Abstract
Free full text

The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis.
Abstract
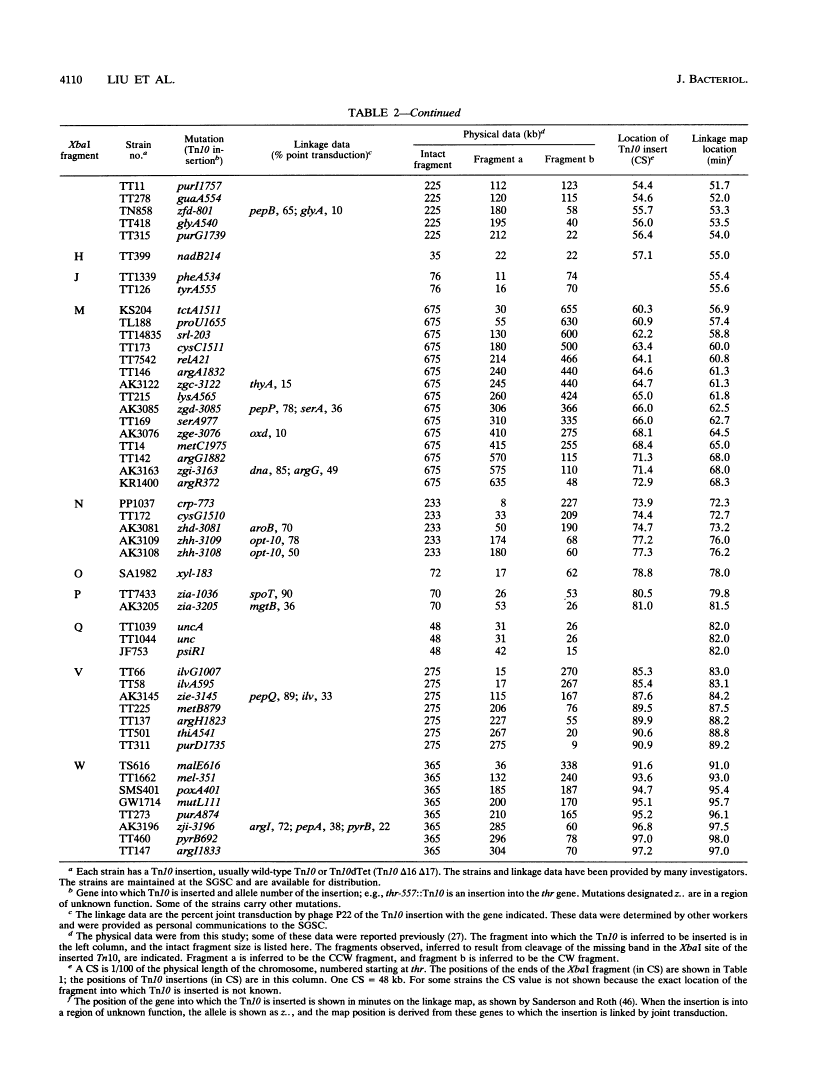
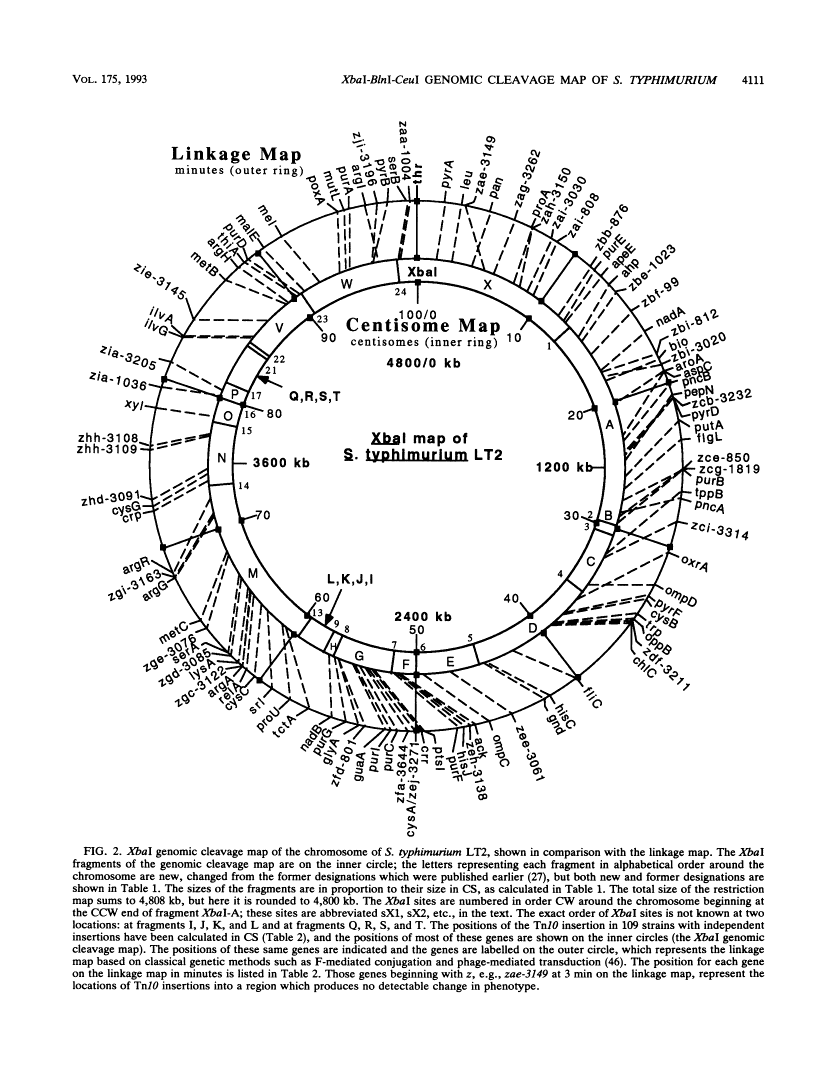
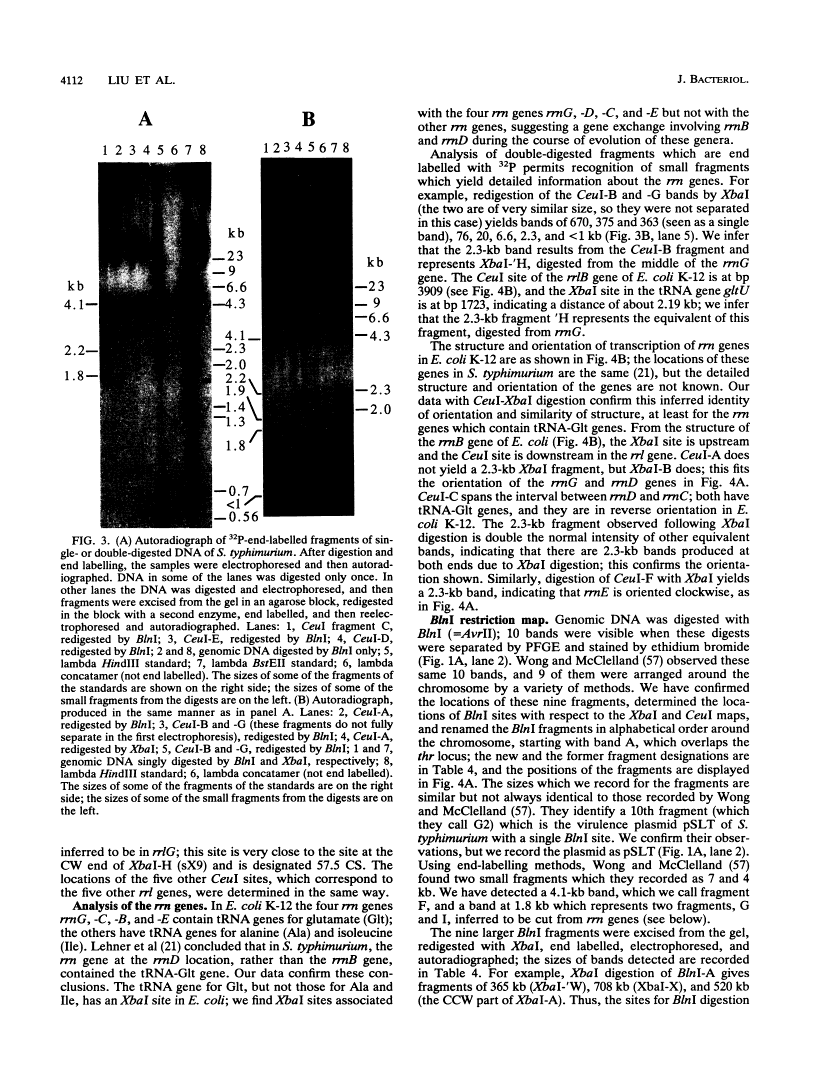
Endonuclease digestion of the 4,800-kb chromosome of Salmonella typhimurium LT2 yielded 24 XbaI fragments, 12 BlnI fragments, and 7 CeuI fragments, which were separated by pulsed-field gel electrophoresis. The 90-kb plasmid pSLT has one XbaI site and one BlnI site. The locations of the fragments around the circular chromosome and of the digestion sites of the different endonucleases with respect to each other were determined by excision of agarose blocks containing fragments from single digestion, redigestion with a second enzyme, end labelling with 32P by using T7 DNA polymerase, reelectrophoresis, and autoradiography. Forty-three cleavage sites were established on the chromosome, and the fragments and cleavage sites were designated in alphabetical order and numerical order, respectively, around the chromosome. One hundred nine independent Tn10 insertions previously mapped by genetic means were located by pulsed-field gel electrophoresis on the basis of the presence of XbaI and BlnI sites in Tn10. The genomic cleavage map was divided into 100 units called centisomes; the endonuclease cleavage sites and the genes defined by the positions of Tn10 insertions were located by centisome around the map. There is very good agreement between the genomic cleavage map, defined in centisomes, and the linkage map, defined in minutes. All seven rRNA genes were located on the map; all have the CeuI digestion site, all four with the tRNA gene for glutamate have the XbaI and the BlnI sites, but only four of the seven have the BlnI site in the 16S rRNA (rrs) gene. Their inferred orientation of transcription is the same as in Escherichia coli. A rearrangement of the rrnB and rrnD genes with respect to the arrangement in E. coli, observed earlier by others, has been confirmed. The sites for all three enzymes in the rrn genes are strongly conserved compared with those in E. coli, but the XbaI and BlnI sites outside the rrn genes show very little conservation.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (3.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann BJ. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. [Europe PMC free article] [Abstract] [Google Scholar]
