Abstract
Free full text

Slit-2 Induces a Tumor-suppressive Effect by Regulating β-Catenin in Breast Cancer Cells*
Abstract
SLIT-2 is considered as a candidate tumor suppressor gene, because it is frequently inactivated in various cancers due to hypermethylation of its promoter region and allelic loss. However, the exact mechanism of its tumor-suppressive effect has not been elucidated. Here, we observed that Slit-2-overexpressing breast cancer cells exhibited decreased proliferation and migration capabilities compared with control cells under in vitro conditions. These results were confirmed in vivo in mouse model systems. Mice injected with MCF-7/Slit-2 cells showed a 60–70% reduction in tumor size compared with mice injected with MCF-7/VC cells both in the absence and presence of estrogen. Upon further elucidation, we observed that Slit-2 mediates the tumor-suppressive effect via a coordinated regulation of the β-catenin and PI3K signaling pathways and by enhancing β-catenin/E-cadherin-mediated cell-cell adhesion. Our study for the first time reveals that Slit-2-overexpressing breast cancer cells exhibit tumor suppressor capabilities through the novel mechanism of β-catenin modulation.
Slit-2, a protein that belongs to the Slit family of large extracellular matrix-secreted glycoproteins, has been shown to exhibit tumor-suppressive effects in various human cancers (1–7). However, the exact mechanism of its tumor suppressor ability is not well characterized. SLIT-2 is located in chromosome 4p15.2 and encodes the human orthologue of the Drosophila Slit-2 protein (8). Slit consists of a family of 3 genes (SLIT-1, SLIT-2, and SLIT-3), and these genes are candidate ligands for the repulsive guidance receptors, members of the ROBO gene family (1, 9–12). Slit-2 has four leucinerich repeats at its N-terminal end, followed by nine epidermal growth factor (EGF)3 repeats, a laminin G domain, and a cysteine-rich C-terminal region (13, 14). Slit-2 interacts with its receptor, Roundabout-1 (Robo-1), through its leucinerich repeat domain (13). Initially, the role of the Slit/Robo complex was described in the nervous system, where it regulated axon guidance, branching, and neural migration (9, 15–18). Recently, the importance of the Slit/Robo pathway was reported during the embryonic development of various organs such as the lung, kidney, and heart and systems such as the CNS (1, 19–23). Slit-2 expression has also been detected in normal skin, as well as in epithelial and endothelial cells (12, 20, 24–27). These studies suggest that Slit/Robo may be involved in the regulation of organogenesis.
SLIT-2 promoter region hypermethylation has been detected in various cancers such as breast cancer, non-small cell lung cancer, small cell lung cancer, colorectal carcinoma, and gliomas (2–7, 28). It has been also shown that, in various cancer types, Robo expression is also altered (19, 29). This indicates that Slit/Robo signaling may play important roles in cancer development. Mutation studies and expression analysis of the SLIT-2 gene have revealed loss of heterozygosity, a couple of missense substitutions in one of the EGF-like domains, and rearrangement at its genomic locus (4p15.2) in a significant proportion of human cancers (3–6). Moreover, primary breast tumors and a majority of breast tumor cell lines have been reported to exhibit reduced or absent SLIT-2 expression (3). Furthermore, analysis of the Slit-2 promoter region in these tumor cells showed the presence of extensive hypermethylation of the SLIT-2 5′-cytosine-guanine (CpG) island (3). Lack or reduced expression of SLIT-2 was correlated with CpG hypermethylation, and further treatment with the demethylating agent 5′-azacytidine restored SLIT-2 expression (3). In addition, Slit-2-overexpressing breast cancer cells or Slit-2-treated breast cancer cells showed decreased colony formation (3).
Although these studies indicate that SLIT-2 can act as a tumor suppressor gene, its tumor-suppressive effect under in vivo conditions and the exact mechanisms of its anti-tumor property are not yet known. One of the pathways through which Slit-2 mediates its function may be by modulating β-catenin/wnt signaling, because the Slit family of proteins were identified as conserved targets of the β-catenin/wnt signaling pathway and interaction between Slit/Robo signaling and wnt signaling was observed during ureteric bud development (30, 31). β-Catenin is a critical regulatory molecule of the wnt signaling pathway and also plays an important role in cadherin-based cell-cell adhesion by indirectly linking cadherins to the actin cytoskeleton (32, 33). In the cytoplasm, serine and threonine phosphorylation regulated the stability of β-catenin by targeting it to GSK-3β/Axin/adenomatous polyposis coli complex-mediated proteasomal degradation or its translocation to the nucleus. In the nucleus, β-catenin interacts with members of the LEF/TCF family of transcriptional activators (34, 35). Several β-catenin/TCF target genes (such as matrix metalloproteases, cytoskeletal proteins, and angiogenic cytokines) have been implicated in cancer progression (36–41). Thus, β-catenin is considered to be an oncogene, and its deregulation or mutational activation can lead to cancer (42–44). Furthermore, up-regulation of β-catenin has been shown in various cancer tissues, including breast cancer (42).
