Abstract
Free full text

The Hypothetical Protein CT813 Is Localized in the Chlamydia trachomatis Inclusion Membrane and Is Immunogenic in Women Urogenitally Infected with C. trachomatis
Abstract
Using antibodies raised with chlamydial fusion proteins, we have localized a protein encoded by hypothetical open reading frame CT813 in the inclusion membrane of Chlamydia trachomatis. The detection of the C. trachomatis inclusion membrane by an anti-CT813 antibody was blocked by the CT813 protein but not unrelated fusion proteins. The CT813 protein was detected as early as 12 h after chlamydial infection and was present in the inclusion membrane during the entire growth cycle. All tested serovars from C. trachomatis but not other chlamydial species expressed the CT813 protein. Exogenously expressed CT813 protein in HeLa cells displayed a cytoskeleton-like structure similar to but not overlapping with host cell intermediate filaments, suggesting that the CT813 protein is able to either polymerize or associate with host cell cytoskeletal structures. Finally, women with C. trachomatis urogenital infection developed high titers of antibodies to the CT813 protein, demonstrating that the CT813 protein is not only expressed but also immunogenic during chlamydial infection in humans. In all, the CT813 protein is an inclusion membrane protein unique to C. trachomatis species and has the potential to interact with host cells and induce host immune responses during natural infection. Thus, the CT813 protein may represent an important candidate for understanding C. trachomatis pathogenesis and developing intervention and prevention strategies for controlling C. trachomatis infection.
Chlamydiae, a family of obligate intracellular bacterial pathogens, consist of many different species imposing different health problems on animals and humans. For example, Chlamydia caviae GPIC strain (the agent of guinea pig inclusion conjunctivitis) is a natural pathogen of the guinea pig which can cause both ocular and genital tract infections (35), while Chlamydia psittaci 6BC strain causes infections mainly in birds (27, 40). These animal pathogens can also accidentally infect humans (54). The human chlamydial species Chlamydia pneumoniae infects mainly the human respiratory tract (19, 30), and C. pneumoniae infection is associated with cardiovascular diseases (29). The species Chlamydia trachomatis consists of more than 15 different serovars designated A to L, including Ba, L1, L2, and L3 plus various subtypes. Different serovars cause different diseases in humans, with serovars A to C infecting human eyes, potentially leading to preventable blindness (62), and serovars D to K infecting the human urogenital tract; if left untreated, this infection can cause severe complications such as ectopic pregnancy and infertility (28, 53). The L, also known as LGV (lymphogranuloma venereum), serovars can occasionally cause outbreak infections in certain human populations (7, 47, 55). The mouse biovar of C. trachomatis, formerly known as mouse pneumonitis agent (designated MoPn), is now classified as a new species, Chlamydia muridarum. However, MoPn has been and is still extensively used to study C. trachomatis pathogenesis and immunobiology in mouse models (6, 10, 32, 36, 38, 39, 66).
Despite the large differences in tissue tropism and disease processes among different chlamydial organisms, all chlamydial species have similar genomes (41, 42, 57) and share a common intracellular biphasic growth cycle (21, 22). Chlamydial pathogenicity is determined mainly by the chlamydial ability to replicate inside a cytoplasmic vacuole of host cells. A typical chlamydial infection starts with the entry of elementary bodies (EBs), the infectious form, into host cells via endocytosis. The internalized EBs within the endosomal vacuole can rapidly differentiate into reticulate bodies, the metabolically active but noninfectious form of chlamydial organisms. After replication, the reticulate bodies can differentiate back into EBs to spread to adjacent cells. Chlamydia can accomplish its entire biosynthesis, particle assembly and differentiation within the cytoplasmic vacuole (also termed inclusion). The chlamydial inclusion membrane serves as both a barrier for protecting the intravacuolar organisms and a gate for chlamydial interactions with host cells. To establish and maintain its intravacuolar growth, chlamydia must exchange both materials and signals with the host cell across the inclusion membrane. Chlamydia not only is able to import nutrients and metabolic intermediates from host cells (9, 23, 24, 48, 59) but also secretes chlamydial factors into host cells (70). Chlamydia can actively manipulate host signal pathways (15, 20, 59, 64). Despite the frequent exchanges in both materials and information between chlamydia and the host cell, the mechanisms by which these exchanges occur across the inclusion membrane are largely unknown. Since proteins localized in the inclusion membrane can potentially play important roles in chlamydial interactions with host cells, identification and characterization of chlamydial inclusion membrane proteins have become an area of intensive investigation.
In the past decade, significant progress has been made in identifying chlamydial inclusion membrane proteins (designated Inc). Since Rockey et al. (44) reported the first chlamydial inclusion membrane protein (designated IncA) from C. caviae (GPIC) in 1995, many Inc homologues have been described for C. trachomatis. For example, the regions of the C. trachomatis genome covering open reading frames (ORFs) CT115 to CT119 (5, 50) and CT222 to CT233 (3, 4) carry numerous inc genes, although not every protein encoded in these regions has been shown to be in the chlamydial inclusion membrane (3). Several other C. trachomatis proteins encoded by genes outside of the above genomic regions were also found in the chlamydial inclusion membrane, including the CT089 (17), CT442 (3, 56), and CT529 (18) proteins. As the chlamydial genome sequences became available and in an attempt to search for more inclusion membrane proteins, both Bannantine et al. (3) and Toh et al. (63) used computer-based methods to predict chlamydial inclusion membrane proteins. Although about 100 chlamydial proteins were predicted to localize in the chlamydial inclusion membrane (3, 63), these computer prediction results have not been validated with experimental evidence. Indeed, some of the predicted inclusion membrane proteins were determined not to be in the inclusion membrane (3). Therefore, it is necessary to use experimental approaches to identify and characterize new inclusion membrane proteins.
Since chlamydial protein localization is a phenotype that can be experimentally tracked with specific reagents, we have initiated an effort to use antibodies raised with chlamydial fusion proteins to localize the endogenous chlamydial proteins. After screening about 300 such antibodies, we identified a new chlamydial inclusion membrane protein that is encoded by the hypothetical ORF CT813. CT813 is unique to C. trachomatis species, including the murine biovar MoPn. There are no homologues of the CT813 protein in other chlamydial species, although there is a low level of homology between the CT813 protein and several proteins from nonchlamydial species (data not shown). The CT813 protein is expressed in cultures by serovars from C. trachomatis but not other chlamydial species. The CT813 protein is also expressed during chlamydial natural infection in humans, since women with C. trachomatis urogenital infection developed high titers of antibodies to the CT813 protein. Thus, the CT813 protein may represent an important target for both understanding C. trachomatis pathogenesis and developing intervention and prevention strategies for controlling C. trachomatis infection.
