Abstract
Free full text

Characterization of the short RNAs bound by the P19 suppressor of RNA silencing in mouse embryonic stem cells
Abstract
Studies of mammalian RNA interference (RNAi) have focused largely on the actions of microRNAs; however, in other organisms, endogenous short-interfering RNAs (siRNAs) are involved in silencing processes. To date, similar molecules have been difficult to characterize in mammalian cells. P19 is a plant suppressor of RNA silencing that binds with high affinity to siRNAs. Here, the short RNAs bound by P19 in mouse embryonic stem (ES) cells have been characterized. We show that P19 selectively immunoprecipitates endogenous short RNAs from ES cells. Cloning of immunoprecipitated RNA reveals a strong selection for short RNAs that are exact matches to ribosomal RNA (rRNA), with particular short rRNA species highly enriched in P19 immunoprecipitates. Complementary strands to the enriched rRNAs were not cloned, which was surprising because P19 was previously thought to bind only siRNAs. We show that P19 binds tightly to a noncanonical dsRNA substrate comprised of a short RNA annealed to a much longer partner, such that the double-stranded region between the two is 19 base pairs long. Binding to similar endogenous species might explain the association of P19 with short rRNAs in ES cells. Finally, we show that the P19-enriched rRNAs are not involved in canonical RNAi, as they exist in the absence of Dicer and do not function as post-transcriptional gene silencers. Our results support the previous observation that endogenous siRNAs are not abundant molecules in mouse ES cells.
INTRODUCTION
Short RNAs play a central role in eukaryotic biology by regulating gene expression through a process called RNA interference (Novina and Sharp 2004). cDNA libraries sampling the short RNA population in mammalian cells reveal predominantly the products of a conserved class of noncoding RNA genes called microRNAs (miRNAs). Mature miRNAs are processed from longer RNA precursors through sequential cleavage by the RNAse III enzymes Drosha and Dicer, generating ~22-nucleotide (nt)-long species with defined 5′ and 3′ ends. It is thought that mammalian miRNAs primarily exert their influence on gene expression post-transcriptionally by binding with imperfect complementarity to the 3′ UTR of their target mRNAs. An accumulating body of evidence suggests miRNAs are key regulators of both developmental transitions and cell-type specification (Ambros 2004; Bartel 2004; Alvarez-Garcia and Miska 2005; Farh et al. 2005; Stark et al. 2005). miRNAs are also misregulated in human cancers, potentially indicating a causal role in tumorigenesis (He et al. 2005; Lu et al. 2005; Voorhoeve et al. 2006).
Almost all of the known functions of endogenous RNAi in mammals are attributed to miRNAs; in other eukaryotes, this is not the case. The cloning of short RNAs from Schizosaccharomyces pombe, Arabidopsis thaliana, Caenorhabditis elegans, and Drosophila melanogaster has identified endogenous short-interfering RNAs (siRNAs) that are encoded by high copy sequences in the genome, such as transposons and retrotransposons. Except in Drosophila, these repeat-associated siRNAs (rasiRNAs) are thought to be processed by Dicer from long, repeat-derived double-stranded RNA (dsRNA). In S. pombe, Arabidopsis, and C. elegans, this dsRNA precursor is generated in part by the action of an RNA-dependent RNA polymerase, while in Drosophila rasiRNA biogenesis is less clear. rasiRNAs map predominantly to heterochromatic regions of the genome, and are proposed to be the guides for an siRNA-mediated transcriptional repression complex that nucleates heterochromatin (Ambros et al. 2003; Baulcombe 2004; Lippman and Martienssen 2004; Sontheimer and Carthew 2005; Lee et al. 2006; Vagin et al. 2006).
Mouse embryonic stem (ES) cells are pluripotent cells derived from the inner cell mass of the blastocyst in the midst of the epigenetic reprogramming that occurs during early development (Jaenisch 1997; Li 2002). ES cells are capable of executing well-studied epigenetic silencing processes, such as X inactivation and silencing of Moloney leukemia viruses (Stewart et al. 1982; Cherry et al. 2000; Plath et al. 2002). These processes are similar to examples of rasiRNA-mediated silencing in other organisms. However, previous cloning efforts from ES cells did not identify repeat-associated siRNAs, potentially because they are too low in abundance compared with miRNAs and other short RNAs (Houbaviy et al. 2003). Developing methods to facilitate the identification of low-abundance RNAi-specific short RNAs will, therefore, be necessary to more completely understand the function of RNAi in mammals.
In this work, we express epitope-tagged versions of the P19 suppressor of RNAi silencing in ES cells in an attempt to identify endogenous siRNAs. The P19 protein is expressed by the Tombusvirus as a counterdefense against the plant RNAi pathway that degrades RNA viruses, and functions by specifically binding and sequestering siRNAs that would otherwise mediate viral RNA destruction (Scholthof 2006). Biochemical and structural studies show that P19 binds with high affinity and specificity to siRNA-like molecules that are 5′ phosphorylated with double-stranded RNA segments 19 base pairs (bp) long. This affinity dramatically decreases if the double-stranded RNA segment is either shorter or longer than 19 bp. P19 also has no measurable affinity for single-stranded RNA or double-stranded DNA (Vargason et al. 2003; Ye et al. 2003; Lakatos et al. 2004). p19 transgenic plants display developmental defects associated with loss of miRNA function, and immunoprecipitations (IPs) of P19 from these plants enrich for miRNA duplexes, showing that P19 inhibits not only virally induced gene silencing but also other endogenous RNAi processes (Silhavy et al. 2002; Chapman et al. 2004; Dunoyer et al. 2004). Together, these observations indicate P19 may be a useful tool to identify endogenous siRNAs present in mammalian cells. Indeed, P19 has previously been shown to inhibit RNAi in human cells, suggesting that its functions carry over to mammals (Dunoyer et al. 2004; Lecellier et al. 2005).
RESULTS
Epitope-tagged P19 binds short RNAs from ES cells
Putative rasiRNAs involved in heterochromatin formation might be localized specifically to the nucleus. For this reason, we constructed two vectors for mammalian P19 expression; both were tagged at the C terminus with a V5–6×His epitope, and one contained two tandem SV40 nuclear localization sequences, increasing its nuclear concentration (hereafter referred to as P19V5 and P19NLS, respectively; Fig. 1A). Immunofluorescence of transiently transfected ES cells shows P19V5 is present in both the cytoplasm and nucleus, while P19NLS is localized almost exclusively to the nucleus (Fig. 1B).
