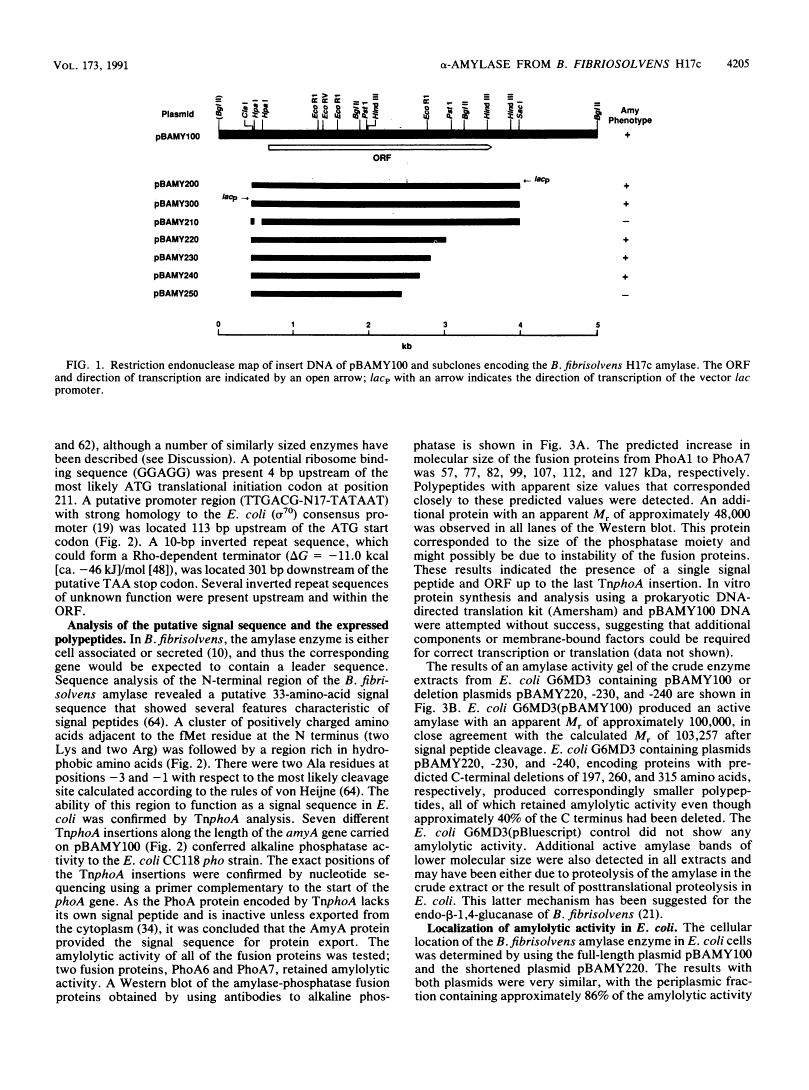
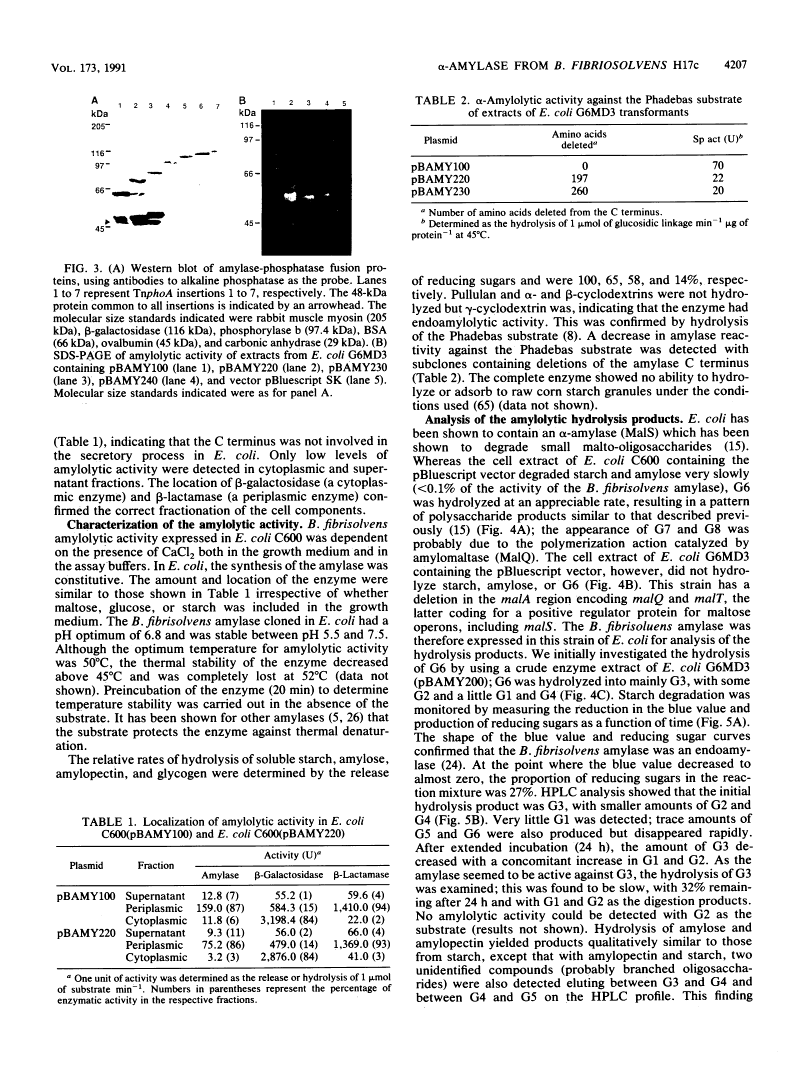
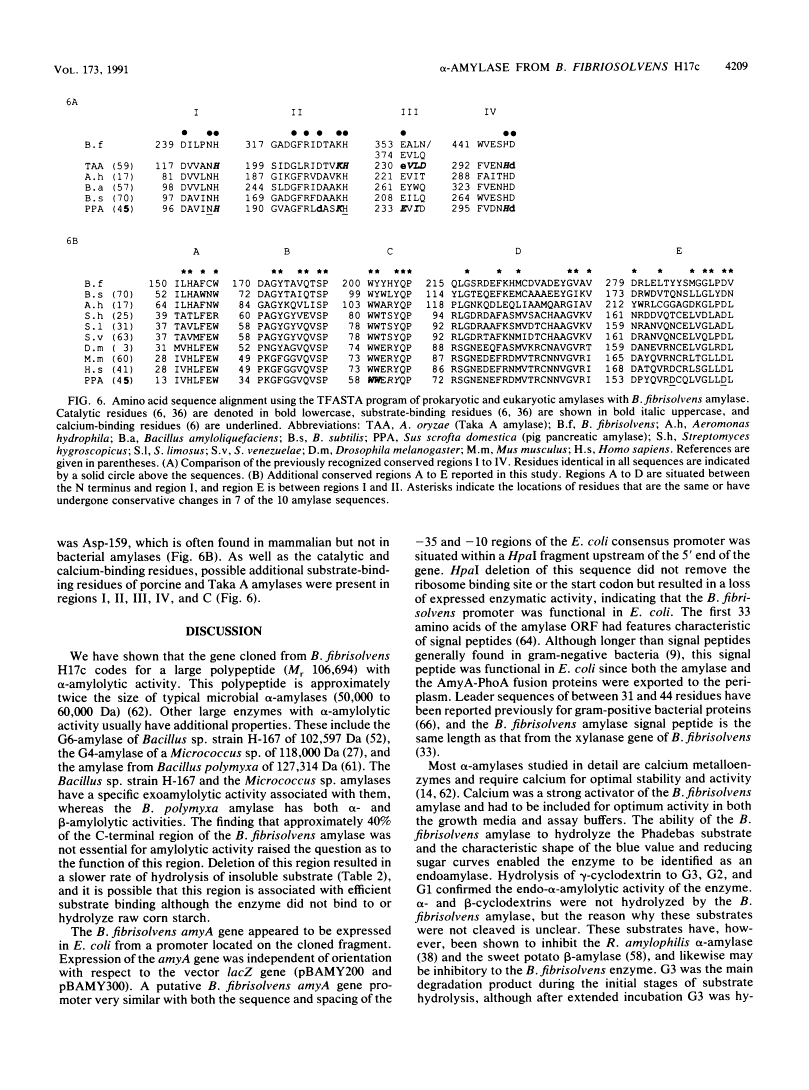
Abstract
Free full text

Cloning, nucleotide sequence, and enzymatic characterization of an alpha-amylase from the ruminal bacterium Butyrivibrio fibrisolvens H17c.