MATERIALS AND METHODS
Chlamydial organisms and infection.
The chlamydial serovars/strains used for the present study include A, B, C, D, E, F, G, I, K, L1, L3, and Ba (obtained from Harlan Caldwell at the Rocky Mountain Laboratory, NIAID, NIH, Hamilton, Montana), 6BC (Thomas Hatch at the University of Tennessee, Memphis) (14), MoPn (Louis De La Maza, University of California, Irvine) (37), and L2 and GPIC (our own stocks). These organisms were grown, purified, and titrated as previously described (20). Aliquots of the organisms were stored at −80°C until use. HeLa cells (ATCC, Manassas, VA) maintained in Dulbecco modified Eagle medium (GIBCO BRL, Rockville, MD) with 10% fetal calf serum (GIBCO BRL) at 37°C in an incubator supplied with 5% CO2 were used for the present study. To prepare chlamydial infection samples for an immunofluorescence assay, HeLa cells were grown on glass coverslips in 24-well plates overnight prior to chlamydial inoculation. Chlamydial organisms diluted in Dulbecco modified Eagle medium with 10% fetal calf serum and 2 μg/ml of cycloheximide (Sigma, St. Louis, MO) were directly inoculated onto the cell monolayers. The infection dose was pretitrated for individual serovars, and an infection rate of ~50% was applied for all serovars. The cell samples were cultured at 37°C in a CO2 incubator and processed at various time points after infection as indicated for the individual experiments. For the Western blot assay, the chlamydial infection was carried out similarly as described above except that the cell samples were grown in 75-cm2 tissue culture flasks and collected via lysis with a 2% sodium dodecyl sulfate (SDS) sample buffer.
Prokaryotic expression of chlamydial fusion proteins and production of antifusion protein antibodies.
The ORFs coding for hypothetical proteins, including the CT813 protein, and various known proteins, including IncG, IncA, MOMP, HSP60, and CPAF from the C. trachomatis serovar D genome (http://www.stdgen.lanl.gov/), were cloned into pGEX vectors (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) and expressed as fusion proteins with glutathione S-transferase (GST) fused to the N terminus of the chlamydial proteins. Expression of the fusion proteins was induced with isopropyl-β-d-thiogalactopyranoside (IPTG; Invitrogen, Carlsbad, CA), and the fusion proteins were extracted by lysing the bacteria via sonication in Triton X-100 lysis buffer (1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 75 units/ml of aprotinin, 20 μM leupeptin, and 1.6 μM pepstatin). After high-speed centrifugation to remove debris, the fusion protein-containing supernatants were either directly used in various assays or further purified using glutathione-conjugated agarose beads (Pharmacia). The bead-bound fusion proteins were also used to deplete antigen-specific antibodies from antiserum samples (see below). For antibody production, the purified fusion proteins were used to immunize mice as described previously (67, 68, 71-73). After the titers of specific antibody reached 1:2,000 or higher, the mice were sacrificed. The mouse sera were collected and stored in 50% glycerol at −20°C until use.
Transient transfection of mammalian cells.
The ORF coding for the CT813 protein from the C. trachomatis serovar D genome was cloned into the pDsRed-Monomer-C1 (BD Biosciences Clontech, San Jose, CA) and pFLAG-CMV-4 (Sigma, St. Louis, MO) mammalian expression vector systems with either a red fluorescence protein (RFP) gene or a FLAG tag coding sequence (24 nucleotides) fused to the 5′ end of CT813. The recombinant plasmids were transfected into HeLa cells by using Lipofectamine 2000 transfection reagent following the protocol recommended by the manufacturer (Invitrogen, Carlsbad, CA). At various time points after transfection, indicated for the individual experiments, CT813 protein expression was visualized via either the fusion tag RFP or mouse anti-D813 antibody labeling.
Immunofluorescence staining.
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde dissolved in phosphate-buffered saline for 20 min at room temperature, followed by permeabilization with 0.1% Triton X-100 for an additional 4 min. After being washed and blocked, the cell samples were subjected to various combinations of antibody and chemical staining. Hoechst (blue) (Sigma) was used to visualize nuclear DNA. A rabbit antichlamydial organism antibody (R1L2, raised with C. trachomatis serovar L2 organisms) (data not shown), anti-CT395 antibody (raised with the CT395 fusion protein; the CT395 protein is a GrpE-related chaperonin with >70% amino acid sequence identity among all chlamydial species) (data not shown), or anti-IncG antibody (kindly provided by Ted Hackstadt at the Rocky Mountain Laboratory, NIAID, NIH, Hamilton, Montana) (50) plus a goat anti-rabbit immunoglobulin G (IgG) secondary antibody conjugated with Cy2 (green) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was used to visualize chlamydial inclusions or the inclusion membrane. The mouse antibodies against the CT813 protein, IncG, MOMP (monoclonal antibody, clone MC22), and CPAF (monoclonal antibody, clone 100a) plus goat anti-mouse IgG conjugated with Cy3 (red) (Jackson ImmunoResearch) were used to visualize the corresponding antigens. In some cases, the primary antibodies were preabsorbed with either the corresponding or heterologous fusion proteins immobilized onto agarose beads (Pharmacia) prior to the staining of cell samples. The preabsorption approach was carried out by incubating the antibodies with bead-immobilized antigens for 1 h at room temperature or overnight at 4°C, followed by pelleting of the beads. The remaining supernatants were used for immunostaining. For the transfected cell samples, the CT813 protein was visualized either by the fusion tag RFP or by costaining with a mouse anti-CT813 antibody. In addition, the transfected cells were also costained with phalloidin conjugated with Alexa 488 (green) (Molecular Probes, Eugene, OR) to visualize F-actin, an anti-α-tubulin antibody (clone B-5-1-2; Sigma) to detect microtubules, and an anti-cytokeratin 8 antibody (clone M20; Sigma) to detect intermediate filaments (IF). The costainings were visualized by goat anti-mouse IgG conjugated with Cy2 (green) (Jackson ImmunoResearch).
Fluorescence and confocal microscopy.