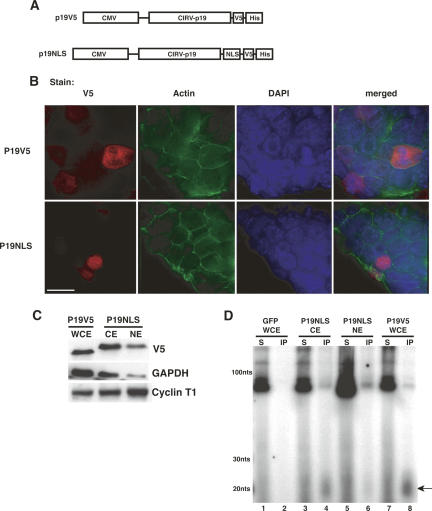
P19 binds endogenous short RNAs when expressed in ES cells. (A) P19 expression constructs used in this study. (B) Subcellular localization of P19V5 and P19NLS in ES cells (bar, 15 μm). (C) Western blots showing protein composition of P19-containing extracts. GAPDH (cytoplasmic) and Cyclin T1 (nuclear) serve as fractionation controls. (WCE) whole cell extract, (CE) cytoplasmic extract, (NE) nuclear extract. (D) P19 constructs bind short RNAs when expressed in ES cells. Cells transfected with GFP, P19V5, or P19NLS were lysed with either WCE or NE buffer, and immunoprecipitations were performed with αV5 Protein G agarose beads. Bound RNAs were 3′-end labeled with 5′-32P cytidine 3′, 5′-bis(phosphate) and resolved on a 12% denaturing polyacrylamide gel. The size markers correspond to a 10-bp DNA ladder. (Arrow) A ~20-nt band observed in the P19 immunoprecipitations.
To test if P19 bound endogenous short RNAs from ES cells, we extracted the nucleic acids that co-immunoprecipitated with our epitope-tagged constructs and radioactively labeled the 3′ end of the bound RNA with 5′-32P cytidine 3′,5′-bis(phosphate). To recover both cytoplasmic and nuclear siRNA molecules, P19V5 was immunoprecipitated from whole cell extracts (WCE). Western blots of the WCE detected both cytoplasmic and nuclear markers, indicating the presence of proteins from both subcellular compartments (Fig. 1C). Comparing the P19V5 bound RNA to the RNA from the WCE supernatant shows a strong enrichment for short RNAs ~20 nt long in the P19V5 immunoprecipitate (Fig. 1D, cf. lanes 7 and 8). This enrichment is P19V5 dependent, as there is no detectable RNA immunoprecipitated from cells transfected with GFP (Fig. 1D, lane 2).
To better enrich for putative low-abundance nuclear siRNAs, we performed an immunoprecipitation of P19NLS from a nuclear extract made under nondenaturing conditions. Western blots comparing the intensities of GAPDH and Cyclin T1 between the cytoplasmic (CE) and nuclear (NE) fractions of the extract show a decrease in GAPDH and an increase in Cyclin T1 in the nuclear as compared with the cytoplasmic extract, indicating an approximately threefold nuclear enrichment (Fig. 1C). Immunoprecipitation of P19NLS from the NE enriches for short RNAs identical in length to those immunoprecipitated from the CE of the P19NLS-transfected cells, as well as the WCE of P19V5-transfected cells (Fig. 1D, lanes 4,6,8).
We cloned the short RNAs bound by the P19 constructs in ES cell extracts using a procedure that selects for 18- to 26-nt-long RNAs that have 5′ phosphates and 2′ or 3′ hydroxyls (Lagos-Quintana et al. 2001; Lau et al. 2001; J.R. Neilson and P.A. Sharp, in prep.). As controls, we also cloned short RNA from the supernatants of the P19-transfected cell extracts. In each case ~300 independent clones were analyzed (Table 1). Sequences were initially annotated as known noncoding RNAs (ncRNAs) if they had an exact match to any annotated non-protein-coding RNA in the Rfam and NONCODE RNA databases, including rRNAs, tRNAs, snRNAs, snoRNAs, and miRNAs, as well as ncRNAs involved in imprinting and other processes (Griffiths-Jones et al. 2003, 2005). Those clones that had no matches in either database were deemed novel and were analyzed against the mouse genome using the UCSC genome browser to identify overlapping genomic features (Kent et al. 2002; Karolchik et al. 2003). Sequences with no exact match to the genome were reannotated as known ncRNAs or novel if they had ≥90% identity with a sequence in either set. Tables Tables11 and and22 are summaries of the cloning data and short RNA annotation.
TABLE 1
Gross statistics of short RNAs cloned from indicated RNA starting material
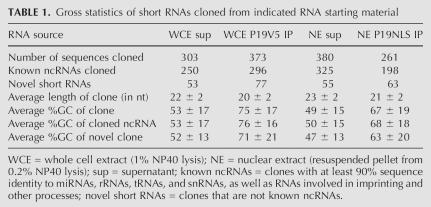
TABLE 2
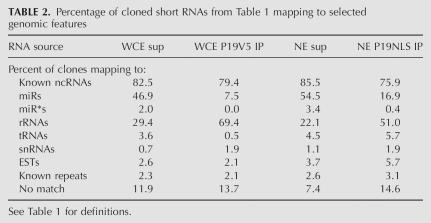
Percentage of cloned short RNAs from Table 1 mapping to selected genomic features
The majority of all sequences cloned were known ncRNAs, regardless of the RNA starting material (Tables (Tables1,1, ,2).2). Short RNAs immunoprecipitated by P19 were on average 2 nt shorter than those cloned from control supernatants (22 versus 20 nt for the WCE supernatant versus IP; 23 versus 21 nt for the NE supernatant versus IP, p < 0.0001 for both; Table 1), consistent with previous observations that P19 has a decreased affinity for double-stranded RNA both shorter and longer than 19 bp (Vargason et al. 2003; Ye et al. 2003). Moreover, the GC content of the P19-bound RNA was significantly higher than control RNA (53% versus 75% for the WCE sup versus IP, and 49% versus 67% for the NE sup versus IP; p < 0.0001 for both; Table 1). This large average difference in GC content was observed for both known ncRNAs and novel RNAs, indicating an overall preference of P19 for GC-rich RNA (Table 1).
A proportion of the novel sequences, between 2% and 3% of all sequences cloned from each population, were exact matches to known repetitive elements cataloged by Repeatmasker (Supplemental Data; Table 2). On average, these short RNAs were slightly shorter than the average miRNA cloned in this study (20.9 versus 22.3 nt for repeat-associated RNAs versus miRNAs; p < 0.0001). Similar repeat-derived short RNAs were not identified in previous ES cell cloning efforts, most likely due to limitations in the depth of short RNAs sequenced (Houbaviy et al. 2003).