Here, we report for the first time that the overexpression of Slit-2 in breast cancer cells inhibits breast cancer growth in two in vivo mouse model systems, both in the absence and presence of estrogen. We have also shown that Slit-2 induces its tumor suppressor activity by a coordinated regulation of the β-catenin/TCF and PI3K/Akt signaling pathways as well as by enhancing β-catenin/E-cadherin-mediated cell-cell adhesion.
EXPERIMENTAL PROCEDURES
Cell Lines—The Slit-2-overexpressing MCF-7 (MCF-7/Slit-2) stable cell line and vector control cell line (MCF-7/VC) were provided by Dr. Farida Latif (University of Birmingham, Birmingham, UK). These cells were maintained in DMEM containing 10% fetal bovine serum, 50 μg/ml Zeocin, and 1% penicillin-streptomycin at 37 °C in 5% CO2 (3). MDA-MB-231 cells were generously provided by Hava Avraham (Beth Israel Deaconess Medical Center, Boston) and maintained in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C in 5% CO2.
Microarray Analysis—Total RNA was collected from both MCF-7/Slit-2 and MCF-7/VC cell lines by using TRIzol reagent (Invitrogen), per the manufacturer's protocol. Microarray analysis was performed at the Beth Israel Deaconess Medical Center genomics core facility by using an Affymetrix Microarray gene chip containing 40,000 human genes.
Proliferation Assay—Cells (5 × 103), seeded on 96-well plates, were cultured in DMEM supplemented with 0.2% fetal bovine serum and 50 ng/ml of EGF. The number of viable cells was quantified at various time points by using the CellTiter 96®Aqueous kit (Promega, Madison, WI), as per the manufacturer's instructions.
Chemotaxis Assay—A chemotaxis assay was performed in 24-well cell culture chambers with 8-μm pore inserts, as described previously (45). Membranes were pre-coated with fibronectin (2.5 μg/ml). Breast cancer cells were suspended in chemotaxis buffer (DMEM/0.1% bovine serum albumin/12 mm HEPES) at 2.5 × 106 cells/ml. 150 μl of cell suspension was loaded onto the upper chamber. 0.6 ml of medium containing CXCL12 (5 nm) was then loaded onto the lower chamber. The plates were incubated for 6 h at 37 °C in 5% CO2. After incubation, the inserts were removed carefully and cells were fixed, stained, and counted using standard procedures.
Soft-agar Colony Formation Assay—Cells (2 × 104) suspended in medium containing 0.4% Ultra pure agarose (Invitrogen) were layered on top of 1 ml of medium containing 0.8% agarose in 6-well culture plates. Cells were incubated with 1 ml of DMEM supplemented with 10% fetal bovine serum. After 2 weeks of incubation, the colonies were stained with 0.005% Crystal Violet for 1 h, and colonies were counted by using a Nikon Diaphot 300 inverted microscope.
Immunoprecipitation and Western Blot Analysis—Immunoprecipitation and Western blot analysis were done as described elsewhere (45). According to the particular experiment, equal amounts of protein (1 mg) were bound with specific antibodies. Then, the immune complexes were precipitated with Protein A-Sepharose beads (Amersham Biosciences) and were Western blotted with the relevant antibodies.
Extractions of Nuclear and Cytoplasmic Fractions—Nuclear and cytoplasmic fractions were collected by using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Pierce), as per the manufacturer's protocol. The purity of fractionation was determined with Oct-1 antibody.