Abstract
A Butyrivibrio fibrisolvens amylase gene was cloned and expressed by using its own promoter on the recombinant plasmid pBAMY100 in Escherichia coli. The amylase gene consisted of an open reading frame of 2,931 bp encoding a protein of 976 amino acids with a calculated Mr of 106,964. In E. coli(pBAMY100), more than 86% of the active amylase was located in the periplasm, and TnphoA fusion experiments showed that the enzyme had a functional signal peptide. The B. fibrisolvens amylase is a calcium metalloenzyme, and three conserved putative calcium-binding residues were identified. The amylase showed high sequence homology with other alpha-amylases in the three highly conserved regions which constitute the active centers. These and other conserved regions were located in the N-terminal half, and no similarity with any other amylase was detected in the remainder of the protein. Deletion of approximately 40% of the C-terminal portion of the amylase did not result in loss of amylolytic activity. The B. fibrisolvens amylase was identified as an endo-alpha-amylase by hydrolysis of the Phadebas amylase substrate, hydrolysis of gamma-cyclodextrin to maltotriose, maltose, and glucose and the characteristic shape of the blue value and reducing sugar curves. Maltotriose was the major initial hydrolysis product from starch, although extended incubation resulted in its hydrolysis to maltose and glucose.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.8M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger E, Jones WA, Jones DT, Woods DR. Cloning and sequencing of an endoglucanase (end1) gene from Butyrivibrio fibrisolvens H17c. Mol Gen Genet. 1989 Oct;219(1-2):193–198. [Abstract] [Google Scholar]
- Boer PH, Hickey DA. The alpha-amylase gene in Drosophila melanogaster: nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res. 1986 Nov 11;14(21):8399–8411. [Europe PMC free article] [Abstract] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. [Abstract] [Google Scholar]
- Brown SH, Costantino HR, Kelly RM. Characterization of Amylolytic Enzyme Activities Associated with the Hyperthermophilic Archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990 Jul;56(7):1985–1991. [Europe PMC free article] [Abstract] [Google Scholar]
- Buisson G, Duée E, Haser R, Payan F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J. 1987 Dec 20;6(13):3909–3916. [Europe PMC free article] [Abstract] [Google Scholar]
- Candussio A, Schmid G, Böck A. Biochemical and genetic analysis of a maltopentaose-producing amylase from an alkaliphilic gram-positive bacterium. Eur J Biochem. 1990 Jul 20;191(1):177–185. [Abstract] [Google Scholar]
- Ceska M, Birath K, Brown B. A new and rapid method for the clinical determination of alpha-amylase activities in human serum and urine. Optimal conditions. Clin Chim Acta. 1969 Dec;26(3):437–444. [Abstract] [Google Scholar]
- Chang S. Engineering for protein secretion in gram-positive bacteria. Methods Enzymol. 1987;153:507–516. [Abstract] [Google Scholar]
- Cotta MA. Amylolytic activity of selected species of ruminal bacteria. Appl Environ Microbiol. 1988 Mar;54(3):772–776. [Europe PMC free article] [Abstract] [Google Scholar]
- Cotta MA, Hespell RB. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1986 Jul;52(1):51–58. [Europe PMC free article] [Abstract] [Google Scholar]
- Dehority BA. Characterization of several bovine rumen bacteria isolated with a xylan medium. J Bacteriol. 1966 May;91(5):1724–1729. [Europe PMC free article] [Abstract] [Google Scholar]