Strikingly, P19 immunoprecipitation selected against miRNAs and enriched for short RNAs matching mature ribosomal RNA (rRNA) species (Supplemental Data; Table 2); 48.9% and 57.9% of the short RNAs cloned from the WCE and NE supernatants mapped to annotated miRNA hairpins (miRNAs and miRNA*s), compared with only 7.5% and 17.3% of those cloned from the immunoprecipitates. Conversely, 69.4% and 51.0% of all sequences cloned from the WCE and NE immunoprecipitates were short rRNAs, compared with 29.4% and 22.1% of sequences cloned from the WCE and NE supernatants, respectively (Table 2).
Interestingly, the immunoprecipitates also lacked the natural complementary sequence of the miRNA duplex, the miRNA* strand (Table 2). The ratio of miRNA to miRNA* strands in the supernatants was roughly 24:1 and about the same level in P19 immunoprecipitates, indicating there was no selection for miRNA duplexes by P19. This was surprising given that P19 binds tightly to double-stranded RNA with almost no affinity for single-stranded RNA substrates, and associates with miRNA duplexes in plants (Silhavy et al. 2002; Vargason et al. 2003; Ye et al. 2003; Chapman et al. 2004; Dunoyer et al. 2004; Lakatos et al. 2004).
The specific miRNAs cloned from the immunoprecipitates were similar to those cloned from the supernatants, perhaps consistent with the majority of P19-associated miRNAs being derived from a nonspecific background. The same group of 20 miRNAs comprised 79% and 83% of all miRNAs cloned from WCE IP and supernatant, respectively, and comprised 78% and 77% of all miRNAs cloned from the NE IP and supernatant, respectively. There was no significant difference in the GC content or length of the miRNAs cloned from the immunoprecipitates compared with those cloned from the supernatants (not shown). If, indeed, the miRNAs present in P19 immunoprecipitates were due to background contamination, then a comparison of the number cloned from each population would suggest that P19 immunoprecipitation gave a sevenfold enrichment of bound short RNAs versus miRNAs from the supernatant of the WCE, and a threefold enrichment of bound short RNAs versus miRNAs from the supernatant of the NE. These are minimal estimates, as some miRNAs might be preferentially bound to P19.
All but one of the short rRNAs cloned from the supernatants and the immunoprecipitations were in the sense orientation relative to the full-length transcribed 45S pre-rRNA and 5S RNA, and 97% of these sequences mapped to the mature 18S, 5.8S, or 28S rRNA. Figure 2A shows a representation of all the short rRNAs cloned, aligned to bases 3900 to 13,000 of the 13,404-base 45S pre-ribosomal RNA. Only 13 out of 564 total cloned short rRNAs did not have an exact match in this region; seven of these were matches to the 5S rRNA, and the remaining six mapped to regions of the 45S precursor not included in Figure 2A. Particular classes of short rRNAs were highly enriched in the P19-bound RNA compared with unbound controls (Fig. 2A, “*”). Mapping these RNAs to established rRNA secondary structure maps shows the enriched short rRNAs are not necessarily from regions of the ribosome that resemble canonical Drosha or Dicer substrates (not shown; Cannone et al. 2002).
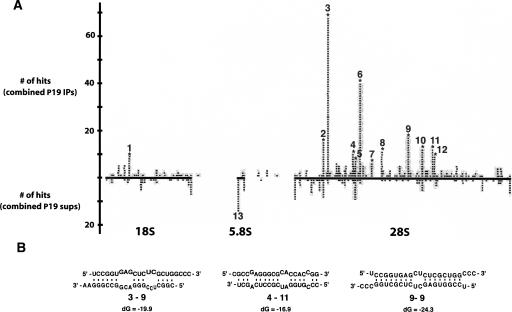
P19 enriches for particular short rRNA species when expressed in ES cells. (A) Specific short rRNAs are highly enriched in P19 immunoprecipitations compared with control supernatants. Shown is a scaled representation of all the short rRNAs cloned, aligned to bases 3900 to 13,000 of the 13,404-bp 45S pre-rRNA. Highlighted in bold along the X axis are the locations of the mature 18S, 5.8S, and 28S rRNA species relative to the full-length 45S pre-rRNA. Each gray bar represents one cloned short rRNA positioned directly above or below its matching sequence in the 45S pre-rRNA. Gray bars above the X axis were cloned from one or both of the immunoprecipitations, and those below the X axis were cloned from one or both of the supernatants. (B) Certain P19-enriched short rRNAs form partial double-stranded RNA structures with themselves. Shown are selected Mfold-predicted dsRNA structures of enriched rRNAs folded against each other.
Because P19 binds almost exclusively to double-stranded RNA, it was surprising that no short RNAs with exact complementarity to P19-enriched short rRNAs were cloned. Speculating that these RNAs could possibly form partial duplexes with other short RNAs, we investigated whether the most abundantly cloned rRNAs in the immunoprecipitations (cloned more than five times) could form bulged duplexes with other abundantly cloned species. We limited our initial analysis to the most abundantly cloned short RNAs, reasoning that abundantly cloned short RNAs could have been likely complements in binding to P19. The short rRNAs, numbered 1 through 12 in Figure 2A, were folded against each other in all possible permutations using the Mfold server (Zuker 2003). Of the 140 predicted structures, we found 14 that when annealed had a double-stranded region of the right size to potentially bind P19 with high affinity, between 18 and 20 bp (Vargason et al. 2003). In all cases, there was at least one bulged region in the RNA duplex predicted by Mfold (for selected examples, see Fig. 2B); however, it has been previously observed that P19 associates with bulged duplexes in plants (Chapman et al. 2004). This partial complementarity between particular abundantly cloned species provides a possible explanation for the enrichment of some short rRNAs in the P19 IP, but not all. Notably, although short rRNA #6 was the second most abundantly cloned short rRNA in the P19 immunoprecipitations, it was not predicted to form any duplexed structures with other abundantly cloned short rRNAs.