Small Interfering RNA (siRNA)-mediated Knockdown—siRNA-mediated knockdown of β-catenin and Robo-1 was performed using β-catenin SMART pool siRNA reagents and ON-TARGETplus SMARTpool Robo-1 siRNA reagents (Dharmacon, Inc., Boulder, CO) respectively, according to the manufacturer's protocol. Briefly, MCF-7 cells were electroporated with 100 nm siRNA using the Amaxa Nucleofector™ device (Amaxa Biosystems, Cologne, Germany), as per the manufacturer's protocol. The respective non-targeted siRNA SMARTpool was used as a control. β-Catenin siRNA-mediated knockdown was estimated by detecting β-catenin expression 48 h after the initial transfection by using Western blot analysis. Robo-1 siRNA-mediated knockdown was estimated by detecting Robo-1 receptor expression 48 h after the initial transfection by using flow cytometry.
Constructs and Transfections—ORF Clone of Homo sapiens slit homolog 2 (Drosophila) (SLIT2) was purchased from OriGene Technologies Inc. (Rockville, MD) and subcloned into pcDNA 3.1/V5-His Vector (Invitrogen). 2 μg of pcDNA 3.1/V5-His-SLIT-2 plasmid was mixed with the cellular suspensions, transferred to a 2.0-mm electroporation cuvette, and nucleofected using the Amaxa Nucleofector™ device (Amaxa Biosystems) as per manufacturer's protocol. Transfection efficiency was monitored by Western blotting procedures by using anti-V5 antibody (Invitrogen).
Confocal Microscopy—MCF-7/VC and MCF-7/Slit-2 cells were cultured in chamber slides. Then slides were fixed in 4% paraformaldehyde for 15 min at room temperature. The cells were washed thrice with PBS, blocked with 5% normal goat serum in PBS/Triton for 60 min, and treated with rabbit anti-β-catenin antibody (Cell Signaling Technology, Beverly, MA), and monoclonal anti-E-cadherin antibody (Chemicon, Temecula, CA) overnight at 4 °C. The slides were washed thrice with PBS and stained with Alexa Fluor 568-labeled anti-rabbit IgG antibodies, Alexa Fluor 568-labeled anti-mouse IgG antibody (Molecular Probes, Eugene, OR), or fluorescein isothiocyanate-labeled anti-rabbit IgG antibody. The cells were washed thrice in PBS, and slides were mounted by using Prolong Gold Antifade with DAPI (Invitrogen). The cells were then examined under a Zeiss confocal microscope, and the images were acquired by using LSM510 software. Nuclear staining was quantitated by using Volocity Software at the Beth Israel Deaconess Medical Center Microscopy Core Facility.
Luciferase Reporter Assay—To determine luciferase reporter activity, TCF luciferase constructs (0.5 μg), containing the wild-type (pTOPFLASH) or mutant (pFOPFLASH; Upstate, Charlottesville, VA) TCF binding sites, were transfected into MCF-7/VC and MCF-7/Slit-2 cells (5 × 105 per well). Transfection experiments were carried out in triplicate using Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. In addition, the cells were cotransfected with an internal control (0.1 μg of pRL-TK Renilla luciferase vector, Promega, Madison, WI). The cells were incubated for 48 h after the transfection and then treated with EGF (100 ng/ml) for 24 h. The cells were washed with PBS and lysed in 1× passive lysis buffer (Dual Luciferase kit, Promega). To measure the activities of firefly and Renilla luciferase, 20-μl aliquots of the supernatant were transferred into a 96-well plate and assayed in a Wallac Victor2 counter luminometer (PerkinElmer Life Sciences) using reagents from the Dual Luciferase kit (Promega). The firefly (TOPFLASH or FOPFLASH) luciferase activity was corrected for Renilla luciferase activity (pRL-TK) to control for transfection efficiency. TOPFLASH activity was also normalized to the FOPFLASH activity. Data are expressed as the mean of triplicate values of the normalized TOPFLASH activity.
In Vivo Tumor Models—Female CB17/SCID and nude (nu/nu) mice, obtained from Charles River Laboratories (Wilmington, MA), were housed under specific pathogenfree conditions. The in vivo experiments were performed in accordance with the guidelines of our Institutional Animal Care and Use Committee. Cells from each stable cell line were concentrated to 3 × 106 per 200 μl and injected subcutaneously into the flank of each mouse. One group of mice was injected subcutaneously with 2.5 μg of β-estradiol 17-valerate in 50 μl of Sesame oil twice a week. Tumor size was assessed once a week, and tumor volume was calculated. At the end of the 4th week for the SCID mice and 6th week for nude mice, the animals were scanned under a Micro CT scanner (GE eXplore Locus Micro CT), and the final tumor volume was assessed. Tumors were excised, minced homogenized in modified RIPA buffer (150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1% Triton X-100, and a mixture protease inhibitors, Roche Applied Science), and clarified by centrifugation at 13,000 × g for 15 min at 4 °C. Supernatants were subjected to Western blot analysis.