After the appropriate immunolabeling, the cell samples were used for image analysis and acquisition with an Olympus AX-70 fluorescence microscope equipped with multiple filter sets (Olympus, Melville, NY) as described previously (15, 20, 65, 70). Briefly, the multicolor-labeled samples were exposed under a given filter set at a time and the single-color images were acquired using a Hamamatsu digital camera. The single-color images were then superimposed with the software SimplePCI to display multiple colors. An Olympus FluoView laser confocal microscope was used to further analyze the costained samples at the UTHSCSA institutional core facility. All microscopic images were processed using the Adobe Photoshop program (Adobe Systems, San Jose, CA).
Western blot assay.
The Western blot assay was carried out as described elsewhere (12, 13, 15, 52, 69). Briefly, the fusion protein, infected whole-cell lysate, or purified chlamydial organism samples were solubilized in 2% SDS sample buffer and loaded to SDS-polyacrylamide gel wells. After electrophoresis, the proteins were transferred to nitrocellulose membranes and the blots were detected with primary antibodies. Primary antibody binding was probed with a horseradish peroxidase-conjugated secondary antibody and visualized with an ECL kit (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The Western blot assay was used for the following purposes. To determine whether the CT813 protein is associated with the purified chlamydial organisms, the chlamydia-infected whole-cell lysate and purified EB samples were compared for their reactivities with the mouse anti-CT813 antibody. To validate the preabsorption efficiency, the mouse anti-CT813 and anti-IncG antibodies were preabsorbed with or without the corresponding or heterologous fusion proteins as described above and then applied to the nitrocellulose membrane. To monitor the time course of CT813 protein expression, the infected HeLa cell samples were harvested at various time points after infection and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). After the cell samples were transferred onto nitrocellulose membranes, the corresponding protein bands were detected with the mouse antibodies recognizing the CT813 protein, MOMP (clone MC22), and host beta-actin (clone Ac-15). To titrate the pooled human sera for reactivity with chlamydial fusion proteins, the purified fusion proteins were loaded at equal amounts to the corresponding lanes of SDS-polyacrylamide gels in multiple sets. One set was stained with brilliant blue R-250 (Sigma) to visualize the total amount of protein in each lane, and the rest of the sets were transferred onto nitrocellulose membrane to assess human antibody binding to the chlamydial fusion proteins after a serial dilution of the sera.
ELISA.
Ten human sera collected from women diagnosed with C. trachomatis urogenital infections (positive sera) and eight human sera collected from women without chlamydial infection (negative sera) were used in the current study. These human sera were measured for their reactivity with the CT813 protein and other chlamydial fusion proteins by using an enzyme-linked immunosorbent assay (ELISA) as described elsewhere (51, 72, 74), except that the fusion proteins were immobilized onto 96-well ELISA microplates (Pierce, Rockford, IL) via the interactions between GST and glutathione precoated onto the microplates. Briefly, bacterial lysates containing the GST fusion proteins were directly added to the glutathione plates. After the plates were washed to remove excess fusion proteins and blocked with 2.5% nonfat milk (in phosphate-buffered solution), the human serum samples were approximately diluted and added to the antigen-immobilized microplates. Serum antibody binding was detected with horseradish peroxidase-conjugated goat anti-human IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in combination with the soluble substrate 2,2′-azinobis(3-ethylbenzothiazoline-6-sulforic acid) (ABTS) diammonium salt (Sigma) and quantitated by reading the absorbance (optical density [OD]) at 405 nm using a microplate reader (Molecular Devices Corporation, Sunnyvale, CA).
RESULTS
Localization of CT813 protein to C. trachomatis inclusion membrane.
To search for new inclusion membrane proteins of C. trachomatis, we used antibodies raised with chlamydial fusion proteins to localize the corresponding endogenous proteins in chlamydia-infected cells in an immunofluorescence assay. After screening ~300 antibodies, we found that the antibody raised with the CT813 fusion protein appeared to label the inclusion membrane (Fig. (Fig.1).1). The anti-CT813 antibody staining pattern (Fig. (Fig.1A,1A, panels a and e) is similar to that of the antibody specifically recognizing IncG (panels b and f), a known chlamydial inclusion membrane protein. As controls, the anti-MOMP antibody detected the intrainclusion organisms (Fig. (Fig.1A,1A, panels c and g) and anti-CPAF antibody labeled the cytosol of chlamydia-infected cells (Fig. (Fig.1A,1A, panels d and h). To more closely compare the staining patterns of anti-D813 and anti-IncG antibodies, the two antibodies were used to costain the same cell samples (Fig. (Fig.1B).1B). Both conventional fluorescence (Fig. (Fig.1B,1B, top row) and laser confocal (Fig. (Fig.1B,1B, bottom row) microscopes revealed that the two antibodies costained the inclusion membrane and that the staining overlapped, confirming that anti-CT813 selectively labeled the C. trachomatis inclusion membrane. In a Western blot assay (Fig. (Fig.1C),1C), the anti-CT813 and anti-IncG antibodies detected the corresponding endogenous proteins in the infected whole-cell lysates but not the purified EBs while an equivalent amount of MOMP was detected in both samples, supporting the notion that the CT813 protein is localized mainly in the inclusion membrane.