P19 binds to regions of short dsRNA
It is possible that P19 also binds substrates with dsRNA regions that are siRNA-like in length, but contain one strand that is significantly longer than the standard siRNA length of 21 nt. Biochemical analysis shows that RNA duplexes without 3′ overhangs bind with slightly higher affinity to P19 than their 3′-overhang-containing counterparts, indicating that the length of the RNA duplex and not the overhang is the key factor in determining P19 binding (Vargason et al. 2003). This hypothesis could explain why some molecules immunoprecipitated by P19 form no canonical siRNA-like duplexes with other cloned RNAs; short RNAs complementary to longer RNAs could be bound by P19, and the iterative size selection in the short RNA cloning protocol would exclude the longer RNA-binding partner from the final set of cloned sequences. The high-GC content of the short RNAs bound by P19 is consistent with this hypothesis (Table 1), allowing a greater promiscuity for stable binding to longer RNA partners. Indeed, in addition to enriching for short RNAs, P19 immunoprecipitation reproducibly pulls down larger RNA species that are the same size as abundant tRNAs, snRNAs, and rRNAs (Figs. 1D, ,4A).4A). While no immunoprecipitated RNAs have exact complements in the set of known tRNAs, snRNAs, and rRNAs, gapped alignments allowing G:U pairing show that P19-enriched RNAs could form duplexes with several ncRNA species, potentially explaining the enrichment for ncRNAs as well as short rRNAs in the immunoprecipitation (not shown).
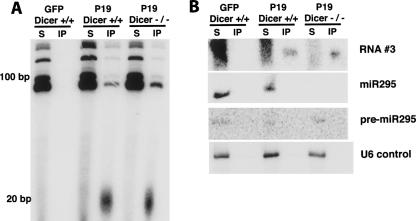
Endogenous short rRNAs exist independently of Dicer. (A) P19 associates with short RNAs in the absence of Dicer. Dicer +/+ or −/− cells were transfected with either P19V5 or GFP as a negative control. Immunoprecipitations using V5 antibody were performed, and the associated RNA was 3′-end labeled and visualized as in Fig. 1D. (B) Similar RNA species associate with P19V5 in the presence or absence of Dicer. Shown is a short RNA Northern blot probing immunoprecipitated and supernatant RNA from A with probes complementary to RNA #3, miR295, or U6 snRNA.
We designed three different RNA duplexes to test the affinity of P19V5 for short RNAs bound to longer RNAs (Fig. 3A). One strand of each duplex was a frequently cloned short rRNA, while the other varied such that its complement was either: in the 5′ region of a 32-nt-long RNA species (5′ complementary), in the 3′ region of a 35-nt-long RNA species (3′complementary), or a 21-nt-long RNA that formed a canonical 19-bp siRNA duplex (siRNA). The length of the double-stranded region in the 5′-complementary RNA was 19 bp and was 21 bp in the 3′-complementary RNA. Since P19 preferentially binds 19-bp duplexes, the latter may be expected to have a lower affinity than the former. In all cases, the dsRNA species had 5′-phosphate and 3′-hydroxyl groups, mimicking endogenous siRNA structure. Binding assays were performed by incubating radiolabeled RNA substrate in increasing concentration with P19V5-Protein G agarose bead complexes (Fig. 3). Control experiments showed that all input RNA was double stranded and of the appropriate dilution (not shown). Under the conditions assayed, there was negligible nonspecific association of RNA with beads, and the P19V5–bead complexes had no affinity for single-stranded RNA (Fig. 3B). The apparent dissociation constant, Kapp, of P19V5 for an siRNA duplex was determined to be 2.8 ± 0.5 nM, nearly identical to previously published binding studies using a P19 C-terminally tagged with GST (Fig. 3C; Lakatos et al. 2004). P19V5 bound the 5′- and 3′-complementary RNAs with Kapp of 7.4 ± 0.7 nM and 27 ± 4 nM, respectively, supporting our hypothesis that P19 binds regions of dsRNA ~19 bp long and not only siRNAs (Fig. 3C).
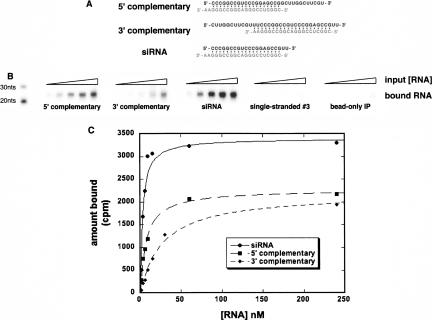
P19 binds with high affinity to RNAs containing 19- to 21-bp double-stranded regions with extended 5′- or 3′-single-stranded segments. (A) Base composition and secondary structure of short RNAs tested for binding to P19V5. In all cases, the strand in gray is short rRNA #3 from Fig. 2A. (B) Representative P19-binding assay. Shown is the RNA bound by P19V5–bead complexes after incubation with increasing concentrations of radiolabeled RNA (1, 5, 10, 50, and 100 nM). (C) Determination of the affinity of P19 for selected RNA species. Shown is the quantitation of a binding assay similar to that in B. The Kapp was determined by fitting the average data points to a fixed endpoint curve using KaleidaGraph data analysis software.
Unknown function of P19 bound short rRNAs
We next tried to determine the function of the short rRNAs enriched in the P19 immunoprecipitation. Short RNAs with exact matches to abundant tRNAs, rRNAs, and snRNAs have been frequently dismissed as nonfunctional degradation products of abundant ncRNAs, particularly in mammalian cloning efforts. While this may be true, it is worth noting that these noncoding RNAs are derived from highly repetitive elements in mammalian genomes, and in this respect are similar to other known targets of RNAi-mediated transcriptional silencing from which rasiRNAs have been cloned (Lander et al. 2001; Waterston et al. 2002).
Studies from S. pombe and Arabidopsis have implicated short rRNAs in the formation of heterochromatin at rDNA repeats, setting a precedent for short rRNA functionality (Xie et al. 2004; Cam et al. 2005). One common feature of these short rRNAs involved in chromatin silencing is that they are both sense and anti-sense to the full-length, transcribed rRNA, supporting the idea that they are generated from processing of a longer dsRNA precursor (Xie et al. 2004; Cam et al. 2005). In contrast, all but one of the short rRNAs cloned in this study were in the sense orientation relative to transcription of the mature rRNAs, suggesting that they arose either by breakdown or processing of mature rRNA sequence. However, this observation does not exclude a possible role for short rRNAs in the chromatin silencing of ES cell rDNA repeats.
Recently, ncRNAs mapping directly upstream of the rDNA transcriptional start site have been shown to direct the nucleolar remodeling complex (NoRC) to transcriptionally silence rDNA repeats in mouse 3T3 cells (Mayer et al. 2006). No short RNAs cloned in this study map to this region of the rDNA repeat, consistent with the authors’ observation that the NoRC-associated RNAs are 150–300 nt long.