RESULTS
In this study, to analyze the tumor suppressor property of Slit-2 gene, we have used myc-tagged human SLIT-2 plasmid stably expressing MCF-7 breast cancer cells. Initially, we confirmed Slit-2 overexpression in two clones of Slit-2-overexpressing MCF-7 cells. Both cell lysates (Fig. 1A, upper panel) showed the overexpression of Slit-2-myc. Clone 2 exhibited more Slit-2-myc expression than clone 1. Because Slit-2 mediates its function by binding to specific receptor roundabout-1 (Robo-1), we also analyzed the expression of this receptor in control and Slit-2-overexpressing clones of MCF 7 and observed that MCF-7 cells and Slit-2-overexpressing MCF-7 cells express moderate level of Robo-1 receptor (Fig. 1A, middle panels). Because Slit-2 is a secretory protein we also confirmed its expression in cell supernatant concentrates of both clones (Fig. 1A, lower panel).
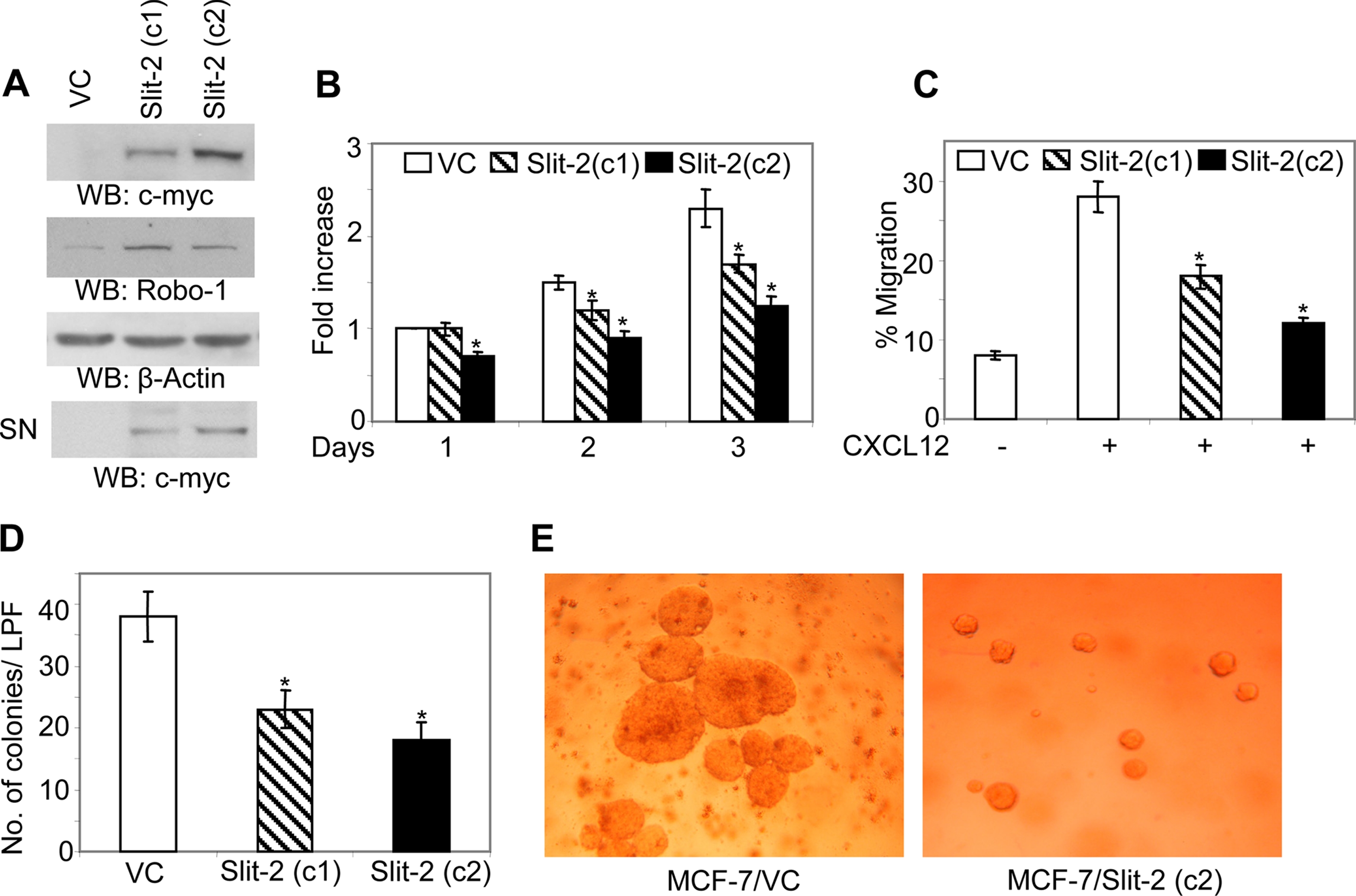
Slit-2-overexpressing MCF-7 cells show decreased proliferation and chemotactic capability. Slit-2 expression (A, upper panel) and Robo-1 expression (A, second panel) in MCF-7/VC and MCF-7/Slit-2 clones were analyzed by Western blotting using anti-c-myc and anti-Robo-1 antibodies. Equal protein was confirmed in each sample by stripping and re-probing the blot with anti-β-Actin antibody (A, third panel). Slit-2-myc expression also analyzed in cell culture supernatant concentrates. Cell culture supernatants were concentrated by using Amicon Ultra-100K filters (A, lower panel). B, cells were subjected to a proliferation assay after seeding in 96-well plates in DMEM supplemented with 0.2% fetal bovine serum and 50 ng/ml EGF. Then the number of viable cells was quantified at various time points by using the CellTiter 96®Aqueous kit (Promega), as per the manufacturer's instructions. C, cells were subjected to chemotaxis assay toward CXCL12 (50 ng/ml) by using chemotaxis chambers as described under “Experimental Procedures.” D and E, for the soft agar colony formation assay, cells were mixed in medium containing 0.4% Ultra pure agarose (Invitrogen) and layered on top of 1 ml of medium containing 0.8% agarose in 6-well culture plates. Then cells were incubated with 1 ml of DMEM supplemented with 10% fetal bovine serum. After 2 weeks of incubation, the colonies were stained with 