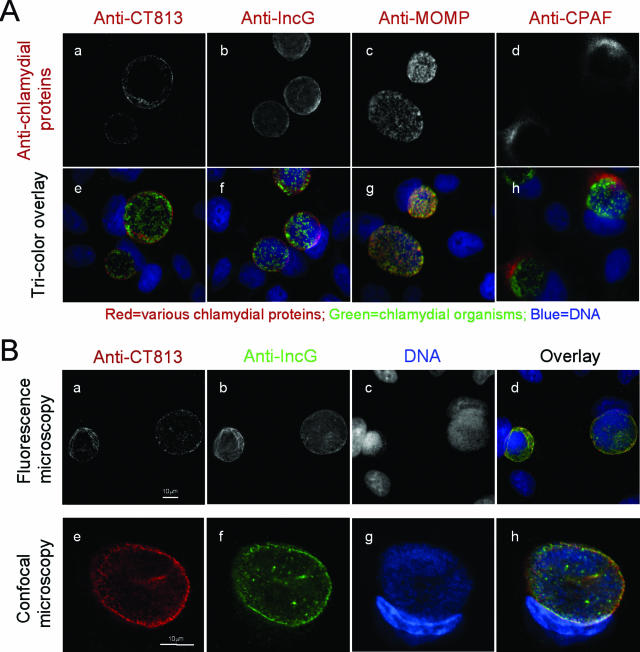
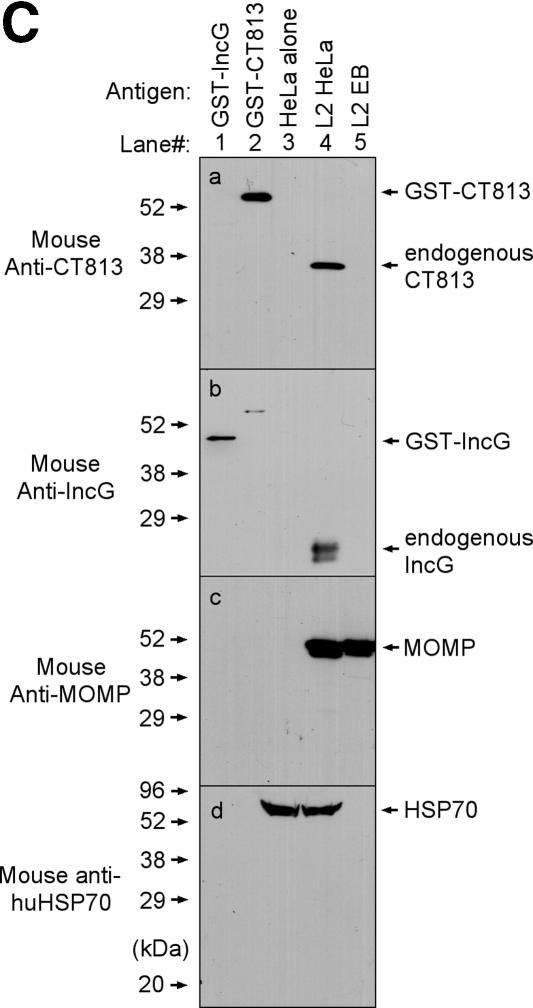
Localization of the CT813 protein in the chlamydial inclusion membrane. HeLa cells infected with C. trachomatis L2 serovar for 40 h were processed for immunofluorescence staining. (A) The rabbit antibody R1L2 was used to detect chlamydial organisms (green), and Hoechst dye was used to detect DNA (blue). The mouse antibodies against the CT813 protein (a and e), IncG (b and f), MOMP (clone MC22) (c and g), and CPAF (clone 100a) (d and h) were used to visualize the corresponding antigens (white in panels a to d and red in e to h). Note that both the anti-CT813 and anti-IncG antibodies labeled the inclusion membrane. (B) Rabbit anti-IncG (green) and mouse anti-CT813 (red) together with the DNA dye Hoechst (blue) were used to costain the same sample. The images were acquired as single color (a to c and e to g) and merged as tricolor (d and h) under either a conventional fluorescence microscope (a to d) or a laser confocal microscope (e to h). Note that the confocal images were amplified 2.5-fold relative to the size of the conventional fluorescence images. The CT813 protein colocalizes with IncG in the inclusion membrane. (C) Reactivities of the anti-CT813 and anti-IncG antibodies with the chlamydia-infected whole-cell lysate (HeLa-L2) and purified chlamydial organism (L2 EB) samples on a Western blot. Both the GST-CT813 and GST-IncG fusion proteins were used as positive controls and normal HeLa cells as negative controls. The same membranes were reprobed with anti-MOMP and anti-human HSP70, respectively.
We next used a preabsorption procedure to evaluate whether inclusion membrane labeling by the anti-CT813 antibody is specific to the CT813 protein (Fig. (Fig.2).2). In this experiment, the anti-CT813 and control anti-IncG antibodies were preabsorbed with or without the corresponding and heterologous fusion proteins, respectively, before being applied to the chlamydia-infected cell samples. Both the anti-D813 and anti-IncG antibodies labeled the chlamydial inclusion membrane (Fig. (Fig.2A,2A, panels a, d, g, and j). Anti-CT813 staining was removed by preabsorption with the GST-CT813 (Fig. (Fig.2A,2A, panels b and e) but not GST-IncG (panels c and f) fusion proteins, while anti-IncG staining was blocked by GST-IncG (panels i and l) but not GST-D813 (panels h and k). The efficiency of the preabsorption procedure was further verified in a Western blot assay (Fig. (Fig.2B).2B). The ability of the anti-D813 and anti-IncG antibodies to recognize both the fusion proteins and endogenous proteins was removed by preabsorption with the homologous but not the heterologous fusion proteins, confirming that the preabsorption was both efficient and specific. Thus, we can conclude that inclusion membrane staining by the anti-CT813 antibody is specific to the CT813 protein.


Specificity of the anti-CT813 antibody. Both the mouse anti-CT813 and anti-IncG antibodies as indicated along the left side of the figure were preabsorbed with or without either the corresponding or heterologous fusion proteins, as indicated at the top of the figure. (A) The mouse anti-CT813 and anti-IncG antibodies with or without preabsorption (red) plus the rabbit antibody R1L2 (green; labeling chlamydial organisms) and Hoechst (blue) were applied to the cells infected with L2 serovar for 40 h. The anti-CT813 and anti-IncG antibody stainings were shown in both single-color (a to c and g to i) and tricolor (d to f and j to l) images. Note that both the anti-CT813 (a and d) and anti-IncG (g and j) antibodies labeled the inclusion membrane. However, anti-CT813 labeling was blocked by preabsorption with GST-CT813 (b and e) but not GST-IncG (c and f) while anti-IncG labeling was blocked by GST-IncG (i and l) but not GST-CT813 (h and k). (B) The mouse anti-CT813 and anti-IncG antibodies with or without preabsorption were applied to nitrocellulose membranes, each blotted with the SDS-PAGE-resolved protein samples (GST-CT813 [lane 1], GST-IncG [lane 2], and L2-infected HeLa cell lysates [lane 3]), as indicated at the top of the figure. Note that anti-CT813 (a) and anti-IncG (d) stained both the corresponding endogenous and fusion protein bands. However, anti-CT813 staining was blocked by preabsorption with GST-CT813 (b) but not GST-IncG (c) while anti-IncG staining was blocked by GST-IncG (f) but not GST-CT813 (e). Anti-IncG also picked up the GST-CT813 band (d) due to anti-IncG's ability to recognize GST, and this staining was removed by GST-CT813 preabsorption (e). Arrows on the left point to the molecular mass markers and, on the right, label the corresponding protein bands. endo., endogenous.