It is also possible that the short rRNAs immunoprecipitated by P19 are involved in post-transcriptional gene silencing (PTGS). If this were true, one might expect complementary sequences to be present in exons or 3′ UTRs of known genes. BLAST analysis of the P19-enriched rRNAs against the mouse genome shows that exactly complementary sequences are not present in known mRNAs, suggesting that these enriched rRNAs are not involved in siRNA-like PTGS. In support of this, no effect was seen on expression of a luciferase reporter when two perfectly complementary binding sites to selected short rRNAs were inserted into its 3′ UTR (Supplemental Fig. 1A).
Finally, we examined whether P19V5 associates with short RNAs in an ES cell line lacking Dicer RNAse III activity. These ES cells were derived from mice homozygous for a conditional allele of Dicer in which the key catalytic residues in the second RNAse III domain are floxed (Harfe et al. 2005). Consistent with previously published results, excision of the floxed region via transient transfection of Cre recombinase results in viable ES cells that do not express miRNAs (Fig. 4B; Murchison et al. 2005; J.M. Calabrese and P.A. Sharp, unpubl.). P19V5 immunoprecipitates short RNAs from Dicer null ES cells as efficiently as from Dicer-containing cells, shown by 3′-end labeling of immunoprecipitated RNA (Fig. 4A). Similarly, short RNA Northern blots probing P19 immunoprecipitates for enriched short rRNA #3 show the same enrichment in the presence and absence of Dicer (Fig. 4B). Reprobing of the same Northern blot shows miR295 is absent from P19 immmunoprecpitates but is present in the supernatants from Dicer-positive cells, confirming the cloning data that show a selection against miRNAs in P19 immunoprecipitates (Fig. 4B). Northern blots to total RNA preparations show that RNA #3 is present in ES cells at the same level in the presence and absence of P19V5, and is not induced nonspecifically by transfection (Supplemental Fig. 1B). A survey of RNA from various mouse tissues indicates that RNA #3 is detectable in several samples, but at levels much lower than those observed in ES cells (Supplemental Fig. 1C). Together, these results indicate that enriched rRNAs are generated independently of Dicer and are not by-products of P19 expression or transfection.
DISCUSSION
We have shown that V5-tagged P19 associates with endogenous short RNAs in mouse ES cells. Cloning of these RNAs from whole cell and nuclear extracts revealed that P19 associates predominantly with short, GC-rich RNAs that are exact matches to portions of the mature 28S and 18S rRNAs. The function of these short rRNAs is not clear. We show that they exist in the absence of Dicer and do not function in endogenous PTGS, suggesting that they are not in the canonical RNAi pathway. In other organisms, short rRNAs have been implicated in the chromatin silencing of rDNA repeats (Xie et al. 2004; Cam et al. 2005); whether the same is true of the short rRNAs identified in this work is not clear. Intriguingly, P19V5 did not immunoprecipitate an easily detectable pool of short RNAs from 293T cells as it did in ES cells, suggesting that similar short rRNAs are not produced to the same extent in this human embryonic kidney cell line, or that they are inaccessible to P19 (Supplemental Fig. 2A). In plants, overexpression of P19 leads to a general accumulation of miRNA* strands, and P19 immunoprecipitates contain both miRNA and miRNA* strands, as detected by short RNA Northern blots to total and P19 immunoprecipitated RNA, respectively (Chapman et al. 2004; Dunoyer et al. 2004). In contrast, P19 expression in mouse ES cells did not lead to accumulation of miRNA* strands in cell extracts, nor were miRNA* strands selectively detected in P19 immunoprecipitates, suggesting that P19 is unable to access miRNA duplexes as efficiently in ES cells as in plants.
Because of our experimental design, we obtained cloning data from a nuclear-enriched extract. We observed few significant differences between the short RNA profiles of nuclear compared with cytoplasmic extracts, suggesting that novel short RNAs are not abundant in the nucleus of ES cells. A surprisingly high proportion of short RNAs in the nuclear extract were miRNAs, given that mature miRNAs are predominantly cytoplasmic (Houbaviy et al. 2005). A simple explanation for these results is that the profile of short RNAs in the nucleus is similar to that in the cytoplasm. It is also possible that most of the short RNAs present in the nuclear extract were from cytoplasmic contamination. Controls testing the extent of fractionation in nuclear extracts suggest a threefold enrichment for components of this compartment. This level of enrichment might not be sufficient for the identification of short, nuclear-localized RNAs if they are low in abundance compared with cytoplasmic short RNAs. Alternatively, nuclear siRNAs might be chemically modified such that they are not identifiable by the cloning methods used here, which require a 5′-phosphate and 2′- or 3′-hydroxyl groups.
Notably, between 2% and 3% of the short RNAs cloned in this study overlapped with repetitive elements cataloged by Repeatmasker. It is unlikely that these RNAs exist predominantly as double-stranded siRNAs in cell extracts, as they were not enriched in P19 immunoprecipitates. Their function is unclear, but they may be analogous to repeat-associated siRNAs identified in S. pombe, C. elegans, Drosophila, and Arabidopsis. Alternatively, because such a large proportion of the mouse genome is annotated as repetitive, overlap of specific short RNAs with repeats may be coincidental and not indicative of novel function (Lander et al. 2001; Waterston et al. 2002).
Recently, repeat-associated short RNAs were identified from mouse oocytes, where they appear to be as abundant as miRNAs. Also, reporter constructs with complementary 3′-UTR sequences were destabilized, suggesting that at this developmental stage, repeat-associated short RNAs are involved at least in the PTGS of complementary transcripts (Watanabe et al. 2006). The short RNAs overlapping repetitive elements identified in this study are probably at a far lower abundance; ~25-fold less abundant than miRNAs. This difference suggests a difference in activity of these sequences in ES cells compared with oocytes.
Previous biochemical studies have shown that P19 dimers bind tightly to siRNA duplexes (Vargason et al. 2003; Ye et al. 2003; Lakatos et al. 2004). Here, we show that compared with an siRNA, P19 binds with roughly threefold-reduced affinity to a dsRNA species containing one strand that extends far beyond the edge of the RNA duplex. We, therefore, conclude the likely reason for the association of P19 with specific short rRNAs in ES cells is that they bind with partial complementarity to larger, abundant noncoding RNAs to form RNA duplexes ~19 bp long. The observed threefold difference in affinity for a 19-bp duplexed siRNA compared with a 19-bp duplex with an extended 3′ strand is small enough that abundant endogenous dsRNAs similar in structure to the latter could compete with endogenous siRNAs for P19 binding.