0.005% Crystal Violet for 1 h and counted by using a Nikon Diaphot 300 inverted microscope. All of the above experiments were repeated three times, and a representative one is shown. LPF = low power field. *, p < 0.05 for all experiments. VC, Vector Control-MCF-7; Slit-2 (c1), Slit-2-overexpressing MCF-7 clone 1; Slit-2 (c2), Slit-2-overexpressing MCF-7 clone 2; SN, cell supernatant concentrate.
Slit-2-overexpressing Cells Exhibit Inhibited Proliferation and Migration—Increased proliferation, migration, and adhesion are important properties of tumorigenesis and the metastasis of cancer cells (46). Dallol et al. (3) have shown that Slit-2-overexpressing breast cancer cells and breast cancer cells treated with Slit-2-conditioned medium have decreased colony-forming activity. When we studied the proliferation rate of Slit-2-overexpressing MCF-7 (MCF-7/Slit-2) breast cancer clones in the presence of EGF, we found that the MCF-7/Slit-2 cells showed significantly decreased proliferation as compared with the MCF-7 vector control (MCF-7/VC) cells (Fig. 1B). These clones also exhibited decreased chemotaxis toward CXCL12 (Fig. 1C). CXCL12 has been shown to play an important role in cancer metastasis. We also observed that the number and size of the colonies formed by the Slit-2-overexpressing cells were drastically reduced compared with the vector control-expressing cells (Fig. 1, D and E). These data support the notion that Slit-2-overexpression in MCF-7 cells significantly inhibits the proliferation of these cells. Consistent with Slit-2 expression, MCF-7/Slit-2 clone 2 exhibited more decreased proliferation and migration properties than clone 1, we have used clone 2 for our further experiments.
Further, to analyze the role of Robo-1 in Slit-2-overexpressing MCF-7 cells, we knocked down Robo-1 by using an siRNA technique and studied the Slit-2-induced effects in MCF-7/Slit-2 cells. As shown in Fig. 2A, ~55–60% knockdown of Robo-1 was observed in the MCF-7/Slit-2 (c2) cells transfected with the Robo-1 siRNA, as compared with cells transfected with the control (non-targeted) siRNA. We found significant increase in proliferation (Fig. 2B) of Robo-1siRNA-transfected MCF-7/Slit-2 cells compared with control-transfected cells. These results indicate that Slit-2 may function in an autocrine manner.