- Emori M, Takagi M, Maruo B, Yano K. Molecular cloning, nucleotide sequencing, and expression of the Bacillus subtilis (natto) IAM1212 alpha-amylase gene, which encodes an alpha-amylase structurally similar to but enzymatically distinct from that of B. subtilis 2633. J Bacteriol. 1990 Sep;172(9):4901–4908. [Europe PMC free article] [Abstract] [Google Scholar]
- Freundlieb S, Boos W. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986 Feb 25;261(6):2946–2953. [Abstract] [Google Scholar]
- Fujita M, Torigoe K, Nakada T, Tsusaki K, Kubota M, Sakai S, Tsujisaka Y. Cloning and nucleotide sequence of the gene (amyP) for maltotetraose-forming amylase from Pseudomonas stutzeri MO-19. J Bacteriol. 1989 Mar;171(3):1333–1339. [Europe PMC free article] [Abstract] [Google Scholar]
- Gobius KS, Pemberton JM. Molecular cloning, characterization, and nucleotide sequence of an extracellular amylase gene from Aeromonas hydrophila. J Bacteriol. 1988 Mar;170(3):1325–1332. [Europe PMC free article] [Abstract] [Google Scholar]
- Gutierrez C, Barondess J, Manoil C, Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J Mol Biol. 1987 May 20;195(2):289–297. [Abstract] [Google Scholar]
- Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. [Europe PMC free article] [Abstract] [Google Scholar]
- Hazlewood G, Dawson RM. Characteristics of a lipolytic and fatty acid-requiring Butyrivibrio sp. isolated from the ovine rumen. J Gen Microbiol. 1979 May;112(1):15–27. [Abstract] [Google Scholar]
- Hazlewood GP, Davidson K, Laurie JI, Romaniec MP, Gilbert HJ. Cloning and sequencing of the celA gene encoding endoglucanase A of Butyrivibrio fibrisolvens strain A46. J Gen Microbiol. 1990 Oct;136(10):2089–2097. [Abstract] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. [Abstract] [Google Scholar]
- Hespell RB, Wolf R, Bothast RJ. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl Environ Microbiol. 1987 Dec;53(12):2849–2853. [Europe PMC free article] [Abstract] [Google Scholar]
- HOBSON PN, MACPHERSON M. Amylases of Clostridium butyricum and a Streptococcus isolated from the rumen of the sheep. Biochem J. 1952 Dec;52(4):671–679. [Europe PMC free article] [Abstract] [Google Scholar]
- Hoshiko S, Makabe O, Nojiri C, Katsumata K, Satoh E, Nagaoka K. Molecular cloning and characterization of the Streptomyces hygroscopicus alpha-amylase gene. J Bacteriol. 1987 Mar;169(3):1029–1036. [Europe PMC free article] [Abstract] [Google Scholar]
- Kainuma K, Wako K, Kobayashi S, Nogami A, Suzuki S. Purification and some properties of a novel maltohexaose-producing exo-amylase from Aerobacter aerogenes. Biochim Biophys Acta. 1975 Dec 18;410(2):333–346. [Abstract] [Google Scholar]
- Kimura T, Horikoshi K. The nucleotide sequence of an alpha-amylase gene from an alkalopsychrotrophic Micrococcus sp. FEMS Microbiol Lett. 1990 Sep 1;59(1-2):35–41. [Abstract] [Google Scholar]
- Lacks SA, Springhorn SS. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J Biol Chem. 1980 Aug 10;255(15):7467–7473. [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Lin LL, Rumbak E, Zappe H, Thomson JA, Woods DR. Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a beta-glucosidase. J Gen Microbiol. 1990 Aug;136(8):1567–1576. [Abstract] [Google Scholar]
- Long CM, Virolle MJ, Chang SY, Chang S, Bibb MJ. alpha-Amylase gene of Streptomyces limosus: nucleotide sequence, expression motifs, and amino acid sequence homology to mammalian and invertebrate alpha-amylases. J Bacteriol. 1987 Dec;169(12):5745–5754. [Europe PMC free article] [Abstract] [Google Scholar]
- Mannarelli BM, Evans S, Lee D. Cloning, sequencing, and expression of a xylanase gene from the anaerobic ruminal bacterium Butyrivibrio fibrisolvens. J Bacteriol. 1990 Aug;172(8):4247–4254. [Europe PMC free article] [Abstract] [Google Scholar]
- Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. [Abstract] [Google Scholar]
- Matsuzaki H, Yamane K, Yamaguchi K, Nagata Y, Maruo B. Hybrid alpha-amylases produced by transformants of Bacillus subtilis. I. Purification and characterization of extracellular alpha-amylases produced by the parental strains and transformants. Biochim Biophys Acta. 1974 Sep 13;365(1):235–247. [Abstract] [Google Scholar]
- McWethy SJ, Hartman PA. Purification and some properties of an extracellular alpha-amylase from Bacteroides amylophilus. J Bacteriol. 1977 Mar;129(3):1537–1544. [Europe PMC free article] [Abstract] [Google Scholar]
- Mountfort DO, Asher RA. Production of alpha-Amylase by the Ruminal Anaerobic Fungus Neocallimastix frontalis. Appl Environ Microbiol. 1988 Sep;54(9):2293–2299. [Europe PMC free article] [Abstract] [Google Scholar]
- Nishide T, Emi M, Nakamura Y, Matsubara K. Corrected sequences of cDNAs for human salivary and pancreatic alpha-amylases [corrected]. Gene. 1984 May;28(2):263–270. [Abstract] [Google Scholar]
- Orpin CG, Mathiesen SD, Greenwood Y, Blix AS. Seasonal changes in the ruminal microflora of the high-arctic Svalbard reindeer (Rangifer tarandus platyrhynchus). Appl Environ Microbiol. 1985 Jul;50(1):144–151. [Europe PMC free article] [Abstract] [Google Scholar]
- Pasero L, Mazzéi-Pierron Y, Abadie B, Chicheportiche Y, Marchis-Mouren G. Complete amino acid sequence and location of the five disulfide bridges in porcine pancreatic alpha-amylase. Biochim Biophys Acta. 1986 Jan 30;869(2):147–157. [Abstract] [Google Scholar]
- Rogers JC. Conserved amino acid sequence domains in alpha-amylases from plants, mammals, and bacteria. Biochem Biophys Res Commun. 1985 Apr 16;128(1):470–476. [Abstract] [Google Scholar]
- Rybicki EP, von Wechmar MB. Enzyme-assisted immune detection of plant virus proteins electroblotted onto nitrocellulose paper. J Virol Methods. 1982 Dec;5(5-6):267–278. [Abstract] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. [Abstract] [Google Scholar]
- Scholle RR, Robb SM, Robb FT, Woods DR. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrase gene (scrB). Gene. 1989 Aug 1;80(1):49–56. [Abstract] [Google Scholar]
- Schwartz M. Location of the maltose A and B loci on the genetic map of Escherichia coli. J Bacteriol. 1966 Oct;92(4):1083–1089. [Europe PMC free article] [Abstract] [Google Scholar]
- Shane BS, Gouws L, Kistner A. Cellulolytic bacteria occurring in the rumen of sheep conditioned to low-protein teff hay. J Gen Microbiol. 1969 Mar;55(3):445–457. [Abstract] [Google Scholar]
- Shirokizawa O, Akiba T, Horikoshi K. Nucleotide sequence of the G6-amylase gene from alkalophilic Bacillus sp. H-167. FEMS Microbiol Lett. 1990 Jul;58(2):131–135. [Abstract] [Google Scholar]
- Slyter LL. Influence of acidosis on rumen function. J Anim Sci. 1976 Oct;43(4):910–929. [Abstract] [Google Scholar]
- Sykes RB, Nordström K. Microiodometric determination of beta-lactamase activity. Antimicrob Agents Chemother. 1972 Feb;1(2):94–99. [Europe PMC free article] [Abstract] [Google Scholar]
- Takkinen K, Pettersson RF, Kalkkinen N, Palva I, Söderlund H, Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [Abstract] [Google Scholar]
- Tosi M, Bovey R, Astolfi S, Bodary S, Meisler M, Wellauer PK. Multiple non-allelic genes encoding pancreatic alpha-amylase of mouse are expressed in a strain-specific fashion. EMBO J. 1984 Dec 1;3(12):2809–2816. [Europe PMC free article] [Abstract] [Google Scholar]