It should be noted that P19V5, as well as a previously published HA epitope-tagged P19 construct, did not inhibit siRNA-mediated knockdown of a reporter gene in 293T cells (Supplemental Fig. 2C). This conflicts with results from HeLa cells, where epitope-tagged P19 was an effective inhibitor of exogenously introduced siRNAs, and with results from 293T cells, where untagged P19 was able to interfere with endogenous miRNA activity (Dunoyer et al. 2004; Lecellier et al. 2005). The reason for these discrepancies is unclear, but it suggests that P19 may not inhibit RNAi in heterologous systems as robustly as previously anticipated.
The original objective of these experiments was to use the affinity of P19 for siRNA duplexes to identify these types of short RNAs in mouse ES cells. Functional and biochemical studies of P19 in plants and animals were consistent with this approach. Analysis of the short RNAs bound by P19 in ES cells did not generate an apparent siRNA fraction in spite of evidence for a strong enrichment of certain RNA species as compared with the supernatants from associated ES cell extracts. RNAs with siRNA structure do bind to P19 with high affinity, and the failure to detect them might indicate their scarcity in these cells. Alternatively, P19 might be denied access to endogenous RNAi-related short RNAs if the majority of siRNAs in ES cells are bound by RNAi pathway components that have a higher affinity for these molecules than P19.
MATERIALS AND METHODS
Plasmid construction
CIRV P19 (obtained from J. Burgyan) was amplified via PCR and cloned into pcDNA3.1 (Invitrogen), using the primers 5′-CACCATGGAACGAGCTATACAAGGA-3′ and 5′-CTCGCTTTCTTTCTTGAAGGTTTCA-3′. To make P19NLS, two SV40 NLS sequences were added to P19V5 by annealing two DNA oligos with an 18-bp overlap, (5′-CCGCTCGAGTGATCCAAAAAAGAAGAGAAAGGTAGATCCAAAA-3′and 5′-CGAACCGCGGTACCTTTCTCTTCTTTTTTGGATCTACCTTTCT-3′), filling in with Taq DNA polymerase, and inserting into the XhoI/SacII sites of pcDNA3.1. pRL-CMV-6xCXCR4 was described previously (Doench et al. 2003). The P19HA construct was obtained from O. Voinnet (Dunoyer et al. 2004). The V5-tagged βGalactosidase control plasmid was obtained from H. Houbaviy. The 3′-UTR reporter constructs were made by inserting 5′-phosphorylated, annealed dsDNA oligos into the XhoI/ApaI sites in the 3′ UTR of pRL-CMV (Promega). The sequences of the DNA oligos used to make the UTR constructs are available upon request. All DNA oligos were purchased from IDT.
Cell culture, transfection, and luciferase assays
J1 ES and 293T cells were grown as in Houbaviy et al. (2003) and Petersen et al. (2006). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For ES cell and 293T immunoprecipitation experiments, 25 μg of plasmid per 10-cm plate were used. Transfection efficiency as assessed by GFP expression was between 60% and 80% for ES cell immunoprecipitation experiments, and >95% for 293T experiments.
Dual luciferase assays were performed essentially as previously described (Doench and Sharp 2004; Petersen et al. 2006). For ES cell assays, 1 ng of each 3′-UTR reporter construct, 100 ng of pGl3 control (Promega), and 700 ng of pWhiteScript carrier DNA were transfected per well of a 24-well plate and lysed 24 h post-transfection. For 293T assays, 4 μg of plasmid (P19V5/NLS/HA or βGal control) was transfected per well of a six-well plate, and split 8 h post-transfection into a 24-well plate, seeding cells at 2 × 105 cells per well. Twenty-four hours later, an additional 0.7 μg of plasmid was cotransfected along with 30 ng of pRL and pGL3 plasmids (Promega) and the appropriate amount of siRNA to attain the concentration indicated in Supplemental Figure 2. Cells were lysed 24 h post-transfection. The siRNA used perfectly targeted the coding sequence of Renilla Luciferase (target sequence: 5′-GCCAAGAAGUUUCCUAAUA-3′).
Western blots and immunohistochemistry
For western analysis, cell extracts were fractionated on 4%–20% SDS-polyacrylamide gradient gels (Biorad) and transferred to Hybond-C membranes (Amersham). Membranes were blocked with 5% milk in PBS and then incubated with the indicated antibody. Antibodies (αGAPDH [Chemicon]; αV5 [Invitrogen]; αCyclin T1 [Abcam]) were detected using HRP-conjugated antisera (Amersham) and chemiluminescence. For immunofluorescent staining, ES cells were transfected on gelatinized cover slips, then fixed with 4% PFA in PBS and permeabilized with 1% Triton X-100. Coverslips were stained with primary and secondary antibodies in PBS for 1 h and affixed to glass slides with Prolong Gold with DAPI (Molecular Probes). Actin was visualized using Alexa 488 Phalloidin (Molecular Probes).
Immunoprecipitations and RNA isolation and visualization
For each immunoprecipitation experiment, three 10-cm plates of ES cells were lysed 2 d post-transfection with WCE buffer (1% NP40, 30 mM HEPES KOH pH 7.5, 100 mM NaCl, 66 mM KCl, 1 mM MgCl2, 1 mM DTT, 1U/μL SUPERaseIn [Ambion], Complete Protease Inhibitor EDTA-free [Roche], and Phosphatase Inhibitors I and II [Sigma]) or NE buffer (0.2% NP-40, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1U/μL SUPERaseIn, Complete Protease Inhibitor EDTA-free, and Phosphatase Inhibitors I and II). The WCE lysate was spun at 20,000 rpm in a tabletop centrifuge for 15 min after lysis to pellet insoluble material. The NE cells were lysed for 5 min at 4°C and spun for 5 min at 1000g. The pellet (containing nuclei) was resuspended in WCE buffer and sonicated on ice three times for 5 sec at power level 2 on a Branson Sonifier 450 sonicator, with a 1-min rest on ice between sonications. These sonication conditions did not disrupt endogenous short RNA binding to P19 (not shown). The sonicated nuclear fraction was spun at 20,000 rpm for 15 min in a tabletop centrifuge to remove particulate. P19 was immunoprecipitated from extracts by adding 20 μL of Protein G Plus agarose beads (Pierce) preconjugated overnight with 2 μg of V5 antibody per 10-cm plate, and rotating for 45 min at 4°C. Beads were washed with 1 mL of WCE buffer three times, switching tubes for the final wash. Beads were then resuspended in 300 μL 1× Proteinase K buffer (Nykanen et al. 2001) with 5 μL of Proteinase K (Roche) and rotated for 30 min at room temperature, then extracted with phenol/chloroform and precipitated. To isolate supernatant RNA, 300 μL of supernatant from the immunoprecipitation was extracted with phenol/chloroform and precipitated. One hundred nanograms of supernatant RNA and one-tenth the volume of immunoprecipitated RNA were 3′-end labeled with T4 RNA ligase (NEB) and 5′-α32P 5′-3′ cytidine bis-phosphate (NEN) overnight at 4°C in 1× RNA ligase buffer and 30% v/v DMSO. Unincorporated radioactivity was removed with G25 microspin columns (Amersham), and one-half of the labeling reaction was resolved on a 12% or 15% denaturing polyacrylamide gel (National Diagnostics). For size markers, 10-bp DNA ladder (Invitrogen) or a 21-bp siRNA was 5′-end labeled using γ32P-ATP (NEN) and T4 PNK (NEB). Gels were wrapped in Saran Wrap and quantitated on a PhosphoImager. Northern blots were performed as in Houbaviy et al. (2003), except hybridization was carried out in Oligo-Hyb (Ambion) at 37°C. Total mouse tissue RNA was purchased from Ambion. The same conditions for the ES cell WCE immunoprecipitations were used for 293T immunoprecipitations.