Expression of CT813 protein in chlamydia-infected cultures.
CT813 protein expression was monitored in cultures over time by both Western blot (Fig. (Fig.3A)3A) and immunofluorescence (Fig. (Fig.3B)3B) assays. The anti-D813 antibody detected a band corresponding to the endogenous CT813 protein in chlamydia-infected but not uninfected cultures 24 h after infection (Fig. (Fig.3A,3A, top panel). The lack of detection of the CT813 protein at earlier time points may be due to an insufficient amount of samples loaded since MOMP, a constitutively expressed major outer membrane protein, was also first detected 24 h after infection (Fig. (Fig.3A,3A, middle panel). The amounts of total cellular protein loaded to the lanes were similar, as indicated by the detection of host beta-actin (Fig. (Fig.3A,3A, bottom panel). The GST-CT813 fusion protein loaded to the last lane was detected only by the anti-CT813 but not by the anti-MOMP or anti-beta-actin antibodies, validating the antibody binding specificity. In an immunofluorescence assay, the CT813 protein was detected as early as 12 h after infection (Fig. (Fig.3B,3B, panels c, o, o1, and o2) and was present in the inclusion membrane throughout the entire infection course (panels c to f). Since the single-cell-based immunofluorescence microscopy assay is known to be sensitive enough to detect other newly synthesized chlamydial proteins as early as 8 h (8) and since MOMP was visualized at 8 h after infection in the same assay, the failure to detect the CT813 protein at 8 h postinfection suggests that it may not be expressed by C. trachomatis at this time point. This conclusion is consistent with the observation that CT813 was not among the immediate-early genes identified by microarray analysis (8). However, more-extensive, -sensitive, and -careful analyses are required for determining the precise time points at which the CT813 protein is first expressed.


Time course of CT813 protein expression. For the Western blot assay (A), the mouse anti-CT813 (top), anti-MOMP (clone MC22) (middle), and anti-beta-actin (clone AC-15) (bottom) antibodies were used to probe the membrane blotted with SDS-PAGE-resolved cell samples with L2 infection harvested at various time points after infection as indicated at the top of the figure (12 h [lane 3], 24 h [lane 5], 48 h [lane 7], and 60 h [lane 9]). The parallel uninfected HeLa cell samples were harvested similarly at the corresponding time points (0 h [lane 1], 12 h [lane 2], 24 h [lane 4], 48 h [lane 6], and 60 h [lane 8]). Lane 10 was loaded with the GST-CT813 fusion protein. Note that both anti-CT813 and anti-MOMP detected the corresponding endogenous proteins in the 24-h sample. For the immunofluorescence assay (B), the mouse anti-CT813 (panels a to f [single color] in white and panels m to r [tricolor] in red) and rabbit antichlamydial organism R1L2 (g to l and m to r; green) antibodies together with Hoechst (blue) were used to costain HeLa cells infected with L2 for various hours after infection as indicated at the top of the figure. Note that although the organisms (green) were detected throughout the infection course, the CT813 protein (red) was first detected in the 12-h sample. To facilitate visualization, the selected inclusions (n, o, and p) were amplified in new windows, n1 and (anti-CT813 plus DNA), o1, n2 and p1 (tricolor), o2, and p2.
By BLAST sequence searching (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi), homologues of the CT813 protein were found only in strains/serovars of C. trachomatis species (data not shown), including the mouse biovar MoPn. We used a mouse polyclonal anti-CT813 antibody to screen cell cultures infected with multiple chlamydial serovars/strains from four different species (Fig. (Fig.4A).4A). The anti-CT813 antibody labeled the inclusion membrane in cells infected with all C. trachomatis human serovars but not other strains. Although DNA sequence analysis determined that the MoPn genome contains a gene (designated Tc0199 [41]) homologous to CT813, the anti-CT813 antibody failed to label the MoPn-infected culture. To test whether this failure is due to the lack of TC0199 protein expression by MoPn or the relative low homology (~36% amino acid identity) between TC0199 and the CT813 protein (thus lack of sufficient cross-reactivity), we used an antibody raised with the CT0199 fusion protein to detect the MoPn-infected culture. The anti-CT0199 antibody labeled the inclusion membrane of MoPn-infected cells (Fig. (Fig.4B),4B), demonstrating that the CT0199 protein is not only expressed but also, like its homologue, the CT813 protein from the C. trachomatis human biovar, localized in the inclusion membrane.

Expression of the CT813 protein by various chlamydial species. (A) HeLa cells infected with C. trachomatis (serovars A, B, Ba, C, D, E, F, G, I, K, L1, and L2 as well as the mouse biovar MoPn), C. caviae (GPIC), C. psittacis (6BC), and C. pneumoniae (AR39) as indicated at the top of each panel were processed ~30 to 50 h after infection for immunofluorescence staining with the mouse anti-CT813 (white in single color and red in tricolor) and rabbit anti-CT395 (green) antibodies plus Hoechst (blue). (B) The MoPn-infected sample was also stained with a mouse anti-TC0199 (the homologue of the CT813 protein in MoPn) antibody (red). The rest of the staining was the same as that shown in panel A. Note that anti-CT813 positively labeled the inclusion membrane in HeLa cells infected with all C. trachomatis serovars but not other strains. Although anti-CT813 failed to detect the MoPn inclusion membrane, the antibody raised with the homologue TC0199 did.
Exogenously expressed CT813 protein displayed a cytoskeleton-like structure.
To evaluate how the CT813 protein behaves in HeLa cells in the absence of chlamydial infection, we expressed the CT813 protein as a fusion protein with an RFP fused to its N terminus (designated RFP-CT813). Surprisingly, the RFP-CT813 fusion protein formed fibers in the transfected cells (Fig. (Fig.5A,5A, panels b, h, n, and t) while the overly expressed RFP alone evenly distributed throughout the cells, including the nuclei (panels e, k, q, and w). The fibrous structure displayed by the RFP-CT813 fusion protein overlapped with anti-CT813 antibody costaining (Fig. (Fig.5A,5A, panel c) but did not overlap with either F-actin (panel i) or microtubules (panel o). Although the pattern of RFP-CT813 fibers looks like the pattern of IF visualized by costaining cytokeratin 8, the two did not overlap (Fig. (Fig.5A,5A, panel u). To confirm the relationship between RFP-CT813 structure and IF, the costained samples were subjected to a confocal microscopy analysis (Fig. (Fig.5B).5B). The RFP-CT813 fibers did not overlap with the intermediate filaments (Fig. (Fig.5B,5B, panel i1). We designated the RFP-CT813 fibers as a cytoskeleton-like structure, which was observed throughout the transfection period (from 6 to 24 h posttransfection) (Fig. (Fig.5C).5C). The cytoskeleton-like structure formed by RFP-CT813 was unlikely caused by the RFP fusion tag, since the RFP tag alone did not form any obvious fibers (Fig. (Fig.5A)5A) and the CT813 protein expressed with only an 8-amino-acid FLAG tag by the pFLAG-CMV vector also displayed a similar cytoskeleton-like structure (data not shown).
