (A) Embryos were injected with RNA encoding hp60 or hp150, or co-injected with hp60 and hp150 as indicated on the left (1 ng each). Images were recorded at stages 9 and 33 of development. Scale bars are indicated at the bottom. (B) Injections were as in (A) except that hp150(620–938) was injected.
hp150 perturbs chromatin assembly coupled to DNA repair in Xenopus extract
To monitor a dominant interference with CAF-1 activity in vitro, we used Xenopus egg extract and hp150 derivatives purified from expressing bacteria (Figure 7A). Their activity was first tested in the complementation assay using human S100 extract. Consistent with Figure 2C using in vitro translated protein, we could show again that the purified recombinant full-length WT hp150 was able to promote chromatin assembly on a UV-damaged DNA (Figure 7B, lane 4). In contrast, the hp150 derivative lacking the dimerization domain [hp150Δ(642–678)] was inefficient in this complementation assay (Figure 7B, lane 6). Remarkably, in the Xenopus high speed extract (HSE), the addition of full-length functional wild-type hp150 interfered with chromatin assembly coupled to DNA repair of UV-damaged plasmid (Figure 7B, left, lanes 2 and 4), whereas the addition of the derivative unable to dimerize [Figure 2B, hp150Δ(642–678)] did not affect the reaction (Figure 7, lanes 6 and 8). Importantly, as in vivo, the amount of hp150 added to the HSE in order to induce this interference is equivalent to the amount of endogenous xp150 in the HSE as estimated by western blot analysis. We reproducibly noted a decrease in DNA repair synthesis. It is remarkable, however, that the low amount of labelled material is closed (but not supercoiled) as seen on a chloroquine gel (Figure 7C, right, lanes 3 and 5), sugggesting that these DNA molecules have been repaired although not assembled into chromatin. Further investigations will be needed to analyse in detail the coupling of the reactions and the interference in vitro. Most importantly, the lack of accumulation of labelled form I in the presence of the functional wild-type hp150, not observed when using the hp150 derivative that is unable to dimerize, argues for a direct interference with CAF-1 activity caused by the dimerization properties of the hp150.
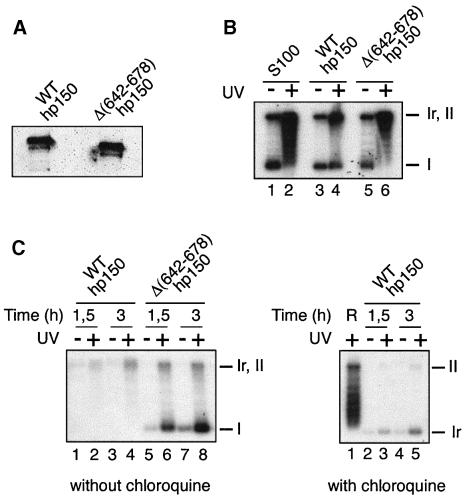
Fig. 7. hp150 interferes in vitro in a Xenopus extract with chromatin assembly coupled to DNA repair. (A) Western blot of purified His6-tagged recombinant wild-type hp150 and hp150Δ(642–678). Proteins were detected using the specific anti-hp150 serum. (B) Wild-type hp150 but not hp150Δ(642–678) complements S100 extracts for chromatin assembly. DNA damaged (+) or not (–) by UV was incubated for 3 h at 37°C in S100 extract (S100), or S100 extract complemented with equal amounts of either purified recombinant wild-type p150 or hp150Δ(642–678) as indicated. The migration of relaxed/nicked (Ir,II) and supercoiled DNA (I) is shown. (C) Left: in vitro interference in a Xenopus HSE; equal amounts of purified recombinant hp150 or hp150Δ(642–678) were added to the HSE; untreated (–) or UV-irradiated (+) pBS was then incubated for 1.5 or 3 h at 23°C as indicated. The migration without chloroquine of relaxed/nicked (Ir,II) and supercoiled DNA (I) is shown. Right: the same sample corresponding to lanes 1–4 was run in the presence of 10 µg/ml chloroquine to resolve the closed relaxed form (Ir) as indicated on the right. R is a reference showing the intermediate topoisomers and the nicked form (II).