Binding assays
293T lysates were used to make P19-containing cell extracts for binding assays because of the lack of association between P19 and endogenous RNAs in these cells (Supplemental Fig. 2). Extracts were made 48 h post-transfection with P19V5 using 400 μL of WCE buffer per plate. Cells were lysed and extracts were spun at top speed in a tabletop centrifuge for 15 min. Protein G Plus agarose beads pre-conjugated overnight with V5 antibody were added to cleared lysate, and P19V5 was immunoprecipitated for 45 min at 4°C. Beads were washed 3× in WCE buffer and resuspended in WCE buffer such that for each binding condition, 30 μg of extract, 10 μL of 50% protein G slurry, and 0.5 μg of V5 antibody were used. Radiolabeled dsRNA was added to P19V5–bead mixtures at the indicated concentrations, and the binding reactions were rotated for 30 min at room temperature. Beads were washed 2× with 500 μL of WCE buffer, and then 2× with 1 mL of WCE buffer, switching tubes for the final wash. The bound RNA was eluted using the Proteinase K phenol/chloroform treatment described above. One-half of each reaction was resolved on a 15% polyacrylamide gel and visualized with a PhosphoImager, or quantitated via scintillation counting. Data points were then fit to a fixed-endpoint curve, (m2)*m0/(m0+m1), using KaleidaGraph software, where m2 is the maximum amount bound, m1 is the apparent dissociation constant, Kapp, and m0 is the [RNA].
To make radiolabeled, duplexed RNA, 5′-phosphorylated RNA oligos (“RNA#3” 5′-CGGCUCCGGGACGGCCGGGAA-3′; “5´-complementary” 5′-CCCGGCCGUCCCGGAGCCGGCUUGGCUUCGU-3′; “3′-complementary” 5′-CUUGGCUUCGUUUCCCGGCCGUCCCGGAGCCGUU-3′; “siRNA complementary” 5′-CCCGGCCGUCCCGGAGCCGUU-3′) were annealed to their complementary strands at 12 μM in a solution containing 10 mM HEPES pH 7.5, 20 mM NaCl, and 1 mM EDTA by heating the RNAs to 95°C and cooling 1°C/min until the samples reached room temperature. Radioactive 5′-end-labeled RNA #3 was spiked into annealing reactions before heating. Dilution series of each dsRNA species were made, and portions of each were run on a 20% native polyacrylamide gel to assess efficiency of annealing and accuracy of dilution. By this analysis, all dsRNA preparations used for binding were >99% dsRNA and had an R2 value for the dilution series of ≥0.99 (not shown).
Short RNA cloning and sequence analysis
Short RNAs were cloned using a procedure modified from Lagos-Quintana et al. (2001) and Lau et al. (2001) (J.R. Neilson and P.A. Sharp, in prep.). Before adaptor ligation, 18- to 26-nt-long RNA was gel purified from 5 μg of supernatant RNA to use as starting material; immunoprecipitated RNA from one 10-cm plate equivalent was used as starting material and was not gel-purified. Short RNA sequences were extracted from concatamers using scripts from Houbaviy et al. (2003). Rfam (http://www.sanger.ac.uk/Software/Rfam/) and NONCODE (http://www.bioinfo.org.cn/NONCODE/) RNA databases were used to define known miRNAs, tRNAs, rRNAs, snRNAs, and snoRNAs. All genome analysis was performed using the August 2005 assembly of the mouse genome (mm7). BLAST was run with a word size of 7 and the gap-opening penalty set to 1. Repeat and EST overlap was determined using the UCSC genome browser (http://genome.ucsc.edu/), Repeatmasker (http://www.repeatmasker.org), and mouse EST tracks. Sequences with multiple repeat overlaps were annotated as the class of repeat that overlapped most frequently with the short RNA in question. p values comparing values between data sets were obtained using a two-sample test for the difference in proportions.
SUPPLEMENTAL DATA
Supplemental information can be found at http://web.mit.edu/sharplab/calabrese_supp/.
ACKNOWLEDGMENTS
We thank J. Burgyan and O. Voinnet for providing P19 plasmids, and are especially grateful to J. Neilson for protocols and help with short RNA cloning. We also thank A. Seila for help with bioinformatics, C. Whittaker for help with Figure 2, A. Leung for help with microscopy, and S. Erkeland, A. Garfinkel, J. Neilson, and A. Seila for critical reading of this manuscript. This work was supported by a Program Project Grant from the National Cancer Institute to P.A.S. and partially by the Cancer Center Support (core) grant from the National Cancer Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.224606.
