Abstract
Free full text

Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication
Abstract
Centrins are calmodulin-like proteins present in microtubule-organizing centers. The Saccharomyces cerevisiae centrin, Cdc31p, was functionally tagged with a single Z domain of protein A, and used in pull-down experiments to isolate Cdc31p-binding proteins. One of these, Sfi1p, localizes to the half-bridge of the spindle pole body (SPB), where Cdc31p is also localized. Temperature-sensitive mutants in SFI1 show a defect in SPB duplication and genetic interactions with cdc31-1. Sfi1p contains multiple internal repeats that are also present in a Schizosaccharomyces pombe protein, which also localizes to the SPB, and in several human proteins, one of which localizes close to the centriole region. Cdc31p binds directly to individual Sfi1 repeats in a 1:1 ratio, so a single molecule of Sfi1p binds multiple molecules of Cdc31p. The centrosomal human protein containing Sfi1 repeats also binds centrin in the repeat region, showing that this centrin-binding motif is conserved.
Introduction
Centrins are conserved calmodulin-like proteins and, like calmodulin, have roles in a number of different cellular processes (Schiebel and Bornens, 1995; Sullivan et al., 1998; Araki et al., 2001; Pulvermuller et al., 2002). In particular, centrins are ubiquitous components of microtubule-organizing centers (MTOCs), and are often located within different parts of the MTOC (Levy et al., 1996), again suggesting multiple roles. Two clear roles have been established. One is in the duplication of the MTOC (Byers, 1981; Middendorp et al., 2000; Salisbury et al., 2002), and the other is as constituents of contractile fibers within and attached to the MTOC, which can contract in response to changes in Ca2+ concentration (Salisbury et al., 1984). A related fibrous structure that also contracts in response to changes in Ca2+ concentration is the vorticellid spasmoneme (Weis-Fogh and Amos, 1972; Moriyama et al., 1999). Its principle constituent, spasmin (Amos et al., 1975), has homology to EF-hand domain proteins such as centrin (Maciejewski et al., 1999). In addition, the spasmoneme also has elastic properties that are not dependent on the Ca2+ concentration (Weis-Fogh and Amos, 1972). Thus, two properties, Ca2+-dependent contraction and Ca2+-independent elasticity, may be characteristic of centrin-containing fibrous structures.
Budding yeast has one centrin, Cdc31p, some of which is localized (Spang et al., 1993) to the half-bridge of the spindle pole body (SPB). The SPB is embedded in the nuclear envelope, and the half-bridge is a specialized rectangular area of the envelope attached to one side of the SPB (Byers and Goetsch, 1974; Adams and Kilmartin, 1999; O'Toole et al., 1999). The half-bridge plays an important role in SPB duplication. First, the new SPB starts to assemble on the distal cytoplasmic side of the half-bridge; and second, the half-bridge allows the transit of the partly assembled SPB across the two lipid bilayers of the nuclear envelope so that a nuclear spindle can be formed (Adams and Kilmartin, 1999). This transit process occurs by fusion of the cytoplasmic and nuclear lipid bilayers at the distal end of the half-bridge, followed by retraction of the half-bridge across the face of the newly assembled duplication plaque. This exposes the plaque to nucleoplasm, thereby allowing assembly of nuclear SPB components and completing SPB assembly (Adams and Kilmartin, 1999). The retraction process may rely on possible elastic properties of the half-bridge.
Cdc31p is essential for SPB duplication: temperature-sensitive mutants arrest with a single large SPB (Byers, 1981) and fail to form a satellite (Winey et al., 1991). Two Cdc31p-binding proteins in the half-bridge have already been identified; Kar1p (Biggins and Rose, 1994; Spang et al., 1995), and more recently, Mps3p/Nep98p (Jaspersen et al., 2002; Nishikawa et al., 2003). This paper describes a protein, Sfi1p, which binds Cdc31p directly via multiple conserved centrin-binding sites, and which has an essential role in SPB duplication. The repeated domain structure of Sfi1p suggests a mechanism for either contraction or elasticity of the half-bridge of the SPB.
Results
A complex containing Cdc31p and Sfi1p
A powerful way of identifying interacting proteins in a complex is tagging one of the proteins with protein A (prA), followed by purification of the complex on an IgG column (Grandi et al., 1993; Knop and Schiebel, 1997). This approach was applied to Cdc31p, but all COOH-terminal and longer NH2-terminal tags failed to rescue a deletion. However, short NH2-terminal tags containing single epitope tags or a single Z domain of prA (ZCdc31p) were functional, and in the case of ZCdc31p, brought down other proteins in a pull-down experiment (Fig. 1 a).
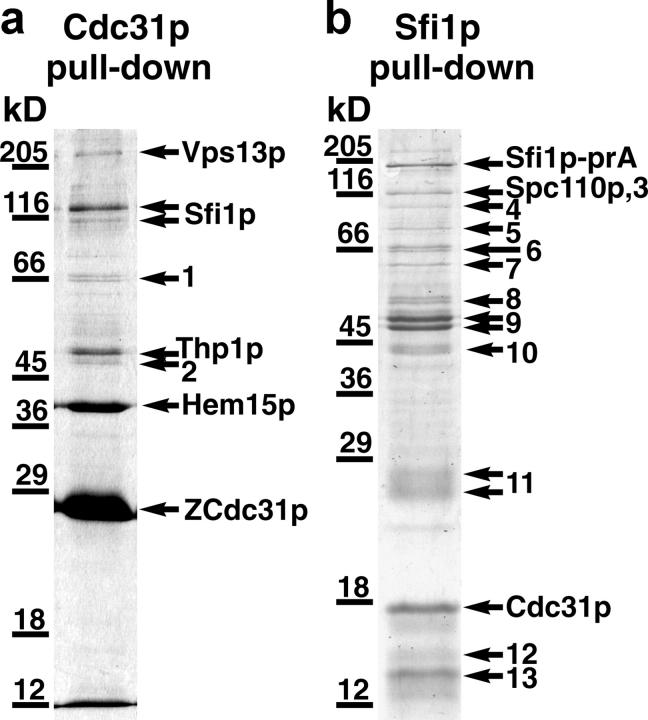
SDS gel of proteins isolated from yeast cells containing ZCdc31p or Sfi1p-prA after binding to IgG-Sepharose. Bands were identified by matrix-assisted laser desorption/ionization mass spectrometry. Numbered bands contained heat shock proteins, proteosome components, ribosomal proteins, keratin, or nonspecific proteins (see Materials and methods).
For these experiments, lysis was performed under low free Ca2+ conditions (5 mM EGTA; Wigge and Kilmartin, 2001). This was because there is evidence that interactions of Cdc31p with proteins other than Kar1p are less Ca2+ sensitive (Geier et al., 1996). In addition, interactions of spasmin with the spasmoneme (Amos, 1971; Amos et al., 1975), and centrin with a 350-kD protein in the Paramecium infraciliary lattice (Klotz et al., 1997), are stable in the presence of EGTA. The proteins that copurified with ZCdc31p were of diverse function: Vps13p is involved membrane traffic (Brickner and Fuller, 1997), Thp1p is involved in mitotic recombination (Gallardo and Aguilera, 2001), and Hem15p is ferrochelatase (Labbe-Bois, 1990). One protein of particular interest, Sfi1p, was initially identified as a suppressor of the heat sensitivity associated with a particular mutation in adenyl cyclase (Ma et al., 1999). When Sfi1p was partly characterized, it was found not to be involved in the adenyl cyclase pathway, but it is an essential protein whose depletion causes a G2/M arrest with failure to form a mitotic spindle (Ma et al., 1999). This is similar to the phenotype of cdc31-1 (Byers, 1981), and indicates a possible function at the SPB.
To confirm that the interaction between Sfi1p and Cdc31p was reciprocal, Sfi1p-prA was used in a pull-down experiment under the same lysis conditions (Fig. 1 b). This pull-down was not as clean, but it did isolate Cdc31p together with another SPB component, Spc110p (Rout and Kilmartin, 1990; Kilmartin et al., 1993). It is not clear whether this reflects a direct interaction with Spc110p or with other SPB components such as Spc42p (Donaldson and Kilmartin, 1996), which are present in a complex with Spc110p (Elliott et al., 1999). At the low levels of protein present in Fig. 1 b, these other SPB components would be difficult to detect.
These experiments show a reciprocal interaction between Sfi1p and Cdc31p. In addition, inspection of the relative intensities of the Coomassie-stained bands for Sfi1p-prA and Cdc31p (the bands were too faint to scan accurately) suggests that Cdc31p is present in a molar ratio >1.
Localization of Sfi1p-GFP
Sfi1p was tagged with GFP and was found to localize to one or two spots that were coincident with nuclear DNA (Fig. 2, a and b). This suggests localization to the SPB region, which was confirmed by immunofluorescence (Fig. 2, c and d). Immuno-EM then defined the localization more precisely to the half-bridge (Fig. 2 e). The immuno-EM staining of Sfi1p in single SPBs appeared somewhat distal to the existing SPB, and in paired SPBs was more centrally located on the bridge compared with other bridge components such as Spc72p (Adams and Kilmartin, 1999). Whether this indicates a more specific localization of Sfi1p on the half-bridge or merely antigen accessibility is not clear. Cdc31p is also localized to the half-bridge (Spang et al., 1993), thus Sfi1p and Cdc31p are located in the same part of the SPB.