Discussion
Chromatin assembly is important during replication and repair for the establishment and inheritance of defined epigenetic states [see minireviews in Cell, 1999, 5], which are fundamental during development as well as for cell cycle progression. We have cloned and identified the Xenopus CAF-1 p150 homologue and discovered a novel homodimerization property of this subunit. We show that the largest subunit of CAF-1 performs an essential role in early Xenopus development. We propose that during this time, the necessity to coordinate replication or repair events tightly to chromatin assembly imposes a dependence on CAF-1 in order to perform mitosis without intervening gap phases. These findings highlight an important role for CAF-1 in vivo and provide new insights into the mechanism and regulation of its activity in a developing organism.
Dimerization of human and Xenopus p150 subunits: a regulatory switch for CAF-1 activity?
By using the yeast two-hybrid system, in vitro binding assays and sedimentation analysis, we demonstrated that the human and Xenopus p150 CAF-1 subunits can homodimerize via their C-terminal domain (Figure 2A and B). This conserved property of p150 proteins requiring 36 homologous amino acids (Figures 1A and and5,5, right) explains interspecies interactions. Deletion of this region in hp150 abolishes the formation of p150 dimers (Figure 2A and B) and, most importantly, impairs its ability to promote chromatin assembly in vitro (Figure 2C). Furthermore, the interference assay shown in Figure 7C argues for the functional importance of a dimer of CAF-1–p150 to participate actively in nucleosome assembly. These data not only provide an explanation for the in vivo effect of hp150 expression but also have strong implications for the mechanism of CAF-1 function.
It is tempting to speculate that the p150 subunit alternates between homodimeric and dimeric forms and that these forms display different interaction properties. One possibility would be that the intrinsic component of the CAF-1 complex, i.e. the p60 and p48 proteins, would not bind equally well to these forms. In this context, it is interesting to note that based on immunoprecipitation experiments (Smith and Stillman, 1991), it was suggested that there might be a fraction of p150 that would not be associated with p60. Other interactions could also be modulated by the monomeric state of p150, with major consequences for the chromatin assembly reaction such as binding to PCNA and to histones H3 and H4. Future experiments to explore whether the monomeric p150 mutants are capable of binding to PCNA and to histones H3 and H4 will thus be very interesting and should provide major insight into the mechanism of CAF-1 function. In any case, a dynamic cycle between these possible p150-containing complexes could be used to regulate CAF-1 activity. The presence of DNA damage (Gaillard et al., 1996; Martini et al., 1998) or the rapid cell divisions of early embryogenesis that increase the requirement for CAF-1 may act on the equilibrium between p150 complexes. The regulation of this equilibrium could be accomplished by the binding of an as yet unidentified factor, or by post-translational modifications such as phosphorylation/dephosphorylation, since various phosphorylated forms of p150 do exist (Smith and Stillman, 1991; Marheineke and Krude, 1998; Martini et al., 1998; Keller and Krude, 2000). As a consequence, interfering with the rapid transition between p150 complexes would compromise CAF-1-dependent chromatin assembly activities in vivo.
A requirement for p150 CAF-1 during the rapid cell divisions of early Xenopus development
Dominant-negative strategies have been applied successfully to assign a specific function to proteins that can oligomerize (Herskowitz, 1987). The dimerization properties of p150 CAF-1 using a unique defined region of 36 amino acids thus presented an attractive possibility for the investigation of the functional importance of CAF-1 in vivo. Exogenous expression of the largest subunit of the human CAF-1 complex, hp150, at levels comparable to its endogenous conterpart (see Figure 3B), arrested developmental progression in the Xenopus embryo prior to MBT (Figure 4A). The specificity of the p150 interference in our experiment was assessed as follow: (i) the interference is only obtained with the p150 forms containing the 36 amino acids dimerization motif (Figure 5A); (ii) this motif could not be found in other proteins by a BLAST search through data banks; (iii) immunoprecipitation experiments in the embryo (Figure 3) effectively pulled-down p150 homo/heterodimers but not other known histone chaperone such as N1 and nucleoplasmin, PCNA or histones themselves; (iv) this interference was not mediated by hp60 (a specific p150 interactor) expression; (v) the co-injection of hp60 with hp150 could neutralize the negative effect mediated by hp150 or hp150(620–938) when injected alone (Figure 6); and (vi) a truncated version of xp150 containing the conserved dimerization motif, but not the full-length xp150, led to a similar effect, hence allowing a dosage effect to be ruled out (see Figure 5B). Considering the critical importance of the p150 CAF-1 subunit for the function of the assembly factor in vitro, our results argue that a CAF-1-dependent chromatin assembly pathway is essential in early development related to either replication or repair events. In support of this hypothesis, similar interference can be obtained in vitro for chromatin assembly on plasmid DNA (Figure 7C). Thus, the most parsimonious explanation of both the in vivo and in vitro data is that nucleosome assembly requires dimerization of the p150 subunit and that the human–Xenopus heterodimer is somehow inactive for nucleosome assembly as shown in Figure Figure77C.