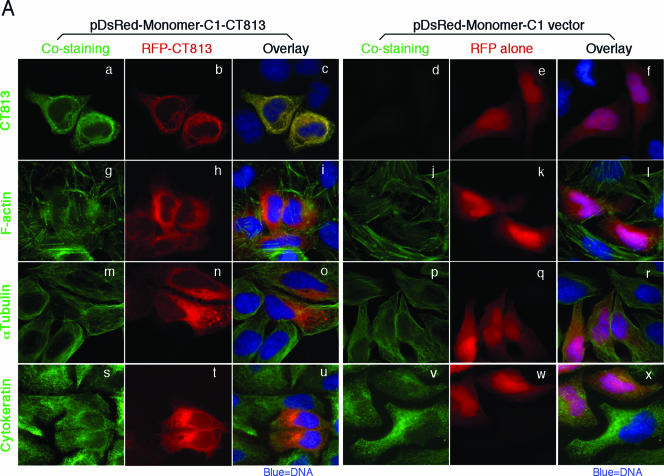

Expression of the CT813 protein in HeLa cells via a transient gene in the absence of chlamydial infection. HeLa cells transiently transfected with a recombinant plasmid encoding D813 as the RFP-CT813 fusion protein were processed for costaining with corresponding antibodies or biochemical dyes. (A) RFP-CT813 encoded by the pDsRed-Monomer-C1-CT813 plasmid (b, h, n, and t) or RFP alone encoded by the pDsRed-Monomer-C1 vector alone (e, k, q, and w) was visualized via RFP (red). Anti-CT813 (a and d), phalloidin-Alexa 488 (g and j), anti-α-tubulin (m and p), and anti-cytokeratin 8 (s and v) were used to costain the samples (all in green). The DNA was visualized with the Hoechst dye (blue). The three-color stainings were overlaid as tricolor images (indicated at the top of the figure). Note that RFP-CT813 formed fibers with a pattern similar to the anti-cytokeratin 8 antibody-stained pattern (designated as a cytoskeleton-like structure) (u) while RFP alone displayed a smooth distribution. (B) The anti-cytokeratin 8-stained samples (left) and the anti-CT813-stained samples (right; as the control for positive overlapping) were analyzed under a laser confocal microscope. Three z-axis serial sections with an interval of 1 μm were analyzed. The selected areas from panels i and l were amplified in new views (i1 and l1) for better visualization. Note that the antikeratin stainings did not overlap with the cytoskeleton-like structure formed by RFP-CT813 (c, I, o, and i1) while anti-CT813 did (f, l, r, and l1). (C) The cytoskeleton-like structure formation by RFP-CT813 was monitored as a function of time posttransfection. HeLa cells transfected with the pDsRed-Monomer-C1-CT813 recombinant plasmid were costained with Hoechst (blue) at various times after transfection as indicated at the top of the figure. The RFP-CT813 fusion was visualized via RFP (red). Note that the cytoskeleton-like structure was formed as early as 6 h and was present up to 24 h.
CT813 protein is both expressed and immunogenic during C. trachomatis infection in humans.
Although we have demonstrated that the CT813 protein is expressed in the inclusion membrane by all C. trachomatis serovars in cultures, it is not known whether it is expressed during chlamydial natural infection in humans. Since it is very difficult to directly detect chlamydial proteins in human samples, we used the human antibody responses to the CT813 protein to indirectly assess its expression by chlamydia in humans. Serum samples from 10 women diagnosed with C. trachomatis urogenital infection were each assayed against chlamydial fusion proteins in an ELISA (Fig. (Fig.6A).6A). The titer of human antibodies specific to the CT813 fusion protein was as high as that of human antibodies specific to the MOMP fusion protein. The observation that the titer of anti-CPAF antibody was the highest is consistent with a previous finding (51). The titers of antibody to both HSP60 and IncA were lower than that to the CT813 protein. No significant background antibody levels were detected against GST alone. This is probably due to the fact that all human serum samples assayed here were preabsorbed with bacterial lysates containing free GST. To confirm the ELISA specificity, the pooled human samples were subjected to preabsorption with HeLa lysates or chlamydia-infected cell lysates in addition to bacterial lysates (Fig. (Fig.6B).6B). Human antibody reactivity with the various chlamydial fusion proteins was blocked by preabsorption with chlamydia-infected HeLa lysates (Fig. (Fig.6B,6B, panel c) but not HeLa-alone lysates (panel b). Furthermore, sera from eight women without chlamydial infection reacted minimally with these chlamydial fusion proteins (Fig. (Fig.6B,6B, panel d). Human antibody reactivity with the CT813 protein was further confirmed in a Western blot assay (Fig. (Fig.7).7). The pooled positive sera recognized all fusion proteins, including GST-CT813 but not GST alone, at a dilution of 1:500 (Fig. (Fig.7,7, panel b). As human serum dilution increased, fewer fusion proteins were recognized (Fig. (Fig.7,7, panels b to d). However, the positive sera still recognized CT813, MOMP, and CPAF fusion proteins at a dilution of 1:62,500 (Fig. (Fig.7,7, panel d) while the pooled negative sera failed to recognize any antigens even at 1:500 (panel e). The Western blot assay result largely supported the above ELISA observation.