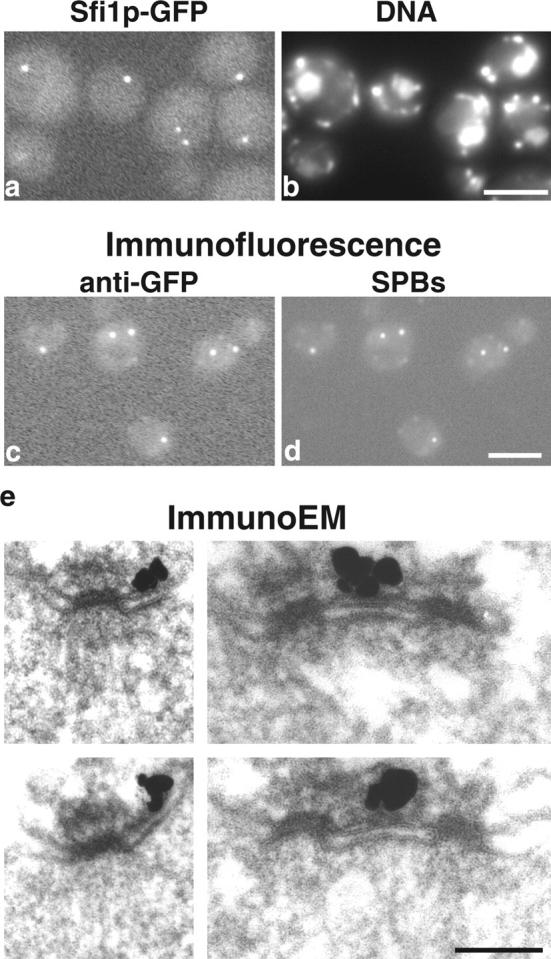
Localization of S. cerevisiae Sfi1p. (a and b) Fluorescence of unfixed cells containing Sfi1p-GFP. GFP fluorescence (a) and DAPI fluorescence for DNA (b) show one or two GFP spots coincident with nuclei. In b, nuclei are the larger areas of staining; the small bright spots are mitochondria. (c and d) Immunofluorescence with anti-GFP (c) and anti-90-kD (Spc98p) mAbs to localize SPBs (d). (e) Immuno-EM of GFP-Sfi1p showing staining localized to the half-bridge. Bars: (a–d) 2 μm; (e) 0.1 μm.
Phenotype of sfi1-3 and sfi1-7
The localization of Sfi1p to the half-bridge of the SPB suggests that it might have a function in SPB duplication. Temperature-sensitive mutants were prepared to check the arrest in more detail, in particular to determine whether SPB duplication had occurred and also to look for genetic interactions with CDC31. Two alleles, sfi1-3 and sfi1-7, were examined by immunofluorescent staining after arrest for 4 h at 36°C, and showed a very similar phenotype to the depletion (Ma et al., 1999), i.e., failure to form a mitotic spindle (unpublished data). However, cells synchronized in G1 with α-factor (10 μg/ml for 150 min at 23°C) and released at 36°C showed interesting differences between the two alleles. At 1.5 h after release, sfi1-7 cells passed through mitosis: 78% of the cells had spindles, and post-anaphase spindles appeared to show normal separating or separated DNA (Fig. 3 a). All of the cells then arrested with single microtubule asters from 2.5 to 5 h (Fig. 3 b). However, sfi1-3 cells (95% of the cells examined), despite budding at the same time as sfi1-7, did not pass through mitosis at 1.5 h, but remained arrested with single microtubule asters between 1.5 and 5 h (Fig. 3 c). These results show that sfi1-3 cells arrest during the first cell cycle, and sfi1-7 cells arrest during the second cell cycle. This behavior is very similar to that of different cdc31 alleles, where some arrest with single microtubule asters in the first cell cycle, and some arrest in the second (Byers, 1981; Vallen et al., 1994).
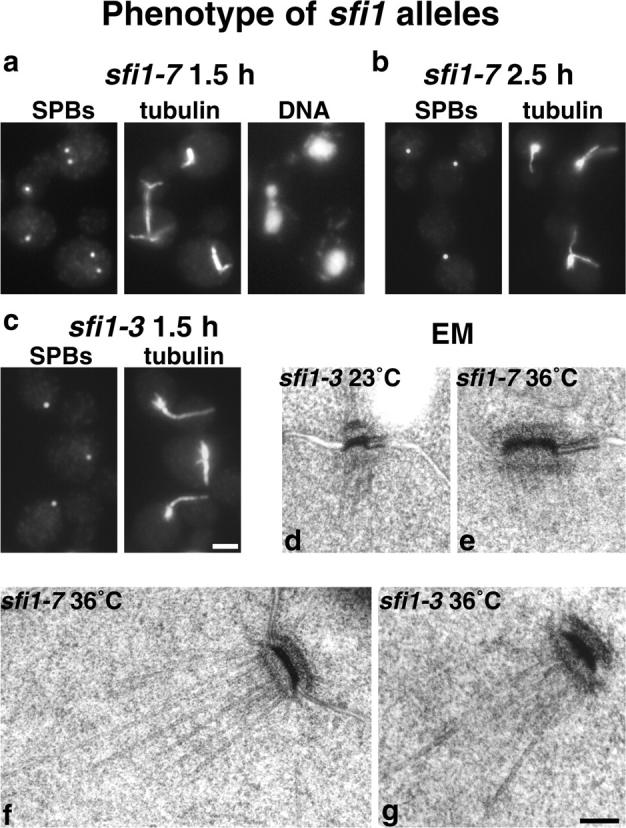
Phenotype of sfi1 alleles. Cells synchronized in G1 with α-factor were released at 36°C. (a) sfi1-7 at 1.5 h, (b) sfi1-7 at 2.5 h, (c) sfi1-3 at 1.5 h, stained with anti-Tub4p for SPBs or anti-tubulin and DAPI for DNA. (d) EM of sfi1-3 at 23°C, (e) sfi1-7 after 5 h at 36°C, (f) sfi1-7 after 4 h, and (g) sfi1-3 after 2.5 h. Bars: (a-c) 2 μm; (d-g) 0.1 μm.
Both alleles were examined by EM to determine whether SPB duplication had occurred. The SPB-containing regions of asynchronous cells after 4 h at 36°C and synchronized cells between 2.5 and 5 h after release were examined by serial sectioning. In a total of 50 cells from both alleles, 49 had only single SPBs (Fig. 3, d–g). The one exception was an sfi1-7 cell with one normal SPB and one small partly assembled SPB still on the cytoplasmic side of the half-bridge, similar to the phenotype of mps2-1 (Winey et al., 1991). About one third of the single SPBs had clear half-bridges (Fig. 3 e), but this is an underestimate, as the half-bridge is difficult to recognize unless it is almost perpendicular to the section. There was a clear increase in SPB size, the central plaque diameter increasing from 0.10 ± 0.02 μm (Fig. 3 d) to 0.16 ± 0.08 μm (Fig. 3, e–g). This is again similar to cdc31-1 (Byers, 1981), although the increase in SPB size is smaller.
In conclusion, the phenotypes of both sfi1-3 and sfi1-7 show striking similarities to cdc31 alleles (Byers, 1981; Vallen et al., 1994). All of these alleles arrest with a single large SPB in either the first or second cell cycle at the restrictive temperature.
Genetic interactions between sfi1-3, sfi1-7, and cdc31-1
The similarity between the phenotypes of sfi1-3, sfi1-7, and cdc31-1 suggest that there might be genetic interactions between SFI1 and CDC31. Overexpression of CDC31 on a 2-μm plasmid suppressed both sfi1-3 (Fig. 4 a) and sfi1-7 at 37°C. There were also synthetic growth defects between both sfi1 alleles and cdc31-1. Only one third of the expected sfi1-7 cdc31-1 double temperature-sensitive spores grew at 23°C, and these all grew extremely slowly, produced a very variable colony size on streaking out, and were not studied further. All except one of the expected sfi1-3 cdc31-1 double temperature-sensitive spores grew at 23°C, but slightly more slowly than the wild type, and failed to grow at all at 30°C. Transformation of these cells with SFI1 restored growth at 30°C, which was dependent on the presence of SFI1 (Fig. 4 b). These experiments show a synthetic growth interaction between the sfi1 alleles and cdc31-1, and both sets of experiments show genetic interactions between SFI1 and CDC31.