During the pre-MBT stages of the Xenopus embryo, the rapid alternation of S and M phases (Kirschner et al., 1985) requires a tight coordination between chromatin assembly and DNA replication or repair events, which is fulfilled using a rapid assembly pathway coupled to DNA synthesis (Almouzni and Wolffe, 1993), possibly through CAF-1. A slower chromatinization process might lead to a mitotic catastrophe (Enoch and Nurse, 1991) in the simple embryonic cell cycle, which lacks control upon completion of S phase to enter into mitosis (Murray and Kirschner, 1989). This mitotic catastrophe scenario could perhaps explain the results in Figure 4B showing that the large cells expressing hp150 fail to stain with DAPI. The embryonic requirement for CAF-1 function is in contrast to observations in a Xenopus somatic cell line (A6 cells) in which our attempts at expression of hp150 did not affect viability (data not shown). These cells, in transient transfection assays, could express h150 for >72 h (two to three successive divisions) without apparent defects when compared with neighbouring cells that did not express hp150 as detected by indirect immunofluorescence (not shown). The context of a somatic cell cycle with a longer division time and gap phases would perhaps be more tolerant of perturbations in chromatin assembly and could use a CAF-1-independent pathway to compensate. To investigate this hypothesis further, other cell lines should be examined. Nevertheless, this hypothesis could be consistent with the mild phenotype of CAF-1 deletion mutants in S.cerevisiae (Enomoto et al., 1997; Kaufman et al., 1997; Monson et al., 1997) in which alternative pathways for chromatin assembly may exist that depend on factors different from CAF-1. One of these factors may be ASF1, which has been shown to cooperate with CAF-1 in the efficient assembly of nucleosomes coupled to DNA replication in vitro (Tyler et al., 1999) and can also facilitate nucleosome formation on non-replicating DNA (Munakata et al., 2000). Although ASF1 proteins remain to be identified in Xenopus, it is possible that homologues could play similar roles. Specialized chaperones that are highly abundant during early development in Xenopus, such as nucleoplasmin, which interacts with histones H2A–H2B (Laskey et al., 1978), and N1/N2, which associates with histones H3 and H4 (Kleinschmidt et al., 1986), could also promote the assembly reaction. Considering their lack of specificity for newly synthesized DNA, these histone chaperones could act as histone donors in an assembly line ending with CAF-1 to ensure a rapid assembly coordinated with DNA synthesis. Alternatively, they may also function independently of CAF-1 for a slow deposition process that would not be compatible with the rapid division of early development. The frog early embryonic cell divisions that are characterized by an extremely stringent need for nucleosome formation in a short time provide the cellular environment most sensitive to a perturbation of CAF-1 function. Since a number of other organisms including Drosophila, sea urchin and zebrafish also undergo rapid embryonic divisions (Yasuda and Schubiger, 1992), it could be anticipated that similar requirements may be imposed during their early development. It is tempting to speculate that the demand for CAF-1 could be directly related to a rapidly proliferating state. In this context and in the absence of proper checkpoint controls, alteration of CAF-1 function would lead to major cellular defects.
Taken together, our results emphasize for the first time the importance of both a developmental and a cellular context to appreciate the requirement for specific chromatin assembly factors. These parameters will surely be equally critical to evaluate the importance of the increasing number of chromatin-modifying factors with redundant functions.