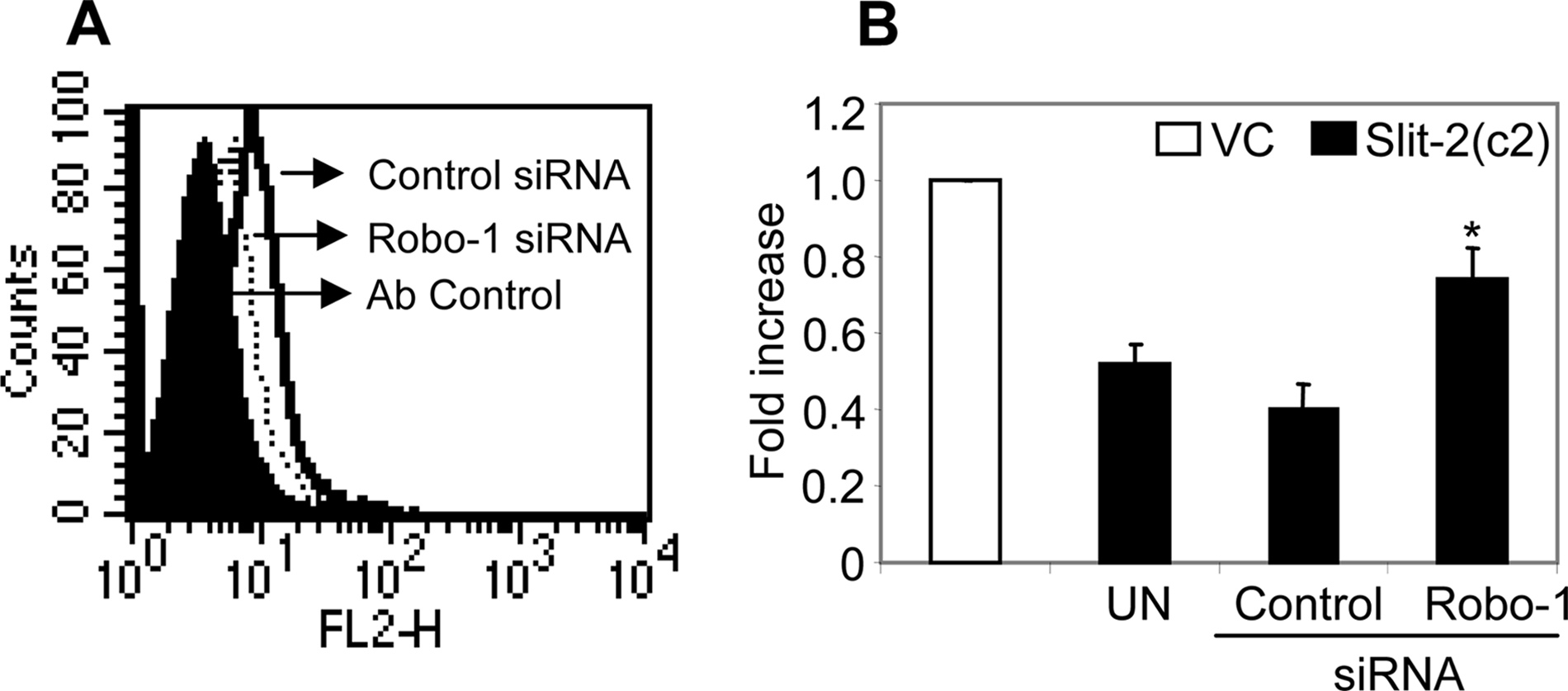
Slit-2 may induce its function through an autocrine manner. A, control siRNA (solid line) and Robo-1 siRNA-transfected (dotted line) MCF-7/Slit-2 (c2) cells were stained using anti-Robo-1 antibody and analyzed by flow cytometry. Cells stained with control IgG (filled area) represent the antibody control. E and F, control siRNA and Robo-1 siRNA-transfected MCF-7/Slit-2 (c2) cells were subjected to a proliferation assay as described under “Experimental Procedures.” The experiments were done in triplicate and are presented as the mean ± S.E. The data are representative of three different experiments. *, p < 0.05 for all experiments.