- Uozumi N, Sakurai K, Sasaki T, Takekawa S, Yamagata H, Tsukagoshi N, Udaka S. A single gene directs synthesis of a precursor protein with beta- and alpha-amylase activities in Bacillus polymyxa. J Bacteriol. 1989 Jan;171(1):375–382. [Europe PMC free article] [Abstract] [Google Scholar]
- Vihinen M, Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol. 1989;24(4):329–418. [Abstract] [Google Scholar]
- Virolle MJ, Long CM, Chang S, Bibb MJ. Cloning, characterisation and regulation of an alpha-amylase gene from Streptomyces venezuelae. Gene. 1988 Dec 30;74(2):321–334. [Abstract] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. [Abstract] [Google Scholar]
- Walker GJ, Hope PM. Degradation of starch granules by some amylolytic bacteria from the rumen of sheep. Biochem J. 1964 Feb;90(2):398–408. [Europe PMC free article] [Abstract] [Google Scholar]
- Watson ME. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang JH, Copeland L. Chemical modification of mitochondria. Uncoupler binding by mitochondria in different metabolic states. Arch Biochem Biophys. 1974 May;162(1):64–72. [Abstract] [Google Scholar]
- Wojciechowicz M, Heinrichova K, Ziołecki A. An exopectate lyase of Butyrivibrio fibrisolvens from the bovine rumen. J Gen Microbiol. 1982 Nov;128(11):2661–2665. [Abstract] [Google Scholar]
- Wood WB. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. [Abstract] [Google Scholar]
- Yang M, Galizzi A, Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. [Europe PMC free article] [Abstract] [Google Scholar]
- Zabeau M, Stanley KK. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1(10):1217–1224. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.173.13.4203-4211.1991
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/173/13/4203.full.pdf
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/173/13/4203
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/173/13/4203
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.173.13.4203-4211.1991
Article citations
Dietary supplementation of ruminant diets with an Aspergillus oryzae α-amylase
Anim Feed Sci Technol, 145(1-4):136-150, 01 Aug 2008
Cited by: 13 articles | AGR: IND44090325
Starch-binding domain affects catalysis in two Lactobacillus alpha-amylases.
Appl Environ Microbiol, 71(1):297-302, 01 Jan 2005
Cited by: 26 articles | PMID: 15640201 | PMCID: PMC544272
Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics.
FEMS Microbiol Rev, 27(5):663-693, 01 Dec 2003
Cited by: 202 articles | PMID: 14638418
Review
Characterization of the L. manihotivorans alpha-amylase gene.
DNA Seq, 12(1):27-37, 01 Jul 2001
Cited by: 13 articles | PMID: 11697143
Distribution and evolution of the xylanase genes xynA and xynB and their homologues in strains of Butyrivibrio fibrisolvens.
Appl Environ Microbiol, 65(8):3660-3667, 01 Aug 1999
Cited by: 10 articles | PMID: 10427063 | PMCID: PMC91548
Go to all (14) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a beta-glucosidase.
J Gen Microbiol, 136(8):1567-1576, 01 Aug 1990
Cited by: 24 articles | PMID: 2262790
Characterization of the Butyrivibrio fibrisolvens glgB gene, which encodes a glycogen-branching enzyme with starch-clearing activity.
J Bacteriol, 173(21):6732-6741, 01 Nov 1991
Cited by: 15 articles | PMID: 1938880 | PMCID: PMC209022
Cloning and sequencing of an endoglucanase (end1) gene from Butyrivibrio fibrisolvens H17c.
Mol Gen Genet, 219(1-2):193-198, 01 Oct 1989
Cited by: 32 articles | PMID: 2615759
Sequencing and expression of a cellodextrinase (ced1) gene from Butyrivibrio fibrisolvens H17c cloned in Escherichia coli.
Mol Gen Genet, 223(2):310-318, 01 Sep 1990
Cited by: 24 articles | PMID: 2250655