Materials and methods
Yeast two-hybrid screen and cloning of Xenopus p150
The bait used in the two-hybrid screen was constructed as follows: the DNA fragment encoding hp150(620–938) was excised from the plasmid pPK8 (Kaufman et al., 1995) and inserted into the pNLX3 vector (Iouzalen et al., 1998) in-frame with the LexA tag. A Xenopus oocyte (stage I–VI) cDNA library cloned into the pGAD-GE vector downstream of the Gal4 activation domain was transformed into the L40 yeast reporter strain (Iouzalen et al., 1998) containing pNLX3-hp150(620–938). Forty-four putative candidates were isolated following standard two-hybrid selection assays. Two clones contained a cDNA encoding an open reading frame (ORF) with 58% amino acid identity and 73% similarity to the portion of the hp150 CAF-1 protein between amino acid 559 and the C-terminal end. The complete xp150 cDNA was obtained with the 5′RACE system (Gibco) using RNA prepared from Xenopus A6 cells (RNAeasy starter kit, Qiagen). DNA sequencing was performed with the Big Dye Terminator kit (PE Applied Biosystem) and the automatic PE sequencer ABI PRISM 310 Genetic Analyzer. The xp150 DDBJ/EMBL/GenBank accession No. is AF222339. Yeast media, growth, transformation, selection and plasmid recovery were as described (Guthrie and Fink, 1993).
Chromatin assembly assays
Analysis of DNA repair and chromatin assembly was performed as described (Gaillard et al., 1997). Briefly, circular pBscript plasmid either mock treated or UV irradiated (500 J/m2) was incubated with Xenopus embryo extract (HSE) (mock or xp150 depleted), S100 human cytosolic extract (Smith and Stillman, 1989) complemented or not by HeLa nuclear extract (Martini et al., 1998), in vitro translated or purified bacterially produced proteins or HSE, in the presence of [α-32P]dCTP for 3 h at 23 (HSE) or 37°C (S100). The extent of supercoiling and DNA synthesis was examined by agarose gel electrophoresis followed by drying of the gel and exposure to X-ray film.
Immunological procedures
Polyclonal anti-xp150 antibody (serum 566) and anti-hp150 antibody (serum 573) were obtained after injections of purified GST–xp150(540– 896) and His6-hp150(620–938) proteins in rabbits (AgroBio, Villeny). Both antibodies were affinity purified by incubating 200 µl of serum with 5–10 µg of purified GST or His6 fusion protein present on nitrocellulose strips (Smith and Fisher, 1984).
For immunodepletion, 50 µl of HSE were mixed with an equal volume of anti-xp150 antibodies (566) bound to protein A–Sepharose for 30 min at 4°C. After centrifugation, the supernatant was subjected to a second round of depletion prior to its use for chromatin assembly reactions.
Embryo extracts were prepared by homogenization in buffer E [20 mM HEPES–KOH pH 7.5, 250 mM sucrose, 50 mM KCl, 2. 5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg/ml pepstatin and leupeptin] and centrifugation at 100 000 g for 30 min at 4°C. For immunoprecipitations, the cleared embryo extract was incubated with an equal volume of anti-xp150 affinity-purified antibody (566) or anti-hp150 affinity-purified antibody (573) for 1 h at 4°C. Protein A–Sepharose (Pharmacia Amersham Biotech) slurry (50 µl) equilibrated in buffer E was used to pull out the antibody.
Proteins were detected on western blots using the polyclonal anti-hp150 573 antibody (1/1000 dilution), the polyclonal anti-xp150 566 antibody (1/1000 dilution), the monoclonal anti-nucleoplasmin PAC35 antibody (1/500 dilution) (Dilworth et al., 1987), the monoclonal anti-PCNA antibody (PC10, DAKO, 1/500 dilution), the polyclonal anti-H4AcK5 antibody (1/500 dilution) (Turner and Fellows, 1989) and the anti-His6 antibody (Santa Cruz, 1/000). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies and the SuperSignal substrate detection kit (Pierce) were used for visualization.