- Bautsch W. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 1988 Dec 23;16(24):11461–11467. [Europe PMC free article] [Abstract] [Google Scholar]
- Benson NR, Goldman BS. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992 Mar;174(5):1673–1681. [Europe PMC free article] [Abstract] [Google Scholar]
- Bhagwat AS, McClelland M. DNA mismatch correction by Very Short Patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 1992 Apr 11;20(7):1663–1668. [Europe PMC free article] [Abstract] [Google Scholar]
- Bouffard G, Ostell J, Rudd KE. GeneScape: a relational database of Escherichia coli genomic map data for Macintosh computers. Comput Appl Biosci. 1992 Dec;8(6):563–567. [Abstract] [Google Scholar]
- Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. [Abstract] [Google Scholar]
- Burgin AB, Parodos K, Lane DJ, Pace NR. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990 Feb 9;60(3):405–414. [Abstract] [Google Scholar]
- Casse F, Pascal MC, Chippaux M. Comparison between the chromosomal maps of Escherichia coli and Salmonella typhimurium. Length of the inverted segment in the trp region. Mol Gen Genet. 1973 Aug 17;124(3):253–257. [Abstract] [Google Scholar]
- Chen HW, Kuspa A, Keseler IM, Shimkets LJ. Physical map of the Myxococcus xanthus chromosome. J Bacteriol. 1991 Mar;173(6):2109–2115. [Europe PMC free article] [Abstract] [Google Scholar]
- Crosa JH, Brenner DJ, Ewing WH, Falkow S. Molecular relationships among the Salmonelleae. J Bacteriol. 1973 Jul;115(1):307–315. [Europe PMC free article] [Abstract] [Google Scholar]
- Daniels DL. The complete AvrII restriction map of the Escherichia coli genome and comparisons of several laboratory strains. Nucleic Acids Res. 1990 May 11;18(9):2649–2651. [Europe PMC free article] [Abstract] [Google Scholar]
- Gauthier A, Turmel M, Lemieux C. A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr Genet. 1991 Jan;19(1):43–47. [Abstract] [Google Scholar]
- Gibson MM, Price M, Higgins CF. Genetic characterization and molecular cloning of the tripeptide permease (tpp) genes of Salmonella typhimurium. J Bacteriol. 1984 Oct;160(1):122–130. [Europe PMC free article] [Abstract] [Google Scholar]
- Heath JD, Perkins JD, Sharma B, Weinstock GM. NotI genomic cleavage map of Escherichia coli K-12 strain MG1655. J Bacteriol. 1992 Jan;174(2):558–567. [Europe PMC free article] [Abstract] [Google Scholar]