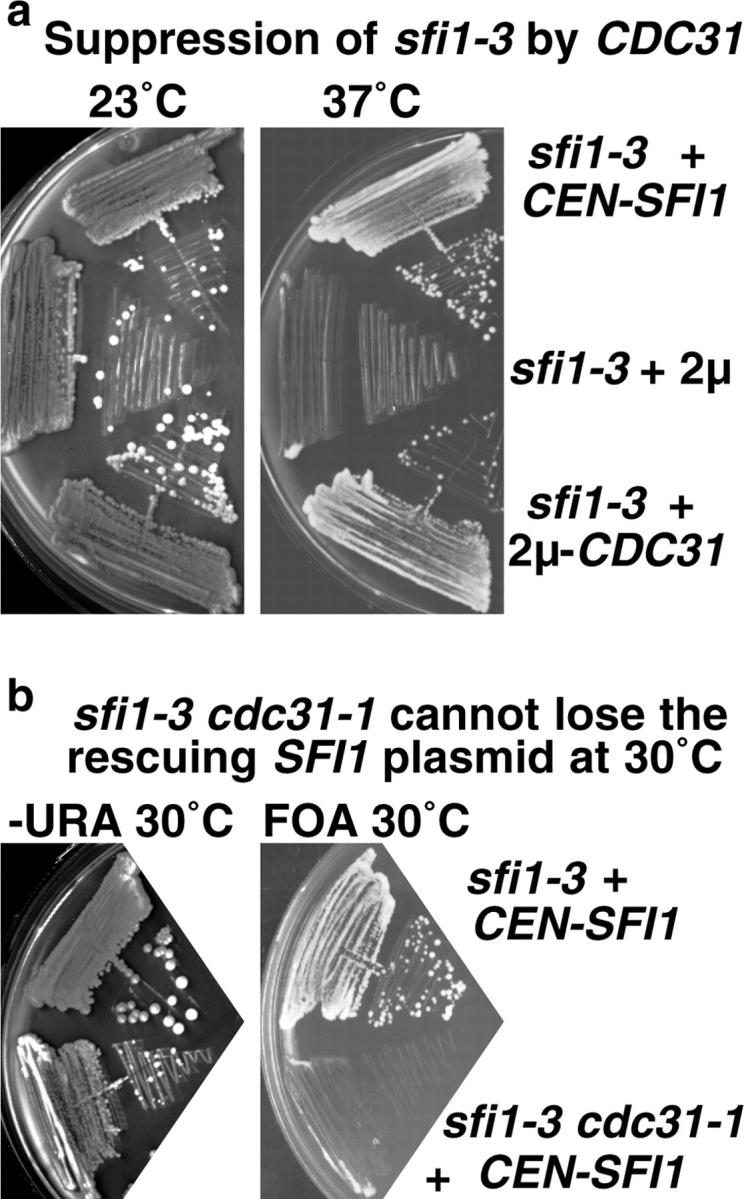
Genetic interactions between SFI1 and CDC31. (a) Suppression of sfi1-3 by CDC31 in an overexpressing 2-μm plasmid. (b) The double temperature-sensitive mutant sfi1-3cdc31-1 does not grow at 30°C and cannot lose the rescuing CEN-URA3-SFI1 on fluoroorotic acid medium (which selects against URA3 plasmids).
Structural features and conservation of Sfi1 repeats
Saccharomyces cerevisiae Sfi1p is a very divergent protein. There is only 24.5% sequence identity (Cliften et al., 2003) between the S. cerevisiae and Saccharomyces castelli proteins (these alignments are displayed at http://db.yeastgenome.org). This degree of divergence is similar or sometimes even worse for most SPB proteins; for example, Spc42p (Donaldson and Kilmartin, 1996) has only 16.5% identity between the two species. However, S. cerevisiae Sfi1p has a series of striking internal repeats (Fig. 5 a) containing a consensus sequence AX7LLX3F/LX2WK/R. This repeat structure is conserved in the other Saccharomyces species, with the possible exception of repeats around W304 and F698, and sometimes the alanine is not as prominent. The gap between the clusters of repeats in S. cerevisiae Sfi1p is between 23 and 35 amino acids.
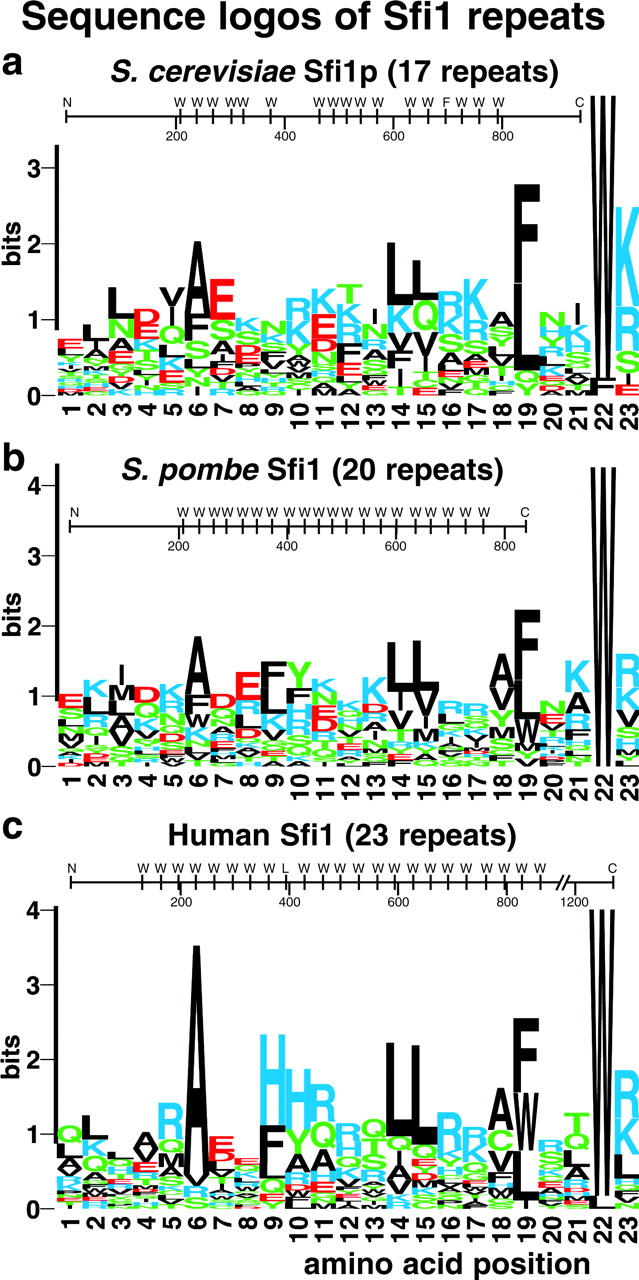
Sequence logos of Sfi1 repeats in S. cerevisiae and potential homologues in S. pombe, SpSfi1, and humans, hSfi1. The domain structure for each protein is also shown with the positions of the conserved tryptophans (position 22) marked.
Proteins with similar repeats to Sfi1p and with similar gaps between the repeats were identified by BLAST searching. These were an S. pombe protein (T40750) called SpSfi1 here, and a human protein (KIAA0542) called hSfi1 here (Fig. 5, b and c). There are at least two versions of hSfi1 with almost identical sequence, but with stop codons after either 968 or 1242 amino acids (see Materials and methods); the version with 1242 amino acids is analyzed here. The repeats in hSfi1 are particularly uniform. Between residues 96 and 862 there are nine continuous 33mer repeats, one 35mer, a further nine 33mer repeats, one 37mer, and finally three 33mer repeats (the position of the conserved tryptophan, W, is shown diagrammatically in Fig. 5). The proline content within the repeat regions for all of the three proteins was low: one for Sfi1p, none for S. pombe Sfi1, and two for human Sfi1. There are no further consensus residues outside of the repeat region shown in Fig. 5, and no homology was detected between the NH2- and COOH-terminal domains of the three proteins.
Are there other proteins in these three organisms that also contain Sfi1 repeats? Because the repeat is quite heterogeneous in sequence, BLAST searching was not suitable, so ScanProsite (Gattiker et al., 2002) was used instead (see Materials and methods). This identified two more human proteins and the partial sequence of another containing multiple copies of this repeat (see Materials and methods). However, these sequences have many fewer repeats than hSfi1, and are thus less similar to the yeast proteins. No further S. cerevisiae proteins were identified. These results suggest that Sfi1 repeats are conserved between fungi and humans, and may be present in multiple proteins within one organism.
Localization of S. pombe Sfi1 and human Sfi1
S. pombe Sfi1 (Fig. 5 b), a potential homologue of S. cerevisiae Sfi1p, was tagged with GFP to see whether it too localized to the SPB. Tagged S. pombe Sfi1 localized to one or two spots per cell coincident with the nuclear DNA (Fig. 6, a and b). Double labeling showed that these spots localized to the ends of the mitotic spindle (Fig. 6, c and d) and were coincident with Sad1 (Fig. 6, e and f), a marker for the SPB (Hagan and Yanagida, 1995). Thus, S. pombe Sfi1, like S. cerevisiae Sfi1p, localizes to the SPB region.
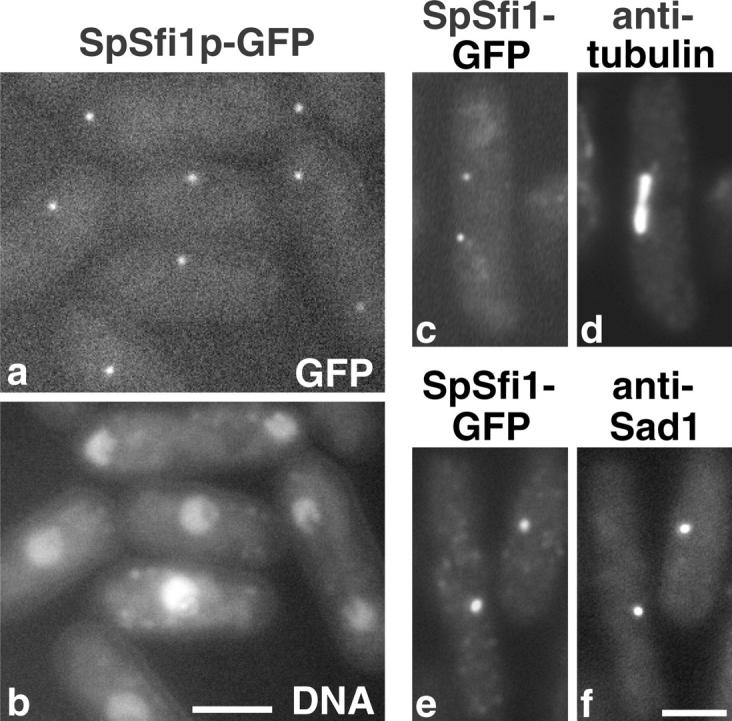
Localization of S. pombe Sfi1 (SpSfi1). (a and b) Fluo-rescence of unfixed cells containing SpSfi1p-GFP. GFP fluorescence (a) and DAPI fluorescence for DNA (b) show one or two GFP spots coincident with nuclei. (c–f) GFP fluorescence and immunofluorescence. GFP fluorescence (c and e) with anti-tubulin (d) to show localization of SpSfi1p to the ends of the mitotic spindle, and with the SPB marker anti-Sad1 (f) to show localization to the SPB region. Bars, 2 μm.