For immunofluorescence, embryos were fixed overnight in 2% paraformaldehyde, 15% sucrose in 0.1× MBS [88 mM NaCl, 1 mM KCl, 0.41 mM CaCl2, 0.33 mM Ca(NO3)2, 0.82 mM MgSO4, 2.4 mM NaHCO3, 10 mM HEPES–KOH pH 7.4] solution, embedded in TissuTek (Sakura) and deep frozen in isopentane cooled by liquid nitrogen. Cryosections of 5 µm were prepared using a cryostat (Leica). The sections were adsorbed on superfrost plus slides and stored at –20°C. Human p150 was detected using mAb1 (1/100 dilution) (Smith and Stillman, 1991) and Texas red-coupled secondary antibody (Martini et al., 1998).
Recombinant proteins, GST pull-down
Recombinant proteins were co-expressed by transforming BL21 (DE3) bacteria (Studier et al., 1990) containing the pET30-hp150(620–938) plasmid or pET30-hp150(678–938) plasmid with pGEX4T-hp150(620– 938), pGEX4T-hp150(678–938) or pGEX4T-xp150(540–896). After induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1.5 h at 30°C, cells were lysed in buffer A (20 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 10 mg/ml pepstatin and leupeptin), sonicated briefly and the soluble fraction incubated with glutathione–Sepharose 4B resin (Pharmacia Amersham Biotech) for 1.5 h at 4°C. After extensive washes in buffer A, the resin was extracted with 1 M NaCl in buffer A. Total proteins bound to the resin were recovered by resuspending the washed resin directly in SDS–PAGE sample buffer (Laemmli, 1970). Bacterial extracts containing wild-type hp150, hp150Δ(642–678) and wild-type xp150 were produced as above. Proteins were purified over a nickel column (Pharmacia) according to the manufacturer’s recommendation and subsequently dialysed against HSE extraction buffer (Gaillard et al., 1997).
In vitro transcription–translation and glycerol gradients
Proteins were translated in vitro from the DNA constructs pβhp150/RN3, pβhp150Δ(642–678)/RN3 and pβxp150/RN3 using the TNT reticulocyte lysate coupled in vitro transcription–translation system (Promega) following the manufacturer’s instructions. Continuous 10–40% glycerol gradients in buffer G (20 mM Tris, 100 mM or 2 M NaCl, 0.025% NP-40, 2 mM EDTA) were poured into 5 ml tubes. A 50 µl aliquot of in vitro translated proteins or bacterial extract to which 150 µl of buffer G were added was loaded on the top of the gradients after a clearing spin of 10 min at 10 000 g at 4°C. Bovine serum albumin (BSA; 66 kDa) and catalase (258 kDa) were mixed and loaded in parallel in each gradient as sedimentation standards. The samples were centrifuged in an SW55 Beckman rotor at 45 000 r.p.m. for 18 h at 4°C. The refraction index was measured for each fraction collected (200 µl) to verify that gradients were identical.
DNA constructs
The plasmid pPK8 (Kaufman et al., 1995) was used to PCR clone the following fragments of hp150 (the first and last amino acids are indicated in parentheses): hp150(1–649) into pET30 vector (His6 tag, Novagen); hp150(678–938) into pGEX4T-1 (GST tag, Pharmacia); and pET30 vectors. hp150(620–938) was excised from plasmid pNLX-hp150(620– 938) (see two-hybrid screen) and inserted into pGEX4T-1 and pET30 vector. The plasmids pPK8 and pED (Kaufman et al., 1995) were used to PCR clone full-length wild-type hp150, hp150(1–649) and hp150ΔED into the pβGFP/RN3 plasmid (Zernicka-Goetz et al., 1996). To produce hp150Δ(642–678), two PCR fragments were generated, one encoding hp150(1–642) and the second hp150(678–938). These PCR products were mixed and used as template for a second PCR with primers annealing 5′ of the first fragment and 3′ of the second. The resulting PCR product, hp150Δ(642–678), was cloned into pβGFP/RN3. The xp150(540–896) retrieved in our two-hybrid screen was cloned into pGEX4T-1. The complete xp150 cDNA was cloned as an EcoRI–NotI fragment into EcoRI–NotI-cleaved pβGFP/RN3P vector, generating pβxp150/RN3. Wild-type hp150, hp150Δ(642–678) and xp150 were subcloned from the pβ/RN3 plasmids into pET30 to generate His6 fusions.