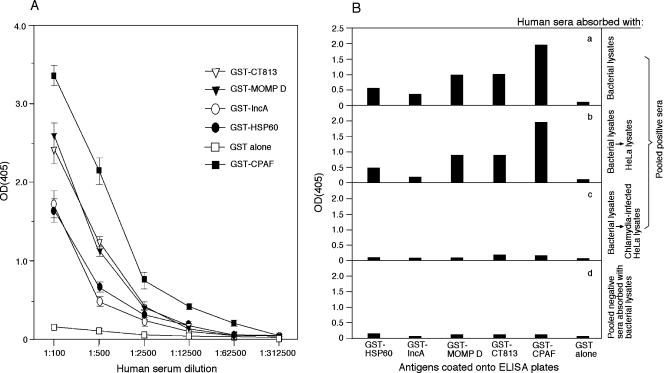
Results of an ELISA of human antibody responses to the CT813 protein. (A) Detection of titers of antibody in individual patients. Serum samples from 10 women diagnosed with urogenital tract infection with C. trachomatis were each serially diluted as shown along the x axis and reacted with GST-CT813, GST-MOMP from serovar D, GST-IncA, GST-HSP60, GST alone, and GST-CPAF immobilized onto the ELISA plates. All dilutions of each serum sample were measured in duplicate. The OD values obtained at the wavelength of 405 nm were expressed as means ± standard deviations, as shown along the y axis. Note that the titers of anti-CT813 were as high as those of anti-MOMP antibody. (B) Specificity of human antibody binding. The same 10 patient sera were pooled together at equal ratios to make the pooled positive sera, while sera from 8 normal individuals were pooled similarly to make the pooled negative sera. Both of the pooled serum samples were subjected to absorption with the bacterial lysates (a and d), while the positive serum samples were further absorbed with either HeLa alone (b) or C. trachomatis serovar D-infected HeLa lysates (c). After a dilution of 1:500 (relative to the initial serum volume), the processed serum samples were reacted with the ELISA plate-immobilized fusion proteins (in duplicate) as indicated along the horizontal axis and the ODs were expressed as the mean values from the duplicate wells as shown along the vertical axis. Note that the pooled positive sera variably reacted with all fusion proteins but not GST alone (a) and that reactivity was blocked by a further absorption with the infected cell lysates (c) but not the uninfected cell lysates (b). The pooled negative sera failed to significantly react with any fusion proteins (d).
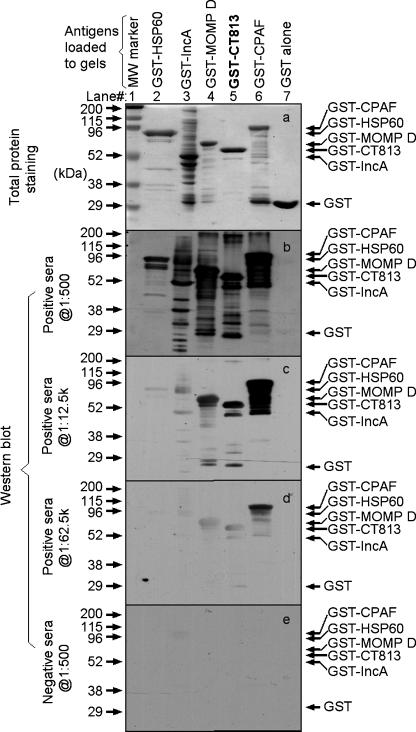
Western blot detection of human antibody recognition of the CT813 protein. The various protein samples, indicated at the top of the figure, were resolved on SDS-PAGE in multiple identical sets, with one set subjected to total protein staining and the rest of the sets transferred onto nitrocellulose membranes for probing human antibody recognition. The pooled sera after bacterial lysate absorption (used for Fig. Fig.6B)6B) were serially diluted as shown along the left side of the figure and were applied to the corresponding membranes. The human antibody reactivity was visualized using a combination of a conjugate secondary antibody and enhanced chemiluminescence. The molecular mass (mw) is marked along the left, and the protein migration bands are indicated along the right side of the figure. Note that the positive sera recognized fusion proteins in all lanes (except the GST lane) at dilutions of 1:500 (b) and 1:12,500 (c). However, at a dilution of 1;62,500 (d), the positive serum sample visibly recognized only GST-CPAF, GST-CT813, and GST-MOMP. The negative sera failed to recognize anything, even at a dilution of 1:500 (e).
DISCUSSION
By using anti-C. trachomatis fusion protein antibodies to localize endogenous chlamydial proteins, we identified a novel chlamydial inclusion membrane protein encoded by the hypothetical ORF CT813. Although antibodies raised with fusion proteins do not always efficiently/specifically recognize the corresponding endogenous proteins, in the current study, we presented several lines of evidence to show that the anti-CT813 staining of the inclusion membrane is specific to the CT813 protein. First, the anti-CT813 antibody labeled the inclusion membrane only in cells infected with serovars from C. trachomatis but not from other chlamydial species, although the chlamydial antigens in the inclusions were visualized in all infected cell samples (Fig. (Fig.4).4). This observation is consistent with the sequence analysis result (data not shown) that the CT813 protein is unique to C. trachomatis and that no significant homologues of the CT813 protein were found in other chlamydial species (except Chlamydia muridarum). Second, C. trachomatis inclusion membrane staining by the anti-CT813 antibody was blocked by preabsorption of the antibody with the CT813 protein, but the control CT118 fusion proteins (Fig. (Fig.2A)2A) and preabsorption with the D813 fusion protein specifically blocked binding of the anti-CT813 antibody to both the endogenous CT813 proteins and CT813 fusion proteins (Fig. (Fig.2B).2B). Third, the anti-CT813 antibody stained the CT813 protein expressed as the RFP-CT813 fusion protein but not RFP alone (Fig. (Fig.5).5). Finally, human antibodies produced during the chlamydial natural infection recognized the CT813 fusion protein and the recognition was blocked by chlamydia-infected cell lysates but not the uninfected cell lysates or the bacterial lysates containing free GST (Fig. (Fig.6),6), suggesting that the CT813 fusion proteins may have the ability to induce antibodies cross-reactive with the endogenous CT813 protein.