Of the four human proteins identified with potential Sfi1 repeats, hSfi1 seemed the most homologous to S. cerevisiae Sfi1p and S. pombe Sfi1. The other human proteins had between 4 and 10 repeats, whereas hSfi1 had 23 repeats, similar to the repeat number in the two yeasts (Fig. 5). In addition, as in the two yeasts, it had the repeats centrally positioned, similarly spaced, and also a low proline content in the repeat region.
hSfi1 was tagged at the NH2 terminus with GFP and was transfected into HeLa cells. A faint signal was detected with anti-GFP consisting of one or two dots within or close to the centrosomal γ-tubulin staining (Fig. 7, a and b). Similar results were found for both the 1242 and 968 amino acid forms of hSfi1, but the transfection efficiency was much higher for the 1242 amino acid form, so this was used for all the experiments. No staining was seen when GFP was placed at the COOH terminus of either form. The staining was almost completely coincident (Fig. 7, c–e) with an mAb, αCetn2 (Hart et al., 2001), against centrin 2, and almost coincident (Fig. 7, f–h) with an mAb, GT335 (Wolff et al., 1992), against glutamylated tubulin and a marker for centrioles (Bobinnec et al., 1998). These results indicate that hSfi1 localizes close to the centriole.
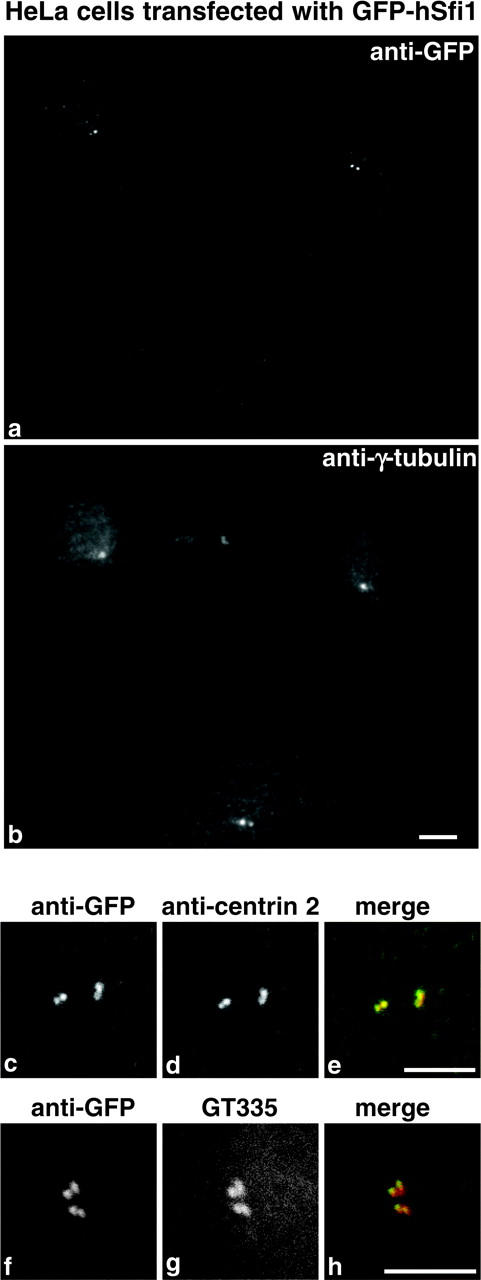
Transfection of HeLa cells with GFP-hSfi1. Cells stained with anti-GFP (a) and anti-γ-tubulin (b). An untransfected cell is shown at the bottom. Staining of the centrosome region with anti-GFP (c and f), anti-centrin 2 (d), GT335 (g), and corresponding merged images (e and h). Anti-GFP is green and anti-centrin 2 and GT335 are red. Bars, 5 μm.
Binding of Cdc31p to Sfi1p
The prA pull-down experiments shown in Fig. 1 suggest the Cdc31p binds directly to Sfi1p. This was tested by GST pull-downs using proteins expressed in Escherichia coli. Because Cdc31p is conserved and the only conserved part of Sfi1p is the repeats, it seemed likely that Cdc31p would bind to this region. Accordingly, Sfi1p was initially divided into five domains (Fig. 8 a); the NH2-terminal domain (fusion 1), three domains containing the repeats (fusions 2–4), and the COOH-terminal domain (fusion 5). The SDS gel of the GST pull-downs (Fig. 8 b) shows the recombinant Cdc31p in the left lane, followed by the fusion protein (F), the supernatant from the pull-down (S), and the washed pellet from the pull-down (P) for each of the five constructs. It is clear that Cdc31p fails to bind to the NH2- and COOH-terminal domains, but does bind to the three fusions containing the repeats (Fig. 8 b). In addition, the relative intensities of the Coomassie-stained bands show that the molar ratio of Cdc31p to fusion is >1. Scanning of the gels showed between 3 and 4 mol of Cdc31p per mol of fusion protein (this number is approximate for fusion 3 because of proteolysis). Increasing the Cdc31p concentration showed some evidence for saturation for fusion 2, where the molar ratio rose to ~5.5 compared with the six predicted repeats (Fig. 8 c).
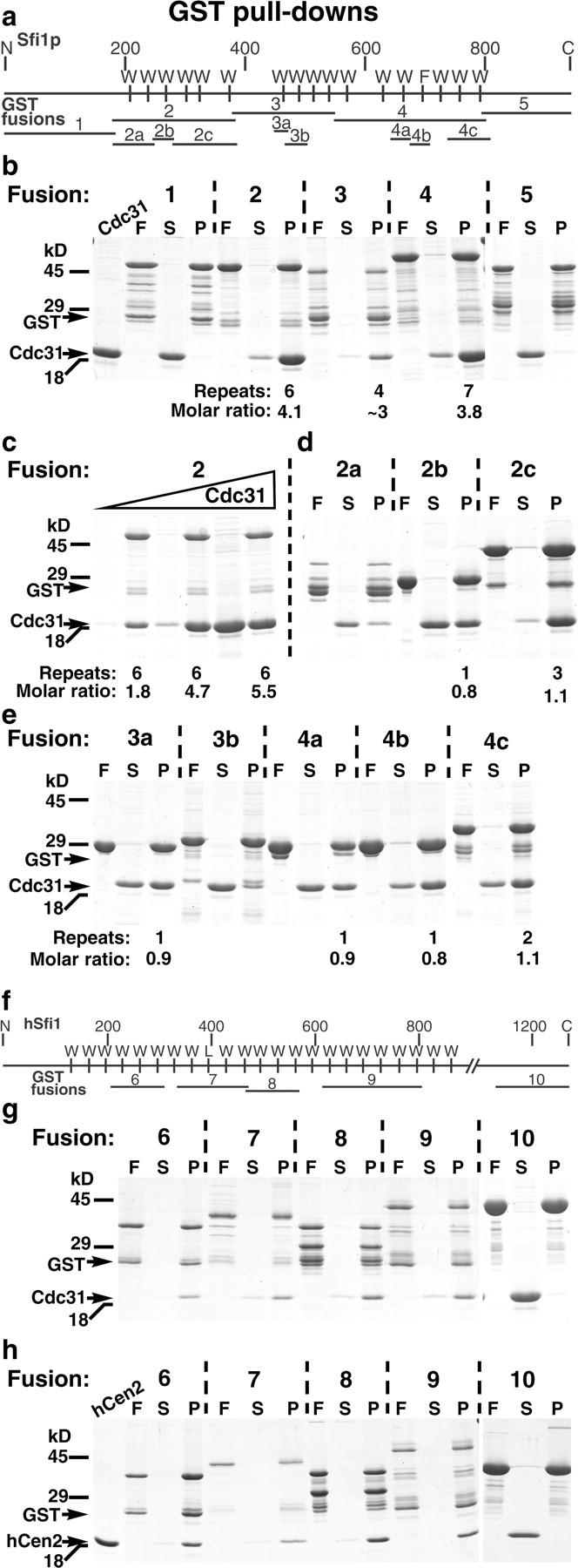
GST pull-downs to localize the centrin-binding sites in Sfi1p and human hSfi1. Maps of the GST fusions are shown (a) for Sfi1p and (f) for hSfi1. Gels were stained with Coomassie blue and show the recombinant Cdc31p (first lane in b) and the recombinant human centrin 2 (first lane in h). In each set of three lanes for the pull-downs, F is the fusion protein, S is the supernatant after the pull-down, and P the pellet (3-μl beads washed 4–5 times with 0.5 ml buffer). (b) GST pull-downs for Cdc31p binding to fusions 1–5 in Sfi1p. (c) Binding of Cdc31p to fusion 2 with increasing Cdc31p concentration. (d and e) Cdc31p binding to Sfi1p fusions containing 1–3 repeats. (g) Cdc31p binding to GST fusions of hSfi1. (h) Human centrin 2 binding to GST fusions of hSfi1.