RNA preparation and microinjection into embryos
For embryo injections, 5′-capped RNAs were prepared using the Riboprobe system (Promega). PCR-amplified fragments from pβGFP/RN3, pβhp150/RN3, pβhp150ΔED/RN3, pβhp150(1–649)/RN3, pβhp150Δ(642–678)/RN3 and pβxp150/RN3 were used as templates for T3 RNA polymerase. RNAs produced from the pβ/RN3 vector contain stabilizing 5′ and 3′ β-globin untranslated regions (Zernicka-Goetz et al., 1996). Linearized pET30-hp150(620–938), pET30-hp150(678– 938), pET30-xp150(540–896) and pPK7 (hp60) (Kaufman et al., 1995) plasmids were used as templates for T7 RNA polymerase. Xenopus embryos were prepared for injection as described in Vize et al. (1991). RNAs in injection buffer (88 mM NaCl, 10 mM HEPES–KOH pH 7.6) were injected into one of the two blastomeres using a nanoject injector (Drummond) adjusted for a delivery of 32.2 nl. Developmental stages of the embryos were determined according to Nieuwkoop and Faber (1994).
Acknowledgements
We thank D.M.J.Roche and F.Sangrado for all their assistance during this work, as well as members of the group. We are grateful to Y.Abraham for his advice on the two-hybrid screen. We thank P.Ridgway, J.Mello, J.Moggs, C.Green and F.Schweisguth for critical reading of the manuscript, and S.Dilworth, B.Turner and B.Stillman for kindly providing us with antibodies. This work was supported by the Association pour la Recherche sur le Cancer and La Ligue Nationale contre le Cancer. P.G. was supported by an EMBO fellowship.
References
- Adams C.R. and Kamakaka,R.T. (1999) Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev., 9, 185–190. [Abstract] [Google Scholar]
- Almouzni G. and Wolffe,A.P. (1993) Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev., 7, 2033–2047. [Abstract] [Google Scholar]
- Dawid I.B. and Sargent,T.D. (1988) Xenopus laevis in developmental and molecular biology. Science, 240, 1443–1448. [Abstract] [Google Scholar]
- Dilworth S.M., Black,S.J. and Laskey,R.A. (1987) Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell, 51, 1009–1018. [Abstract] [Google Scholar]
- Enoch T. and Nurse,P. (1991) Coupling M phase and S phase: controls maintaining the dependence of mitosis on chromosome replication. Cell, 65, 921–923. [Abstract] [Google Scholar]
- Enomoto S. and Berman,J. (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev., 12, 219–232. [Europe PMC free article] [Abstract] [Google Scholar]
- Enomoto S., McCune-Zierath,P., Geraminejad,M., Sanders,M. and Berman,J. (1997) Rlf2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev., 11, 358–363. [Abstract] [Google Scholar]
- Gaillard P.H., Martini,E.M., Kaufman,P.D., Stillman,B., Moustacchi,E. and Almouzni,G. (1996) Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell, 86, 887–896. [Abstract] [Google Scholar]
- Gaillard P.-H., Moggs,J.G., Roche,D.M.J., Quivy,J.-P., Becker,P.B., Wood,R.D. and Almouzni,G. (1997) Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J., 16, 6281–6289. [Europe PMC free article] [Abstract] [Google Scholar]
- Game J. and Kaufman,P. (1999) Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics, 151, 485–497. [Europe PMC free article] [Abstract] [Google Scholar]
- Guthrie G. and Fink,G.R. (eds) (1993) Guide to yeast genetics and molecular biology. Methods Enzymol., 194, 1–863. [Abstract] [Google Scholar]
- Herskowitz I. (1987) Functional inactivation of genes by dominant negative mutations. Nature, 329, 219–222. [Abstract] [Google Scholar]
- Iouzalen N., Camonis,J. and Moreau,J. (1998) Identification and characterization in Xenopus of XsmgGDS, a RalB binding protein. Biochem. Biophys. Res. Commun., 250, 359–363. [Abstract] [Google Scholar]
- Ito T., Tyler,J.K. and Kadonaga,J.T. (1997) Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells, 2, 593–600. [Abstract] [Google Scholar]
- Kamakaka R.T., Bulger,M., Kaufman,P.D., Stillman,B. and Kadonaga,J.T. (1996) Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor I. Mol. Cell. Biol., 16, 810–817. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaufman P.D. and Almouzni,G. (2000) Chromatin assembly, DNA replication and repair. In Workman,J. and Elgin,S. (eds), Chromatin structure and Gene Expression. Frontiers in Molecular Biology, 2nd edn. Oxford University Press, Oxford, UK, pp. 24–48.