Slit-2 Overexpression Inhibits Tumor Growth in a Xenograft Model—To further evaluate the tumor-suppressive effects of Slit-2, we used a xenograft model for tumor growth. MCF-7/Slit-2 (Clone 2) and MCF-7/VC cells (3 × 106) were injected subcutaneously into the flanks of female CB17/SCID mice. After the second week of injection, tumor volume was measured every week for up to 5 weeks. A dramatic decrease in tumor size was observed in mice, which were injected with Slit-2-overexpressing cells compared with mice injected with vector control cells (Fig. 3A). At the end of the fifth week, animals were scanned under a Micro CT scanner to calculate the exact tumor volume. Tumor volume in mice injected with the MCF-7/Slit-2 cells was ~175–220 mm3 compared with mice injected with the MCF-7/VC cells, which had a tumor volume of ~650–800 mm3. Because MCF-7 cells are estrogen receptor-positive, we further examined whether Slit-2 can overcome the tumor-sustaining effects of estrogen. For this study, we injected mice with 2.5 μg of β-estradiol 17-valerate twice weekly starting from 1 week prior to cell injection. Although tumor volume in mice injected with the MCF-7/VC cells was high (~1050–1200 mm3), the tumor volume in mice injected with the MCF-7/Slit-2 cells was much lower (250–320 mm3). We confirmed these results by repeating the experiments in nude mice (nu/nu) (Fig. 3B). SCID and nude mice injected with the MCF-7/Slit-2 cells also showed a drastic decrease in tumor size even in the presence of estrogen as compared with mice injected with the control cells (MCF-7/VC) (Fig. 3, C–G).
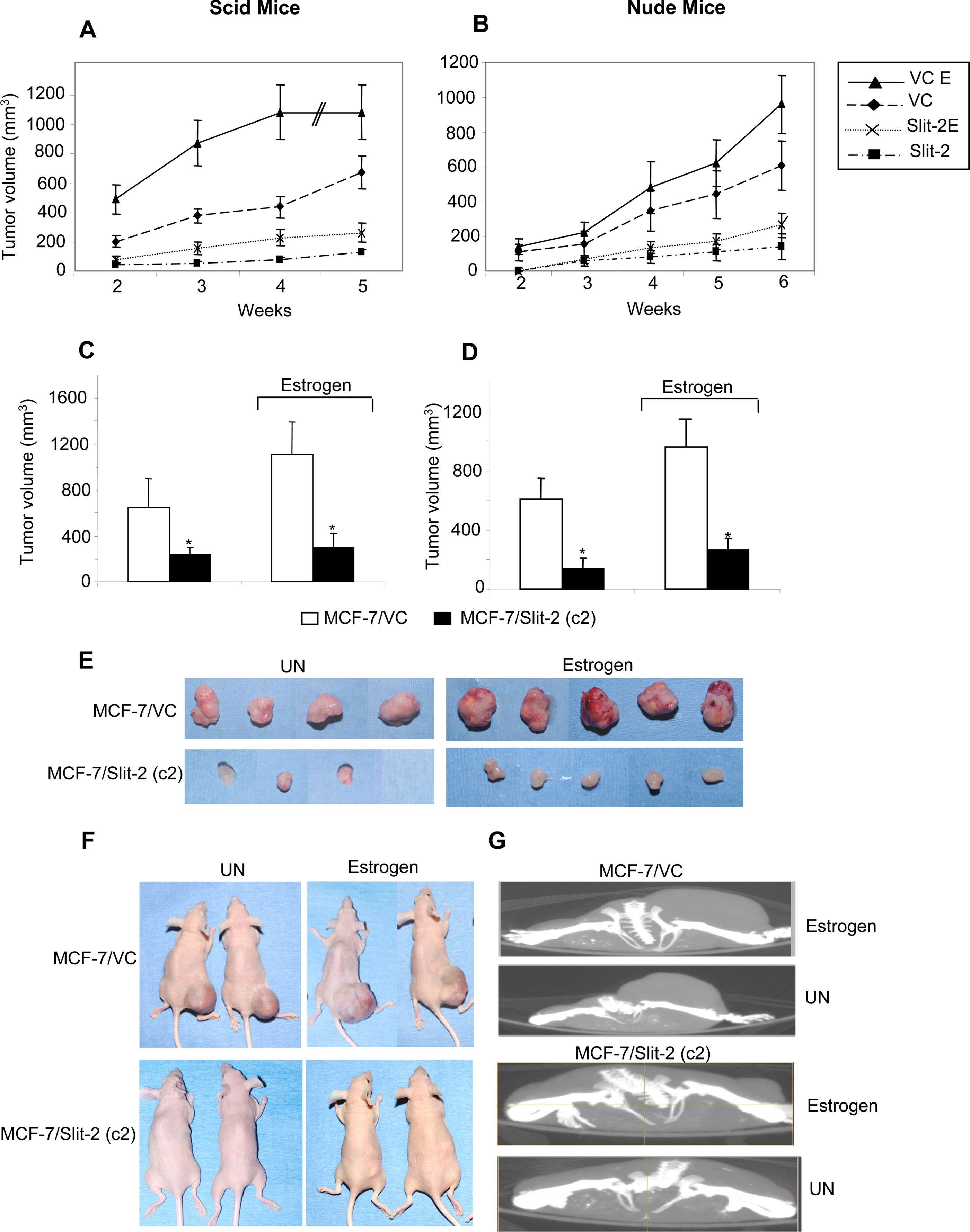
Slit-2 overexpression suppresses the tumorigenic capability of MCF-7 breast cancer cells in in vivo models. MCF-7/VC and MCF-7/Slit-2 (c2) cells were injected (3 × 106 cells) into the flanks of female SCID mice (n = 5). One group of mice was injected subcutaneously with 2.5 μg of β-estradiol 17-valerate in 50 μl of Sesame oil twice a week. Tumor size was measured by using digital calipers once a week, and volume was determined according to the formula (W2 × L)/2. A, the graph represents the tumor volume in mice injected with MCF-7/VC or MCF-7/Slit-2 (c2) cells with or without estrogen treatment. Tumor volume was assessed up until 5 weeks after injection. B, the experiments in panel A were repeated in nude mice with or without estrogen treatment. Tumor size was measured by using digital calipers once a week for 6 weeks, and volume was determined as indicated above. At the end of the fifth week in the case of SCID mice (C) and at the end of the sixth week in the case of nude mice (D), the tumor volume was analyzed