Next, individual and smaller groups of repeats were examined. In designing the fusions for the individual repeats, it was assumed that the conserved tryptophan was the critical element, so it was placed centrally. All of these fusions were positive for Cdc31p binding (Fig. 8, d and e), although for 3b the affinity was low. The other fusions containing single repeats (2b, 3a, 4a, and 4b) showed molar ratios close to 1, which did not rise above 1.1 over a 10-fold range of Cdc31p concentration (unpublished data). For the fusions with 2 to 3 repeats, 2a unfortunately suffered from proteolysis, 2c gave a ratio of 1.1 in Fig. 8 d, which rose to 2.5 when the Cdc31p concentration was increased (unpublished data), compared with the predicted three repeats. The ratio for 4c containing two repeats never rose above 1.1 when the Cdc31p concentration was increased (unpublished data), suggesting that one of the repeats is inactive. As expected from the low free Ca2+ conditions of the yeast lysate pull-down (Fig. 1 a), the binding of Cdc31p to the GST fusions did not appear to be Ca2+-sensitive because it was unaffected by removal of Ca2+ with 5 mM EGTA (unpublished data). Together, these data suggest that most of the Sfi1 repeats in Sfi1p can bind Cdc31p.
Sfi1 repeats have been found in several human proteins (see Fig. 5 c and Materials and methods), so fusions were also prepared for hSfi1 (Fig. 8 f) and tested for binding to Cdc31p and human centrin 2 (Lee and Huang, 1993). The fusions containing Sfi1 repeats bound both Cdc31p (Fig. 8 g) and human centrin 2 (Fig. 8 h), whereas fusion 10 from the COOH terminus, containing no detectable Sfi1 repeats, did not bind either protein (Fig. 8, g and h). Human centrins 1 (Errabolu et al., 1994) and 3 (Middendorp et al., 1997) and mouse centrin 4 (Gavet et al., 2003) gave similar binding to fusions 6–9, but not fusion 10, with slight variations between the different centrins (unpublished data). Again, the binding of Cdc31p was unaffected by 5 mM EGTA, though the binding of centrin 2 was slightly decreased (unpublished data).
These results show that Sfi1p binds multiple molecules of Cdc31p directly through a series of conserved repeats. A human homologue of Sfi1p, hSfi1 containing similar repeats, also binds Cdc31p as well as mammalian centrins directly at these repeats.
Discussion
This paper describes a protein, Sfi1p, isolated from a pull-down using a functional tagged version of the budding yeast centrin, Cdc31p. Like Cdc31p (Byers, 1981; Spang et al., 1993), Sfi1p localizes to the half-bridge of the SPB and has an essential function in SPB duplication. SFI1 and CDC31 show genetic interactions, suggesting they interact in vivo. Cdc31p binds directly to multiple binding sites in Sfi1p that consist of a series of 17 novel repeats in the central region of the protein. This now brings the number of Cdc31p-binding proteins in the half-bridge to three; Kar1p (Biggins and Rose, 1994; Spang et al., 1995), Mps3p/Nep98p (Jaspersen et al., 2002; Nishikawa et al., 2003), and Sfi1p. This paper shows that the Sfi1 repeats are also present in two previously uncharacterized proteins, one from the evolutionally distant yeast S. pombe, and another from humans. The human protein, hSfi1, binds centrin at the repeats, and all three proteins localize to the spindle pole.
Sfi1 repeats have some resemblance to WD repeats (Neer et al., 1994), but are clearly distinct because Sfi1 repeats lack the conserved aspartic acid (Garcia-Higuera et al., 1998) in WD repeats (that would be at position 16 in Fig. 5). Also, WD repeats lack the consensus leucine at position 14 (Fig. 5) of Sfi1 repeats. Both temperature-sensitive alleles of Sfi1p have mutations within the first and second repeats (see Materials and methods), and indeed, sfi1-3 has a mutation (F208C) in the consensus phenylalanine at position 19 in the first repeat. These results suggest that disabling one or two repeats may also affect the function of the rest of the repeats. Sfi1 repeats are apparently absent from the other proteins isolated in the Cdc31p pull-down (Fig. 1), and are not present in the three other proteins that directly bind to Cdc31p, Kar1p (Biggins and Rose, 1994; Spang et al., 1995), Kic1p (Sullivan et al., 1998), and Mps3p/Nep98p (Jaspersen et al., 2002; Nishikawa et al., 2003). The Cdc31p-binding site in Kar1p has been identified (Biggins and Rose, 1994; Spang et al., 1995), but appears different from Sfi1 repeats. This heterogeneity in binding site motifs is very similar to the large variations between calmodulin-binding sites (Hoeflich and Ikura, 2002).
The most analogous calmodulin-binding domain to the centrin-binding Sfi1 repeats might be the IQ domains in the neck region of unconventional myosins (Cheney and Mooseker, 1992). Like the Sfi1 repeats, the myosin IQ domains show Ca2+-independent binding and are present in multiple continuous copies. They are spaced at a slightly shorter distance apart (23–26 residues), though other multiple IQ domains are spaced more widely, for example, 28–32 residues in the IQGAP-related protein Iqg1p (Terrak et al., 2003). Myosin IQ domains are thought to be present in a helical conformation between the NH2- and COOH-terminal lobes of calmodulin (Houdusse et al., 1996); this model is based on the structure of scallop myosin with two calmodulin-like light chains bound (Xie et al., 1994; Houdusse and Cohen, 1996). The spacing of 23–26 residues between adjacent IQ domains is compatible with direct connections between the calmodulin molecules, which can explain some of the Ca2+-dependent regulation of the activity of unconventional myosins. Here, addition of Ca2+ could cause conformational changes in the already bound calmodulin. This may regulate the function of the motor domain because the first bound calmodulin may be directly in contact with that domain (Houdusse et al., 1996).
The Sfi1 repeat regions of the proteins described here generally lack prolines, and thus might adopt a helical conformation like the IQ repeats. The wider spacing between adjacent Sfi1 repeats (usually between 23 and 33 residues) may also be compatible with direct connections between the centrins, given the characteristic larger NH2-terminal domains of centrins compared with calmodulin (Bhattacharya et al., 1993). This type of structure may have some relevance to other EF-hand domain or centrin-containing organelles such as the spasmoneme (Amos et al., 1975) and striated flagellar roots (Salisbury et al., 1984), which rapidly contract on addition of Ca2+. Spasmin or centrin are the main protein components of these organelles, so it was originally thought that they themselves might form the 2–7-nm filaments present in these organelles (Amos, 1975; Schiebel and Bornens, 1995). This now seems unlikely, given the homology of these proteins to EF-hand domain proteins or calmodulin. However, if a protein with multiple Sfi1 repeats were present in these organelles, then it could form a template for what would effectively be a short filament of spasmin or centrin. These filaments could then be made longer by end-to-end or staggered side-to-side interactions. Such a protein would be difficult to identify because it would be faintly stained in Coomassie-stained SDS gels of the organelle, due to the high molar ratio of spasmin or centrin to the Sfi1p-like protein. The association of at least spasmin with such a protein is likely to be Ca2+-independent because spasmonemes are glycerinated in 4 mM EGTA (Amos, 1971). Addition of Ca2+ to spasmin or centrin already bound to an Sfi1p-like protein could cause conformational changes, in the same way that addition of Ca2+ to calmodulin already bound to a K+ channel opens the gate (Schumacher et al., 2001). In the case of centrin bound to an Sfi1p-like protein, similar conformational changes could lead to a rapid shortening of a filament. A simple analogy might be the concertina part of a child's flexible drinking straw. The extended form would be when the concertinas were stretched out, the conformational change would be equivalent to folding individual concertinas, thus leading to shortening.
What would the role of Sfi1p be in SPB duplication? Clearly, if Sfi1p does form filaments, these might be important structural elements of the half-bridge. In addition, they may play a role in the proposed extension and retraction of the half-bridge during SPB insertion (Adams and Kilmartin, 1999). At present, there is no evidence for Ca2+ pulses at this point in the cell cycle, which might be necessary to expand and contract an Sfi1p-Cdc31p filament. However, an additional feature of centrin-binding filaments, at least in the case of the spasmoneme (Weis-Fogh and Amos, 1972), is elasticity, which is not dependent on the Ca2+ concentration. Elastic proteins generally have multiple repeats (Tatham and Shewry, 2000), and in the well-studied example of titin, the stretching occurs by unfolding of tandem Ig-like repeats (Li et al., 2000). Possibly, Sfi1p-Cdc31p might be stretched during duplication plaque assembly by unfolding of Sfi1 repeats, with possible concomitant dissociation of some of the Cdc31p. After completion of assembly, tension would be released in some way and retraction would be driven by reassociation of Cdc31p.
Clearly, the multiple binding of centrin to the Sfi1 repeats is an interesting structural problem, and using some of the recombinant proteins described here, should be amenable to crystallographic investigation. In addition, using longer versions of these recombinant proteins, it should be possible to test any elastic properties or Ca2+-dependent contraction.