Although the CT813 protein was predicted to be in the chlamydial inclusion membrane by computer-based methods (3, 63), no experimental evidence has ever been presented to validate the prediction. After having screened antibodies against about 300 chlamydial proteins, we are presenting the first experimental evidence showing that the CT813 protein is a new Inc protein in addition to the Inc proteins that have previously been demonstrated, including numerous proteins encoded in the gene clusters covering ORFs CT115-119 (5, 50) and CT222-233 (3, 4) and the CT442 (3) and CT529 (18) proteins. Since not every computer-predicted Inc can be experimentally shown to be in the inclusion membrane (3) and since not all experimentally demonstrated Inc proteins, such as the CT529 protein (due to lack of the signature hydrophobicity motif [45]), were predicted by computer programs (3, 18, 63), it is still necessary to experimentally identify new Inc proteins. As more Inc proteins are experimentally demonstrated, we may be able to more comprehensively understand their structural and biological features, which may allow the development of more-reliable prediction models. At a genomic level, CT813 is located near the end of the C. trachomatis genome (containing a total of ~900 ORFs) and far away from the Inc ORF-enriched regions (CT115-119 and CT222-233). Among all known inc genes, CT529 is the closest to CT813. The identification of the CT813 protein as an Inc protein suggests that Inc ORFs are scattered across the entire genome. Like the CT529 protein, the CT813 protein may also be an orphan Inc since neither the CT812 protein nor the CT814/CT814.1 protein was detected in the inclusion membrane (data not shown). However, unlike the CT529 protein, which has homologues in other species, the CT813 protein is unique to C. trachomatis (including the mouse biovar MoPn).
The research interest in Inc proteins stems mainly from their potential roles in mediating the interactions between chlamydia and host cells. Indeed, some Inc proteins have been shown to be modified by (43) and to interact with (46, 49) host cell components. We found that the CT813 protein was able to form a cytoskeleton-like structure when exogenously expressed in HeLa cells (Fig. (Fig.5).5). These observations suggest that the CT813 protein is able to either polymerize or interact with host cell structures. Functionally, expression or secretion of IncA was correlated with the chlamydial ability to fuse between inclusions in the same infected cells (16, 25, 61), which may be due to the ability of IncA to oligomerize and mediate interaction between facing membranes (11). It has also been shown that preexisting IncA from C. caviae can render the transfected host cells more resistant to subsequent chlamydial infection (1). The potential effects of the CT813 protein on chlamydial inclusion development and fusion are under further investigation. Due to lack of genetic manipulation tools for chlamydial genomes, it has been difficult to precisely pinpoint the function of Inc proteins or any other chlamydial proteins. What we can do now, at least, is to experimentally identify more Inc proteins and carefully characterize them in cell-free and other heterologous systems, which may allow us to indirectly assess how various Inc proteins contribute to chlamydial pathogenesis.
The findings that chlamydial inclusion membrane and secreted proteins are immunogenic in infected animals and humans (5, 44, 51, 70) have demonstrated that not all immunogenic proteins expressed by chlamydia during natural infections can be adequately represented in the purified chlamydial organisms. This is because chlamydial Inc proteins and secreted proteins are often present in very small amounts or even absent in mature organisms (Fig. (Fig.1C)1C) (44, 70). Interestingly, the first inclusion membrane protein was identified by skillfully taking advantage of the unique antibodies that are produced only in response to live chlamydial infection but not in response to immunization with purified organisms (44). In the current study, a significant level of the CT813-specific antibodies was detected in women urogenitally infected with C. trachomatis. The human antibody recognition of fusion proteins was blocked by preabsorption of the human sera with chlamydial serovar D-infected HeLa lysates but not HeLa lysates alone (Fig. (Fig.6),6), demonstrating that the fusion protein antigens specifically detected human antibodies cross-reactive with endogenous chlamydial antigens. Although more-careful analyses are required for determining the relative immunodominance of the CT813 protein, we can now conclude that it is both expressed and immunogenic during chlamydial infection in humans. In addition to antibody responses (2, 34, 58), T-cell responses are also important arms of host defense against chlamydial infection (31, 33). While MOMP is enriched with epitopes that can be presented to CD4+ T cells (26, 60), the Inc proteins appear to possess epitopes that can be presented to CD8+ T cells, probably due to its accessibility to the host cell cytosol (18, 56). Since among the chlamydial pathogens that infect humans, only C. trachomatis serovars express the CT813 protein (Fig. (Fig.4)4) and since the CT813 protein is immunogenic during human chlamydial infection (Fig. (Fig.66 and and7),7), we are in the process of further evaluating the role of the CT813 protein in protection against C. trachomatis infection in animal models.
Acknowledgments
This work was supported in part by grants (to G. Zhong) from the U.S. National Institutes of Health.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.00081-06
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1539634?pdf=render
Citations & impact
Impact metrics
Article citations
The acetylase activity of Cdu1 regulates bacterial exit from infected cells by protecting Chlamydia effectors from degradation.
Elife, 12:RP87386, 15 Feb 2024
Cited by: 4 articles | PMID: 38358795 | PMCID: PMC10942603
The serodiagnositic value of Chlamydia trachomatis antigens in antibody detection using luciferase immunosorbent assay.
Front Public Health, 12:1333559, 27 Feb 2024
Cited by: 0 articles | PMID: 38476494 | PMCID: PMC10927828
The emerging complexity of Chlamydia trachomatis interactions with host cells as revealed by molecular genetic approaches.
Curr Opin Microbiol, 74:102330, 27 May 2023
Cited by: 5 articles | PMID: 37247566
Review
Hypothetical protein FoDbp40 influences the growth and virulence of Fusarium oxysporum by regulating the expression of isocitrate lyase.
Front Microbiol, 13:1050637, 28 Nov 2022
Cited by: 1 article | PMID: 36519161 | PMCID: PMC9742485
The bacterial effector GarD shields Chlamydia trachomatis inclusions from RNF213-mediated ubiquitylation and destruction.
Cell Host Microbe, 30(12):1671-1684.e9, 08 Sep 2022
Cited by: 21 articles | PMID: 36084633 | PMCID: PMC9772000
Go to all (47) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins.
Infect Immun, 74(3):1490-1499, 01 Mar 2006
Cited by: 73 articles | PMID: 16495519 | PMCID: PMC1418620
Localization and characterization of the hypothetical protein CT440 in Chlamydia trachomatis-infected cells.
Sci China Life Sci, 54(11):1048-1054, 01 Nov 2011
Cited by: 3 articles | PMID: 22173312
[Localization of the hypothetical protein CT249 in the Chlamydia trachomatis inclusion membrane].
Wei Sheng Wu Xue Bao, 47(4):645-648, 01 Aug 2007
Cited by: 6 articles | PMID: 17944365
[Effector proteins of Clamidia].
Mol Biol (Mosk), 43(6):963-983, 01 Nov 2009
Cited by: 4 articles | PMID: 20088373
Review