- Kaufman P.D., Kobayashi,R., Kessler,N. and Stillman,B. (1995) The p150 and p60 subunits of chromatin assembly factor I—a molecular link between newly synthesized histones and DNA replication. Cell, 81, 1105–1114. [Abstract] [Google Scholar]
- Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor I. Genes Dev., 11, 345–357. [Abstract] [Google Scholar]
- Keller C. and Krude,T. (2000) Requirement of cyclin/Cdk2 and protein phosphatase 1 activity for chromatin assembly factor 1-dependent chromatin assembly during DNA synthesis. J. Biol. Chem., 275, 35512–35521. [Abstract] [Google Scholar]
- Kirschner M., Newport,J. and Gerhart,J. (1985) The timing of early developmental events in Xenopus. Trends Genet., 2, 41–47. [Google Scholar]
- Kleinschmidt J.A., Dingwall,C., Maier,G. and Franke,W.W. (1986) Molecular characterization of a karyophilic, histone-binding protein: cDNA cloning, amino acid sequence and expression of nuclear protein N1/N2 of Xenopus laevis. EMBO J., 5, 3547–3552. [Europe PMC free article] [Abstract] [Google Scholar]
- Kleinschmidt J.A., Seiter,A. and Zentgraf,H. (1990) Nucleosome assembly in vitro: separate histone transfer and synergistic interaction of native histone complexes purified from nuclei of Xenopus laevis oocytes. EMBO J., 9, 1309–1318. [Europe PMC free article] [Abstract] [Google Scholar]
- Kornberg R.D. and Lorch,Y. (1999) Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev., 9, 148–151. [Abstract] [Google Scholar]
- Krude T. (1995) Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res., 220, 304–311. [Abstract] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [Abstract] [Google Scholar]
- Laskey R.A., Honda,B.M., Mills,A.D. and Finch,J.T. (1978) Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature, 275, 416–420. [Abstract] [Google Scholar]
- Marheineke K. and Krude,T. (1998) Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem., 273, 15279–15286. [Abstract] [Google Scholar]
- Martini E., Roche,D.M.J., Marheineke,K., Verreault,A. and Almouzni,G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin following UV irradiation of human cells. J. Cell Biol., 143, 563–575. [Europe PMC free article] [Abstract] [Google Scholar]
- Moggs J.G., Grandi,P., Quivy,J., Jonsson,Z.O., Huebscher,U., Becker,P.B. and Almouzni,G. (2000) A CAF-1/PCNA mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol., 20, 1206–1218. [Europe PMC free article] [Abstract] [Google Scholar]
- Monson E.K., de Bruin,D. and Zakian,V.A. (1997) The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl Acad. Sci. USA, 94, 13081–13086. [Europe PMC free article] [Abstract] [Google Scholar]
- Munakata T., Adachi,N., Yokoyama,N., Kuzuhara,T. and Horikoshi,M. (2000) A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells, 5, 221–233. [Abstract] [Google Scholar]
- Murray A.W. and Kirschner M.W. (1989) Dominoes and clocks: the union of two views of the cell cycle. Science, 246, 614–621. [Abstract] [Google Scholar]
- Nieuwkoop P. and Faber,P. (1994) Normal Table of Xenopus laevis. North-Holland Publishing, Amsterdam, The Netherlands.