Materials and methods
S. cerevisiae strains
All S. cerevisiae strains were prepared in K699 (W303a) background or the isogenic diploid K842 (Nasmyth et al., 1990), and yeast vectors used were the pRS series (Sikorski and Hieter, 1989). The cdc31-1 allele was backcrossed to K699 five times, and the sequence change in this allele, A48T (Vallen et al., 1994), was verified by PCR sequencing.
The construct to make ZCdc31p was prepared by first placing a NcoI site by PCR at the initiator methionine of CDC31; this would cause the change S2G, and did not appear to affect the function of the protein because this construct was able to rescue a deletion. A single Z domain of prA was amplified from pEZZ18 (GenBank/EMBL/DDBJ accession no. M74186) between NcoI and BspLUIII sites and was inserted in the correct orientation into the NcoI site placed in CDC31. This replaced the initiator methionine in Cdc31p with the sequence: MGSVDNKF…AQAPKHM, a total of 63 amino acids with the sequence between VDNKF and AQAPK corresponding to the synthetic 58-amino acid Z domain (Nilsson et al., 1987). The HA-Cdc31p construct was prepared from linker oligos again using NcoI and BspLUIII sites and inserted into the sequence MGSYPYDVPDYAHM in place of the initiator methionine. Both constructs in the CEN vector pRS314 were able to complement a deletion of CDC31 with the cells showing an apparently normal growth rate.
SFI1 was cloned by Vent PCR and by gap repair. Both constructs showed the same amino acid changes compared with the previously published sequence (Ma et al., 1999); these are probably due to strain differences and were: L96P, F278L, D541G, I623V, and G935E.. Temperature-sensitive mutants were prepared by slight modifications of the gap repair procedure (Muhlrad et al., 1992). For sfi1-3, the primers for the mutagenic PCR were 97–119 and 2820–2800, and the plasmid was gapped between AgeI (242) and StyI (2676) sites; for sfi1-7, the primers were 540–561 and 2820–2800, and the plasmid was gapped between MscI (748) and the same StyI site. All base pair numbers start from the A of the codon of the presumed initiator methionine. The PCR conditions were those for colony PCR (Akada et al., 2000) with 7.5 mM MgCl2, 0.2 mM dATP and dGTP, and 1 mM dCTP and dTTP. Transformants were replica plated onto fluoroorotic acid (FOA) medium (Boeke et al., 1984) to remove the wild-type URA3 plasmid, then onto warm (37°C) and ambient YEPD plates. Temperature-sensitive colonies were selected, and the plasmid was recovered. Plasmids conferring a temperature-sensitive phenotype were transferred to the URA3 plasmid pRS306 and transformed into K699 to replace the wild-type gene with the temperature-sensitive allele by the popin-popout procedure (Boeke et al., 1987). Ura+ transformants were patched onto FOA and replica plated onto warm and ambient YEPD plates to select temperature-sensitive colonies. Both the popin parental strain and the popout temperature-sensitive alleles were checked by PCR to confirm the presence of the plasmid at the wild-type locus and its removal after FOA treatment. Finally, the temperature-sensitive mutant strains were transformed with SFI1 to show rescue and backcrossed to show the presence of a single temperature-sensitive mutation. The amino acid changes found in the plasmid used to make sfi1-3 were H207Q, F208C and Y247C, and for sfi1-7, N218D and D242A.
Isolation of complexes and mass spectrometry
Complexes were isolated as before (Wigge and Kilmartin, 2001) from 10 l of yeast cells harvested at 5 × 107 cells/ml, with most of the sample applied to a single SDS gel lane. Proteins in SDS gel bands (Fig. 1) were identified by mass spectrometry (Wigge and Kilmartin, 2001), except that the entire National Center for Biotechnology Information (NCBI) database was searched using the Mascot search engine (Matrix Science, Ltd.), between 40–60 ppm, allowing for methionine oxidation and up one missed tryptic cleavage. All identifications were the top match, and the numbers of peptides identified and the sequence percentage covered by these peptides were as follows: in the Cdc31p pull-down gel (Fig. 1 a), Vps13p: 26, 11%; Sfi1p (top band): 29, 33%; Sfi1p (bottom band): 19, 21%; band 1, Ssa1p: 8, 18%; Ssa2p: 10, 24% and Ssb2p: 8, 22%; Thp1p: 12, 34%; band 2, Ydj1p: 9, 29%; Hem15p: 13, 32% and ZCdc31p: 9, 52%. In the Sfi1p-prA pulldown, the bands identified were: Sfi1p-prA: 28, 29%; band 3, Rpn1p: 24, 25%; Spc110p: 17, 18%; band 4, Rpn2p: 13, 15%; band 5, Sse1p: 8, 15%; band 6, Ssa2p: 13, 29% and Ssb2p: 13, 31%; band 7, Hsp60p: 25, 55%; band 8, Rvb1p: 12, 35% and Rvb2p: 10, 27%; band 9, Ydj1p: 17, 40% and Tef2p: 12, 32%; band 10, Scj1p: 18, 45%; band 11, 32 peptides from human keratin type I and II; Cdc31p: 10, 61%; band 12, Rps18ap: 9, 50%; and band 13, Rpl23ap: 10, 42%. Rpn1p and Rpn2p are proteosome components; Ssa1p, Ssa2p, Ssb2p, Ydj1p, Sse1p, Hsp60p, and Scj1p are heat shock proteins; Rvb1p and Rvb2p are components of a chromatin remodeling complex; Tef2p is translation elongation factor and Rps18ap and Rpl23ap are ribosome subunits. Further information on these proteins can be found at http://db.yeastgenome.org. All of these proteins are probably nonspecific because they have been found in pull-downs with functionally unrelated prA fusions.
Identification of other proteins with multiple Sfi1 repeats
A BLASTP search with S. cerevisiae Sfi1p was not successful, so the Candida albicans homologue (listed at http://genolist.pasteur.fr/CandidaDB; gene name IPF5574) was used instead. A BLASTP search with this Candida homologue identified S. cerevisiae Sfi1p (E value 2e−14), an S. pombe protein (GenBank accession no. T40750; E value 1e−4) called here SpSfi1, and a human protein of 1212 amino acids KIAA0542 (BAA25468; E value 0.096) called here hSfi1. All of these proteins have an unusual repeat structure (Fig. 5). The hSfi1 (KIAA0542) clone was sequenced and found actually to correspond to a splicing variant (T00322) containing 968 amino acids (apart from the substitution H322Y). An Image EST clone (Image 5273112, BI463640) was sequenced and found to contain a full-length insert of the database sequence of hSfi1 with an additional insert of YFCFRALKDNVTHAHLQQIRRNLAHQQHGVT between amino acids 385 and 386, and the substitutions L407I and P1056L. This protein has 1242 amino acids. Apart from the above 31 amino acid insert and the substitution I438L, both versions of hSfi1 are identical, with the 968 amino acid form having an earlier stop codon. Both of these ORFs could be followed continuously over a 120-kb region of human chromosome 22. Mice also have homologues (AAH46305 and AAH26390) that terminate at positions equivalent to 968 and 1242.
Other human proteins that might contain Sfi1 repeats were identified by ScanProsite (Gattiker et al., 2002) using the sequence AX12[F/L]X2W[K/R/H] and calling for at least three hits in the ORF. Out of >800,000 entries, this identified only Sfi1p, it's S. pombe, Aspergillus, mouse, and human homologues, together with the complete sequence of one human protein, AAM34297 (five repeats between residues 757–879) and the partial sequence of another, CAD38978 (10 repeats between residues 152–423), however CAD38978 is quite tryptophan rich in parts of this region. Further searching with [F/W/L]X2W[K/R/H]X21–34[F/W/L]X2W[K/R/H] and calling for two hits in the ORF identified human BAC05092 (four repeats between residues 25–120). Logos were obtained from Jan Gorodkin (The Royal Veterinary and Agricultural University, Frederiksberg, Denmark; Gorodkin et al., 1997).
Tagging of S. cerevisiae Sfi1p, S. pombe Sfi1, and human Sfi1 with GFP and antibody staining
S. cerevisiae Sfi1p was tagged at the COOH terminus by recombinant PCR (Wach et al., 1997; Wigge et al., 1998). Cells were fixed for 2 min (Kilmartin et al., 1993), and were stained with rabbit anti-GFP and pooled anti-90-kD mAbs (Rout and Kilmartin, 1990). The anti-GFP staining was very sensitive to formaldehyde fixation in both S. cerevisiae and human cells.
S. pombe Sfi1 was tagged at the COOH terminus with GFP using recombinant PCR (Bähler et al., 1998; Wigge and Kilmartin, 2001), and cells were fixed and stained as before (Wigge and Kilmartin, 2001).
Human Sfi1 was inserted into peGFP-C1 (U55763) by PCR. HeLa cells were transfected with FuGENE™ 6 (Roche). After 48 h, cells were fixed with methanol/acetone at −20°C, stained with affinity-purified rabbit anti-GFP, mouse mAbs GTU-88 against γ-tubulin (Sigma-Aldrich), αCetn2 against centrin 2 (Hart et al., 2001), and GT335 against glutamylated tubulin (Bobinnec et al., 1998). Secondary antibodies were Alexa® 488 anti–rabbit IgG and Alexa® 594 anti–mouse IgG (Molecular Probes, Inc.).