- Hennecke F, Kolmar H, Bründl K, Fritz HJ. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991 Oct 24;353(6346):776–778. [Abstract] [Google Scholar]
- Jiang XM, Neal B, Santiago F, Lee SJ, Romana LK, Reeves PR. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991 Mar;5(3):695–713. [Abstract] [Google Scholar]
- Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. [Abstract] [Google Scholar]
- Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee JJ, Smith HO, Redfield RJ. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989 Jun;171(6):3016–3024. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehner AF, Harvey S, Hill CW. Mapping and spacer identification of rRNA operons of Salmonella typhimurium. J Bacteriol. 1984 Nov;160(2):682–686. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehner AF, Hill CW. Involvement of ribosomal ribonucleic acid operons in Salmonella typhimurium chromosomal rearrangements. J Bacteriol. 1980 Jul;143(1):492–498. [Europe PMC free article] [Abstract] [Google Scholar]
- Lehner AF, Hill CW. Merodiploidy in Escherichia coli-Salmonella typhimurium crosses: the role of unequal recombination between ribosomal RNA genes. Genetics. 1985 Jul;110(3):365–380. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu SL, Sanderson KE. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992 Mar;174(5):1662–1672. [Europe PMC free article] [Abstract] [Google Scholar]
- MacLachlan PR, Kadam SK, Sanderson KE. Cloning, characterization, and DNA sequence of the rfaLK region for lipopolysaccharide synthesis in Salmonella typhimurium LT2. J Bacteriol. 1991 Nov;173(22):7151–7163. [Europe PMC free article] [Abstract] [Google Scholar]
- McClelland M, Jones R, Patel Y, Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. [Europe PMC free article] [Abstract] [Google Scholar]
- Michiels T, Popoff MY, Durviaux S, Coynault C, Cornelis G. A new method for the physical and genetic mapping of large plasmids: application to the localisation of the virulence determinants on the 90 kb plasmid of Salmonella typhimurium. Microb Pathog. 1987 Aug;3(2):109–116. [Abstract] [Google Scholar]
- Perkins JD, Heath JD, Sharma BR, Weinstock GM. SfiI genomic cleavage map of Escherichia coli K-12 strain MG1655. Nucleic Acids Res. 1992 Mar 11;20(5):1129–1137. [Europe PMC free article] [Abstract] [Google Scholar]