- Philpott A., Krude,T. and Laskey,R.A. (2000) Nuclear chaperones. Cell Dev. Biol., 11, 7–14. [Abstract] [Google Scholar]
- Ridgway, P. and Almouzni,G. (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci., 113, 2647–2658. [Abstract] [Google Scholar]
- Shibahara K. and Stillman,B. (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell, 96, 575–585. [Abstract] [Google Scholar]
- Smith D.E. and Fisher,P.A. (1984) Identification, developmental regulation and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J. Cell Biol., 99, 20–28. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith S. and Stillman,B. (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell, 58, 15–25. [Abstract] [Google Scholar]
- Smith S. and Stillman,B. (1991) Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J. Biol. Chem., 266, 12041–12047. [Abstract] [Google Scholar]
- Studier F.W., Rosenberg,A., Dunn,J.J. and Dubendoff,J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [Abstract] [Google Scholar]
- Taddei A., Roche,D., Sibarita,J., Turner,B. and Almouzni,G. (1999) Duplication and maintenance of heterochromatin domains. J. Cell Biol., 147, 1153–1166. [Europe PMC free article] [Abstract] [Google Scholar]
- Turner B.M. and Fellows,G. (1989) Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur. J. Biochem., 179, 131–139. [Abstract] [Google Scholar]
- Tyler J.K., Adams,C.R., Chen,S., Kobayashi,R., Kamakaka,T.R. and Kadonaga,J.T. (1999) The RCAF complex mediates chromatin assembly during replication and repair. Nature, 402, 555–560. [Abstract] [Google Scholar]
- Verreault, A. (2000) De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev., 14, 1430–1438. [Abstract] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [Abstract] [Google Scholar]
- Vize P.D., Melton,D.A., Hemmati-Brivanlou,A. and Harland,R. (1991) Assays for gene function in developing Xenopus embryos. Methods Cell Biol., 36, 367–387. [Abstract] [Google Scholar]
- Wolffe A. (1998) Chromatin: Structure and Function, 3rd edn. Academic Press, London, UK.
- Workman J.L. and Kingston,R.E. (1998). Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [Abstract] [Google Scholar]
- Yasuda G.K. and Schubiger,G. (1992) Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet., 8, 124–127. [Abstract] [Google Scholar]
- Zernicka-Goetz M., Pines,J., Ryan,K., Siemering,K.R., Haseloff,J., Evans,M.J. and Gurdon,J.B. (1996) An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development, 122, 3719–3724. [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1093/emboj/20.8.2015
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1093/emboj/20.8.2015
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/126255840
Article citations
Roles of Histone H2B, H3 and H4 Variants in Cancer Development and Prognosis.
Int J Mol Sci, 25(17):9699, 07 Sep 2024
Cited by: 0 articles | PMID: 39273649 | PMCID: PMC11395991
Review Free full text in Europe PMC
Replication-coupled inheritance of chromatin states.
Cell Insight, 3(6):100195, 23 Aug 2024
Cited by: 0 articles | PMID: 39391004 | PMCID: PMC11462216
Review Free full text in Europe PMC
The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance.
Biology (Basel), 13(8):572, 29 Jul 2024
Cited by: 0 articles | PMID: 39194510 | PMCID: PMC11351467
Review Free full text in Europe PMC
The cell-cycle choreography of H3 variants shapes the genome.
Mol Cell, 83(21):3773-3786, 20 Sep 2023
Cited by: 5 articles | PMID: 37734377 | PMCID: PMC10621666
Review Free full text in Europe PMC
Unorthodox PCNA Binding by Chromatin Assembly Factor 1.
Int J Mol Sci, 23(19):11099, 21 Sep 2022
Cited by: 4 articles | PMID: 36232396 | PMCID: PMC9570017
Go to all (75) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - AF222339
- (1 citation) ENA - AJ132771
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I.
Genes Dev, 11(3):345-357, 01 Feb 1997
Cited by: 270 articles | PMID: 9030687
Biochemical and Structural Insights into the Winged Helix Domain of P150, the Largest Subunit of the Chromatin Assembly Factor 1.
Int J Mol Sci, 23(4):2160, 15 Feb 2022
Cited by: 3 articles | PMID: 35216276 | PMCID: PMC8874411
A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage.
Mol Cell Biol, 20(4):1206-1218, 01 Feb 2000
Cited by: 205 articles | PMID: 10648606 | PMCID: PMC85246
Chromatin assembly during DNA replication in somatic cells.
Eur J Biochem, 263(1):1-5, 01 Jul 1999
Cited by: 52 articles | PMID: 10429179
Review