Microscopy
Light microscopy was done with either a microscope (Axioskop; Carl Zeiss MicroImaging, Inc.) with Plan Neofluar 100× NA 1.3 or Plan Apochromat 63× NA 1.4 objectives and a MicroMAX CCD camera (RTE/CCD-1300-Y; Princeton Instruments), or for Fig. 7 (c–h) only, with a confocal microscope (Radiance 2100; Bio-Rad Laboratories) on a microscope (Axiophot; Carl Ziess MicroImaging, Inc.) with the 63× Apochromat objective above. The CCD camera used IP Labs software, and for faint images (Fig. 2, a and c; Fig. 6 a), interference lines were removed by application of the IP Labs linear filter to the whole image; the confocal microscope used Lasersharp 2000 software. The whole of the images were then adjusted for brightness and contrast in Adobe Photoshop®. EM and immuno-EM were performed as before (Adams and Kilmartin, 1999).
GST pull-downs
Centrins were obtained as Image clones: human centrin 1 (Image 5163552), human centrin 2 (Image 3836808), human centrin 3 (Image 4657309), and mouse centrin 4 (Image 3657877). Centrins and Cdc31p were expressed as GST fusions (Pulvermuller et al., 2002) with a single HA tag at the NH2 terminus as for HA-Cdc31p (see above). All bacterial manipulations were at 25°C and induction was for 4 h at 25°C; GST was removed by thrombin digestion. GST fusions of Sfi1p were all insoluble when expressed under the same conditions, but remained soluble when the method of Frangioni and Neel (1993) was followed. Fusions of Sfi1p and hSfi1 shown in Fig. 7 were in pGEX-3X (GenBank/EMBL/DDBJ accession no. U13852), and were designed to express the amino acids shown in brackets, for Sfi1p: fusion 1 (1–187), fusion 2 (181–386), fusion 3 (379–552), fusion 4 (551–803), fusion 5 (796–947), fusion 2a (181–250), fusion 2b (246–281), fusion 2c (279–386), fusion 3a (451–474), fusion 3b (469–507), fusion 4a (643–677), fusion 4b (676–710), and fusion 4c (739–811); for hSfi1: fusion 6 (205–309), fusion 7 (332–470), fusion 8 (464–612), fusion 9 (613–805), and fusion 10 (1100–1242). Pull-downs were in a total volume of 40 μl binding buffer (50 mM Tris-Cl, pH 7.4, 0.15 M NaCl, and 0.05% Tween 20 with either 1 mM CaCl2 or 10 mM EGTA) with 3 μl packed beads and a centrin concentration between 1 and 100 μM. For the saturation of fusion 2 (Fig. 8 c), the Cdc31p concentration was between 10 and 100 μM. Beads were rotated for 3–4 h at RT, washed four or five times with 0.5 ml buffer, and equivalent amounts of supernatant and beads were boiled in SDS sample buffer. SDS gels were scanned with a densitometer (Personal Densitometer SI; Molecular Dynamics) and processed with ImageQuant™ 1.2 software (Amersham Biosciences). Bands in the free GST position were ignored when scanning the gels because these failed to react with Cdc31p in overlays.
Acknowledgments
I am particularly grateful to Sew Yeu Peak-Chew and Farida Begum for the mass spectrometry, Jeffrey Salisbury (The Mayo Foundation, Rochester, MN) for αCetn2, Bernard Eddé (CNRS, Montpellier, France) for GT335, Iain Hagan (Paterson Institute for Cancer Research, Manchester, UK) for anti-Sad1, Keith Gull (University of Oxford, Oxford, UK) for TAT1, and to the late Douglas Kershaw for cutting the serial thin sections. Thanks are also due to M.S. Robinson for discussion.
Notes
Abbreviations used in this paper: prA, protein A; SPB, spindle pole body; ZCdc31p, Cdc31p containing a single Z domain of protein A at the NH2 terminus.
References
- Adams, I.R., and J.V. Kilmartin. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809–823. [Europe PMC free article] [Abstract] [Google Scholar]
- Akada, R., T. Murakane, and Y. Nishizawa. 2000. DNA extraction method for screening yeast clones by PCR. Biotechniques. 28:668–674. [Abstract] [Google Scholar]
- Amos, W.B. 1971. Reversible mechanochemical cycle in the contraction of Vorticella. Nature. 229:127–128. [Abstract] [Google Scholar]
- Amos, W.B. 1975. Contraction and calcium binding in the vorticellid ciliates. Soc. Gen. Physiol. Ser. 30:411–436. [Abstract] [Google Scholar]
- Amos, W.B., L.M. Routledge, and F.F. Yew. 1975. Calcium-binding proteins in a Vorticellid contractile organelle. J. Cell Sci. 19:203–213. [Abstract] [Google Scholar]
- Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Ohkuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276:18665–18672. [Abstract] [Google Scholar]
- Bähler, J., J.-Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie, III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [Abstract] [Google Scholar]
- Bhattacharya, D., J. Steinkotter, and M. Melkonian. 1993. Molecular cloning and evolutionary analysis of the calcium-modulated contractile protein, centrin, in green algae and land plants. Plant Mol. Biol. 23:1243–1254. [Abstract] [Google Scholar]
- Biggins, S., and M.D. Rose. 1994. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125:843–852. [Europe PMC free article] [Abstract] [Google Scholar]
- Bobinnec, Y., M. Moudjou, J.P. Fouquet, E. Desbruyeres, B. Edde, and M. Bornens. 1998. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton. 39:223–232. [Abstract] [Google Scholar]
- Boeke, J.D., F. Lacroute, and G.R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346. [Abstract] [Google Scholar]
- Boeke, J.D., J. Trueheart, G. Natsoulis, and G.R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164–175. [Abstract] [Google Scholar]
- Brickner, J.H., and R.S. Fuller. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36. [Europe PMC free article] [Abstract] [Google Scholar]
- Byers, B. 1981. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. Molecular Genetics in Yeast. D. von Wettstein, J. Friis, M. Kielland-Brand, and A. Stenderup, editors. Alfred Benzon Symposium 16, Munksgaard, Copenhagen. 119–131.
- Byers, B., and L. Goetsch. 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 38:123–131. [Abstract] [Google Scholar]
- Cheney, R.E., and M.S. Mooseker. 1992. Unconventional myosins. Curr. Opin. Cell Biol. 4:27–35. [Abstract] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B.A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 301:71–76. [Abstract] [Google Scholar]
- Donaldson, A.D., and J.V. Kilmartin. 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 132:887–901. [Europe PMC free article] [Abstract] [Google Scholar]
- Elliott, S., M. Knop, G. Schlenstedt, and E. Schiebel. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA. 96:6205–6210. [Europe PMC free article] [Abstract] [Google Scholar]
- Errabolu, R., M.A. Sanders, and J.L. Salisbury. 1994. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 107:9–16. [Abstract] [Google Scholar]
- Frangioni, J.V., and B.G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179–187. [Abstract] [Google Scholar]
- Gallardo, M., and A. Aguilera. 2001. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 157:79–89. [Europe PMC free article] [Abstract] [Google Scholar]
- Garcia-Higuera, I., C. Gaitatzes, T.F. Smith, and E.J. Neer. 1998. Folding a WD repeat propeller. Role of highly conserved aspartic acid residues in the G protein beta subunit and Sec13. J. Biol. Chem. 273:9041–9049. [Abstract] [Google Scholar]
- Gattiker, A., E. Gasteiger, and A. Bairoch. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 1:107–108. [Abstract] [Google Scholar]
- Gavet, O., C. Alvarez, P. Gaspar, and M. Bornens. 2003. Centrin4p, a novel mammalian centrin specifically expressed in ciliated cells. Mol. Biol. Cell. 14:1818–1834. [Europe PMC free article] [Abstract] [Google Scholar]
- Geier, B.M., H. Wiech, and E. Schiebel. 1996. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 271:28366–28374. [Abstract] [Google Scholar]
- Gorodkin, J., L.J. Heyer, S.S. Brunak, and G.D. Stormo. 1997. Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci. 13:583–586. [Abstract] [Google Scholar]
- Grandi, P., V. Doye, and E.C. Hurt. 1993. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 12:3061–3071. [Europe PMC free article] [Abstract] [Google Scholar]
- Hagan, I., and M. Yanagida. 1995. The product of the spindle formation gene sad1 + associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047. [Europe PMC free article] [Abstract] [Google Scholar]
- Hart, P.E., G.M. Poynter, C.M. Whitehead, J.D. Orth, J.N. Glantz, R.C. Busby, S.L. Barrett, and J.L. Salisbury. 2001. Characterization of the X-linked murine centrin Cetn2 gene. Gene. 264:205–213. [Abstract] [Google Scholar]
- Hoeflich, K.P., and M. Ikura. 2002. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 108:739–742. [Abstract] [Google Scholar]
- Houdusse, A., and C. Cohen. 1996. Structure of the regulatory domain of scallop myosin at 2 Å resolution: implications for regulation. Structure. 4:21–32. [Abstract] [Google Scholar]
- Houdusse, A., M. Silver, and C. Cohen. 1996. A model of Ca2+-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure. 4:1475–1490. [Abstract] [Google Scholar]
- Jaspersen, S.L., T.H. Giddings, Jr., and M. Winey. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. [Europe PMC free article] [Abstract] [Google Scholar]