REFERENCES
- Alvarez-Garcia, I., Miska, E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. [Abstract] [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. [Abstract] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., Jewell, D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr. Biol. 2003;13:807–818. [Abstract] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [Abstract] [Google Scholar]
- Baulcombe, D. RNA silencing in plants. Nature. 2004;431:356–363. [Abstract] [Google Scholar]
- Cam, H.P., Sugiyama, T., Chen, E.S., Chen, X., FitzGerald, P.C., Grewal, S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. [Abstract] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3 [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Dev. 2004;18:1179–1186. [Europe PMC free article] [Abstract] [Google Scholar]
- Cherry, S.R., Biniszkiewicz, D., van Parijs, L., Baltimore, D., Jaenisch, R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 2000;20:7419–7426. [Europe PMC free article] [Abstract] [Google Scholar]
- Doench, J.G., Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. [Europe PMC free article] [Abstract] [Google Scholar]
- Doench, J.G., Petersen, C.P., Sharp, P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. [Europe PMC free article] [Abstract] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. [Abstract] [Google Scholar]
- Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. [Abstract] [Google Scholar]
- Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. [Europe PMC free article] [Abstract] [Google Scholar]
- Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., Bateman, A. Rfam: Annotating noncoding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. [Europe PMC free article] [Abstract] [Google Scholar]
- Harfe, B.D., McManus, M.T., Mansfield, J.H., Hornstein, E., Tabin, C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. [Europe PMC free article] [Abstract] [Google Scholar]
- He, L., Thomson, J.M., Hemann, M.T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S.W., Hannon, G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. [Abstract] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. [Google Scholar]
- Houbaviy, H.B., Dennis, L., Jaenisch, R., Sharp, P.A. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. [Europe PMC free article] [Abstract] [Google Scholar]
- Jaenisch, R. DNA methylation and imprinting: Why bother? Trends Genet. 1997;13:323–329. [Abstract] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., Haussler, D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. [Europe PMC free article] [Abstract] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. [Abstract] [Google Scholar]
- Lakatos, L., Szittya, G., Silhavy, D., Burgyan, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. [Europe PMC free article] [Abstract] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. [Abstract] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. [Abstract] [Google Scholar]
- Lecellier, C.H., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A., Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. [Abstract] [Google Scholar]
- Lee, R.C., Hammell, C.M., Ambros, V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans . RNA. 2006;12:589–597. [Europe PMC free article] [Abstract] [Google Scholar]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. [Abstract] [Google Scholar]
- Lippman, Z., Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. [Abstract] [Google Scholar]
- Lu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. [Abstract] [Google Scholar]
- Mayer, C., Schmitz, K.M., Li, J., Grummt, I., Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. [Abstract] [Google Scholar]
- Murchison, E.P., Partridge, J.F., Tam, O.H., Cheloufi, S., Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. 2005;102:12135–12140. [Europe PMC free article] [Abstract] [Google Scholar]
- Novina, C.D., Sharp, P.A. The RNAi revolution. Nature. 2004;430:161–164. [Abstract] [Google Scholar]
- Nykanen, A., Haley, B., Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. [Abstract] [Google Scholar]
- Petersen, C.P., Bordeleau, M.E., Pelletier, J., Sharp, P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. [Abstract] [Google Scholar]
- Plath, K., Mlynarczyk-Evans, S., Nusinow, D.A., Panning, B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002;36:233–278. [Abstract] [Google Scholar]
- Scholthof, H.B. The Tombusvirus-encoded P19: From irrelevance to elegance. Nat. Rev. Microbiol. 2006;4:405–411. [Abstract] [Google Scholar]
- Silhavy, D., Molnar, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., Burgyan, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. [Europe PMC free article] [Abstract] [Google Scholar]
- Sontheimer, E.J., Carthew, R.W. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. [Abstract] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. [Abstract] [Google Scholar]
- Stewart, C.L., Stuhlmann, H., Jahner, D., Jaenisch, R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl. Acad. Sci. 1982;79:4098–4102. [Europe PMC free article] [Abstract] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. [Abstract] [Google Scholar]
- Vargason, J.M., Szittya, G., Burgyan, J., Tanaka Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. [Abstract] [Google Scholar]
- Voorhoeve, P.M., le Sage, C., Schrier, M., Gillis, A.J., Stoop, H., Nagel, R., Liu, Y.P., van Duijse, J., Drost, J., Griekspoor, A., et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. [Abstract] [Google Scholar]
- Watanabe, T., Takeda, A., Tsukiyama, T., Mise, K., Okuno, T., Sasaki, H., Minami, N., Imai, H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. [Europe PMC free article] [Abstract] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. [Abstract] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2 [Google Scholar]
- Ye, K., Malinina, L., Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. [Abstract] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from RNA are provided here courtesy of The RNA Society
Full text links
Read article at publisher's site: https://doi.org/10.1261/rna.224606
Read article for free, from open access legal sources, via Unpaywall:
http://rnajournal.cshlp.org/content/12/12/2092.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102665359
Article citations
Short Repeat Ribonucleic Acid Reduces Cytotoxicity by Preventing the Aggregation of TDP-43 and Its 25 KDa Carboxy-Terminal Fragment.
JACS Au, 4(10):3896-3909, 24 Sep 2024
Cited by: 0 articles | PMID: 39483234 | PMCID: PMC11522920
An unnatural enzyme with endonuclease activity towards small non-coding RNAs.
Nat Commun, 14(1):3777, 24 Jun 2023
Cited by: 2 articles | PMID: 37355703 | PMCID: PMC10290691
Computational meta-analysis of ribosomal RNA fragments: potential targets and interaction mechanisms.
Nucleic Acids Res, 49(7):4085-4103, 01 Apr 2021
Cited by: 19 articles | PMID: 33772581 | PMCID: PMC8053083
Cytosolic delivery of siRNA by ultra-high affinity dsRNA binding proteins.
Nucleic Acids Res, 45(13):7602-7614, 01 Jul 2017
Cited by: 6 articles | PMID: 28641400 | PMCID: PMC5570165
A Novel p19 Fusion Protein as a Delivery Agent for Short-interfering RNAs.
Mol Ther Nucleic Acids, 5:e303, 05 Apr 2016
Cited by: 6 articles | PMID: 27045207 | PMCID: PMC5014518
Go to all (25) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Studies of the interaction of the viral suppressor of RNA silencing protein p19 with small RNAs using fluorescence polarization.
Biochemistry, 47(31):8130-8138, 03 Jul 2008
Cited by: 13 articles | PMID: 18597480
Enhanced specificity of the viral suppressor of RNA silencing protein p19 toward sequestering of human microRNA-122.
Biochemistry, 50(36):7745-7755, 17 Aug 2011
Cited by: 10 articles | PMID: 21819044
Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities.
Plant Cell Physiol, 48(7):1050-1060, 13 Jun 2007
Cited by: 124 articles | PMID: 17567638
Small RNAs derived from longer non-coding RNAs.
Biochimie, 93(11):1905-1915, 09 Aug 2011
Cited by: 88 articles | PMID: 21843590
Review