- Römling U, Tümmler B. The impact of two-dimensional pulsed-field gel electrophoresis techniques for the consistent and complete mapping of bacterial genomes: refined physical map of Pseudomonas aeruginosa PAO. Nucleic Acids Res. 1991 Jun 25;19(12):3199–3206. [Europe PMC free article] [Abstract] [Google Scholar]
- Rudd KE, Miller W, Werner C, Ostell J, Tolstoshev C, Satterfield SG. Mapping sequenced E.coli genes by computer: software, strategies and examples. Nucleic Acids Res. 1991 Feb 11;19(3):637–647. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanderson KE, Hall CA. F-prime factors of Salmonella typhimurium and an inversion between S. typhimurium and Escherichia coli. Genetics. 1970 Feb;64(2):215–228. [Europe PMC free article] [Abstract] [Google Scholar]
- Sanderson KE, Roth JR. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. [Europe PMC free article] [Abstract] [Google Scholar]
- Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987 May;4(3):222–230. [Abstract] [Google Scholar]
- Sharp PM, Shields DC, Wolfe KH, Li WH. Chromosomal location and evolutionary rate variation in enterobacterial genes. Science. 1989 Nov 10;246(4931):808–810. [Abstract] [Google Scholar]
- Skurnik M, Toivanen P. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991 Mar;5(3):585–593. [Abstract] [Google Scholar]
- Smith CL, Condemine G. New approaches for physical mapping of small genomes. J Bacteriol. 1990 Mar;172(3):1167–1172. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith CL, Econome JG, Schutt A, Klco S, Cantor CR. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. [Abstract] [Google Scholar]
- Strauch KL, Lenk JB, Gamble BL, Miller CG. Oxygen regulation in Salmonella typhimurium. J Bacteriol. 1985 Feb;161(2):673–680. [Europe PMC free article] [Abstract] [Google Scholar]
- Tinge SA, Curtiss R., 3rd Conservation of Salmonella typhimurium virulence plasmid maintenance regions among Salmonella serovars as a basis for plasmid curing. Infect Immun. 1990 Sep;58(9):3084–3092. [Europe PMC free article] [Abstract] [Google Scholar]
- Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. [Abstract] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong KK, McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong KK, McClelland M. Dissection of the Salmonella typhimurium genome by use of a Tn5 derivative carrying rare restriction sites. J Bacteriol. 1992 Jun;174(11):3807–3811. [Europe PMC free article] [Abstract] [Google Scholar]