- Kilmartin, J.V., S.L. Dyos, D. Kershaw, and J.T. Finch. 1993. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J. Cell Biol. 123:1175–1184. [Europe PMC free article] [Abstract] [Google Scholar]
- Klotz, C., N. Garreau de Loubresse, F. Ruiz, and J. Beisson. 1997. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil. Cytoskeleton. 38:172–186. [Abstract] [Google Scholar]
- Knop, M., and E. Schiebel. 1997. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16:6985–6995. [Europe PMC free article] [Abstract] [Google Scholar]
- Labbe-Bois, R. 1990. The ferrochelatase from Saccharomyces cerevisiae. Sequence, disruption, and expression of its structural gene HEM15. J. Biol. Chem. 265:7278–7283. [Abstract] [Google Scholar]
- Lee, V.D., and B. Huang. 1993. Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA. 90:11039–11043. [Europe PMC free article] [Abstract] [Google Scholar]
- Levy, Y.Y., E.Y. Lai, S.P. Remillard, M.B. Heintzelman, and C. Fulton. 1996. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil. Cytoskeleton. 33:298–323. [Abstract] [Google Scholar]
- Li, H., A.F. Oberhauser, S.B. Fowler, J. Clarke, and J.M. Fernandez. 2000. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA. 97:6527–6531. [Europe PMC free article] [Abstract] [Google Scholar]
- Ma, P., J. Winderickx, D. Nauwelaers, F. Dumortier, A. De Doncker, J.M. Thevelein, and P. Van Dijck. 1999. Deletion of SFI1, a novel suppressor of partial Ras-cAMP pathway deficiency in the yeast Saccharomyces cerevisiae, causes G2 arrest. Yeast. 15:1097–1109. [Abstract] [Google Scholar]
- Maciejewski, J.J., E.J. Vacchiano, S.M. McCutcheon, and H.E. Buhse. 1999. Cloning and expression of a cDNA encoding a Vorticella convallaria spasmin: an EF-hand calcium-binding protein. J. Eukaryot. Microbiol. 46:165–173. [Abstract] [Google Scholar]
- Middendorp, S., A. Paoletti, E. Schiebel, and M. Bornens. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 94:9141–9146. [Europe PMC free article] [Abstract] [Google Scholar]
- Middendorp, S., T. Kuntziger, Y. Abraham, S. Holmes, N. Bordes, M. Paintrand, A. Paoletti, and M. Bornens. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148:405–416. [Europe PMC free article] [Abstract] [Google Scholar]
- Moriyama, Y., H. Okamoto, and H. Asai. 1999. Rubber-like elasticity and volume changes in the isolated spasmoneme of giant Zoothamnium sp. under Ca2+-induced contraction. Biophys. J. 76:993–1000. [Europe PMC free article] [Abstract] [Google Scholar]
- Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast. 8:79–82. [Abstract] [Google Scholar]
- Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 62:631–647. [Abstract] [Google Scholar]
- Neer, E.J., C.J. Schmidt, R. Nambudripad, and T.F. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature. 371:297–300. [Abstract] [Google Scholar]
- Nilsson, B., T. Moks, B. Jansson, L. Abrahmsen, A. Elmblad, E. Holmgren, C. Henrichson, T.A. Jones, and M. Uhlen. 1987. A synthetic IgG-binding domain based on Staphylococcal protein A. Protein Eng. 1:107–113. [Abstract] [Google Scholar]
- Nishikawa, S., Y. Terazawa, T. Nakayama, A. Hirata, T. Makio, and T. Endo. 2003. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 278:9938–9943. [Abstract] [Google Scholar]
- O'Toole, E.T., M. Winey, and J.R. McIntosh. 1999. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2017–2031. [Europe PMC free article] [Abstract] [Google Scholar]
- Pulvermuller, A., A. Giessl, M. Heck, R. Wottrich, A. Schmitt, O.P. Ernst, H.W. Choe, K.P. Hofmann, and U. Wolfrum. 2002. Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol. Cell. Biol. 22:2194–2203. [Europe PMC free article] [Abstract] [Google Scholar]
- Rout, M.P., and J.V. Kilmartin. 1990. Components of the yeast spindle and spindle pole body. J. Cell Biol. 111:1913–1927. [Europe PMC free article] [Abstract] [Google Scholar]
- Salisbury, J.L., A. Baron, B. Surek, and M. Melkonian. 1984. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 99:962–970. [Europe PMC free article] [Abstract] [Google Scholar]
- Salisbury, J., K. Suino, R. Busby, and M. Springett. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287–1292. [Abstract] [Google Scholar]
- Schiebel, E., and M. Bornens. 1995. In search of a function for centrins. Trends Cell Biol. 5:197–201. [Abstract] [Google Scholar]
- Schumacher, M.A., A.F. Rivard, H.P. Bachinger, and J.P. Adelman. 2001. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 410:1120–1124. [Abstract] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [Europe PMC free article] [Abstract] [Google Scholar]
- Spang, A., I. Courtney, U. Fackler, M. Matzner, and E. Schiebel. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123:405–416. [Europe PMC free article] [Abstract] [Google Scholar]
- Spang, A., I. Courtney, K. Grein, M. Matzner, and E. Schiebel. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128:863–877. [Europe PMC free article] [Abstract] [Google Scholar]
- Sullivan, D.S., S. Biggins, and M.D. Rose. 1998. The yeast centrin, Cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143:751–765. [Europe PMC free article] [Abstract] [Google Scholar]
- Tatham, A.S., and P.R. Shewry. 2000. Elastomeric proteins: biological roles, structures and mechanisms. Trends Biochem. Sci. 25:567–571. [Abstract] [Google Scholar]
- Terrak, M., G. Wu, W.F. Stafford, R.C. Lu, and R. Dominguez. 2003. Two distinct myosin light chain structures are induced by specific variations within the bound IQ motifs-functional implications. EMBO J. 22:362–371. [Europe PMC free article] [Abstract] [Google Scholar]
- Vallen, E.A., W. Ho, M. Winey, and M.D. Rose. 1994. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics. 137:407–422. [Europe PMC free article] [Abstract] [Google Scholar]
- Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 13:1065–1075. [Abstract] [Google Scholar]
- Weis-Fogh, T., and W.B. Amos. 1972. Evidence for new mechanisms of cell motility. Nature. 236:301–304. [Abstract] [Google Scholar]
- Wigge, P.A., O.N. Jensen, S. Holmes, S. Souès, M. Mann, and J.V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977. [Europe PMC free article] [Abstract] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [Europe PMC free article] [Abstract] [Google Scholar]
- Winey, M., L. Goetsch, P. Baum, and B. Byers. 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114:745–754. [Europe PMC free article] [Abstract] [Google Scholar]
- Wolff, A., B. de Nechaud, D. Chillet, H. Mazarguil, E. Desbruyeres, S. Audebert, B. Edde, F. Gros, and P. Denoulet. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432. [Abstract] [Google Scholar]
- Xie, X., D.H. Harrison, I. Schlichting, R.M. Sweet, V.N. Kalabokis, A.G. Szent-Gyorgyi, and C. Cohen. 1994. Structure of the regulatory domain of scallop myosin at 2.8 Å resolution. Nature. 368:306–312. [Abstract] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.200307064
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jcb/article-pdf/162/7/1211/1310447/jcb16271211.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102379923
Article citations
The core spindle pole body scaffold Ppc89 links the pericentrin orthologue Pcp1 to the fission yeast spindle pole body via an evolutionarily conserved interface.
Mol Biol Cell, 35(8):ar112, 10 Jul 2024
Cited by: 0 articles | PMID: 38985524 | PMCID: PMC11321043
The relationship between intraflagellar transport and upstream protein trafficking pathways and macrocyclic lactone resistance in Caenorhabditis elegans.
G3 (Bethesda), 14(3):jkae009, 01 Mar 2024
Cited by: 0 articles | PMID: 38227795 | PMCID: PMC10917524
The Chlamydia trachomatis type III-secreted effector protein CteG induces centrosome amplification through interactions with centrin-2.
Proc Natl Acad Sci U S A, 120(20):e2303487120, 08 May 2023
Cited by: 9 articles | PMID: 37155906 | PMCID: PMC10193975
An Sfi1-like centrin-interacting centriolar plaque protein affects nuclear microtubule homeostasis.
PLoS Pathog, 19(5):e1011325, 02 May 2023
Cited by: 4 articles | PMID: 37130129 | PMCID: PMC10180636
Giant proteins in a giant cell: Molecular basis of ultrafast Ca2+-dependent cell contraction.
Sci Adv, 9(8):eadd6550, 22 Feb 2023
Cited by: 5 articles | PMID: 36812318 | PMCID: PMC9946354
Go to all (129) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 6 of 6)
- (2 citations) ENA - CAD38978
- (1 citation) ENA - M74186
- (1 citation) ENA - BAA25468
- (1 citation) ENA - U13852
- (1 citation) ENA - T00322
- (1 citation) ENA - AAM34297
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J Cell Biol, 173(6):867-877, 01 Jun 2006
Cited by: 86 articles | PMID: 16785321 | PMCID: PMC2063913
Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p.
J Cell Biol, 159(6):945-956, 16 Dec 2002
Cited by: 115 articles | PMID: 12486115 | PMCID: PMC2173979
Duplication of the Yeast Spindle Pole Body Once per Cell Cycle.
Mol Cell Biol, 36(9):1324-1331, 15 Apr 2016
Cited by: 25 articles | PMID: 26951196 | PMCID: PMC4836218
Review Free full text in Europe PMC
Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function.
Mol Biol Cell, 18(6):2047-2056, 28 Mar 2007
Cited by: 25 articles | PMID: 17392514 | PMCID: PMC1877113