- Youderian P, Sugiono P, Brewer KL, Higgins NP, Elliott T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988 Apr;118(4):581–592. [Europe PMC free article] [Abstract] [Google Scholar]
- ZINDER ND, LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.175.13.4104-4120.1993
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/175/13/4104.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/175/13/4104
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/175/13/4104
Citations & impact
Impact metrics
Citations of article over time
Article citations
E. coli diversity: low in colorectal cancer.
BMC Med Genomics, 13(1):59, 06 Apr 2020
Cited by: 6 articles | PMID: 32252754 | PMCID: PMC7133007
Genetic boundaries delineate the potential human pathogen Salmonella bongori into discrete lineages: divergence and speciation.
BMC Genomics, 20(1):930, 04 Dec 2019
Cited by: 4 articles | PMID: 31801462 | PMCID: PMC6894293
Distinct evolutionary origins of common multi-drug resistance phenotypes in Salmonella typhimurium DT104: a convergent process for adaptation under stress.
Mol Genet Genomics, 294(3):597-605, 01 Feb 2019
Cited by: 0 articles | PMID: 30710177
Efficacy of Pulsed-Field Gel Electrophoresis and Repetitive Element Sequence-Based PCR in Typing of Salmonella Isolates from Assam, India.
J Clin Microbiol, 56(5):e02043-17, 25 Apr 2018
Cited by: 1 article | PMID: 29491016 | PMCID: PMC5925735
Fingerprint Analysis and Identification of Strains ST309 as a Potential High Risk Clone in a Pseudomonas aeruginosa Population Isolated from Children with Bacteremia in Mexico City.
Front Microbiol, 8:313, 01 Mar 2017
Cited by: 16 articles | PMID: 28298909 | PMCID: PMC5331068
Go to all (72) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The XbaI-BlnI-CeuI genomic cleavage map of Salmonella paratyphi B.
J Bacteriol, 176(4):1014-1024, 01 Feb 1994
Cited by: 26 articles | PMID: 8106312 | PMCID: PMC205152
Genomic cleavage map of Salmonella typhi Ty2.
J Bacteriol, 177(17):5099-5107, 01 Sep 1995
Cited by: 38 articles | PMID: 7665489 | PMCID: PMC177289
The XbaI-BlnI-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2.
Mol Microbiol, 10(3):655-664, 01 Nov 1993
Cited by: 38 articles | PMID: 7968542
Chromosomal rearrangements in enteric bacteria.
Electrophoresis, 19(4):569-572, 01 Apr 1998
Cited by: 19 articles | PMID: 9588803
Review