and calculated by using the Micro CT scan imaging system. *, p < 0.05 for all experiments. E, photographs of the tumors derived from SCID mice 5 weeks after the injection of MCF-7/VC or MCF-7/Slit-2 9 (c2) cells in the presence or absence of estrogen. F, photographs of representative nude mice 6 weeks after the injection of MCF-7/VC or MCF-7/Slit-2 (c2) cells in the presence or absence of estrogen. G, Micro CT-scanned photographs of SCID mice 5 weeks after the injection of MCF-7/VC or MCF-7/Slit-2 (c2) cells in the presence or absence of estrogen. UN = untreated.
Microarray Analysis—To investigate the mechanisms involved in the Slit-2-mediated tumor-suppressive effect, we performed microarray analysis by using a gene chip containing 40,000 human genes. Among the various genes that are down-regulated in MCF-7/Slit-2 cells compared with control cells (MCF-7/VC), we selected the β-catenin/TCF family of transcription factors for our further investigation and then confirmed these results by using Western blot analysis.
Slit-2-overexpressing MCF-7 Cells Show Increased Degradation of β-Catenin—β-catenin/LEF/TCF transcriptional activity has been shown to regulate the expression of a subset of genes that promote tumor establishment, growth, and invasion (34–38, 47). Increased levels of β-catenin within the cytoplasm and/or in the nucleus were observed in breast cancer cells (48, 49). Further mammalian Slit family proteins have also been identified as evolutionarily conserved targets of the wnt/β-catenin signaling pathway (30). When we analyzed the expression of β-catenin in the total cell lysates of both vector control and Slit-2-overexpressing MCF-7 clones, we observed a decrease in β-catenin expression in the Slit-2-overexpressing MCF-7 clones (Fig. 4A). Because we found drastic down-regulation of β-catenin in clone 2, we used clone 2 for our further experiments. Upon further analysis of the mechanisms involved in the decreased expression of β-catenin, we observed an increase in β-catenin phosphorylation at the serine 45 site (Fig. 4B). It has been shown that β-catenin phosphorylation at serine 45 initiates the phosphorylation of other serine and threonine residues, which are necessary for the ubiquitination and proteasomal-mediated degradation of β-catenin (50). Our coimmunoprecipitation study revealed an increase in both the association of GSK-3β with β-catenin and in the ubiquitination of β-catenin in MCF-7/Slit-2 cells compared with MCF-7/VC cells (Fig. 4, C and D). These results indicate that there is increased proteasomal degradation of β-catenin in the Slit-2-overexpressing cells compared with vector control cells.
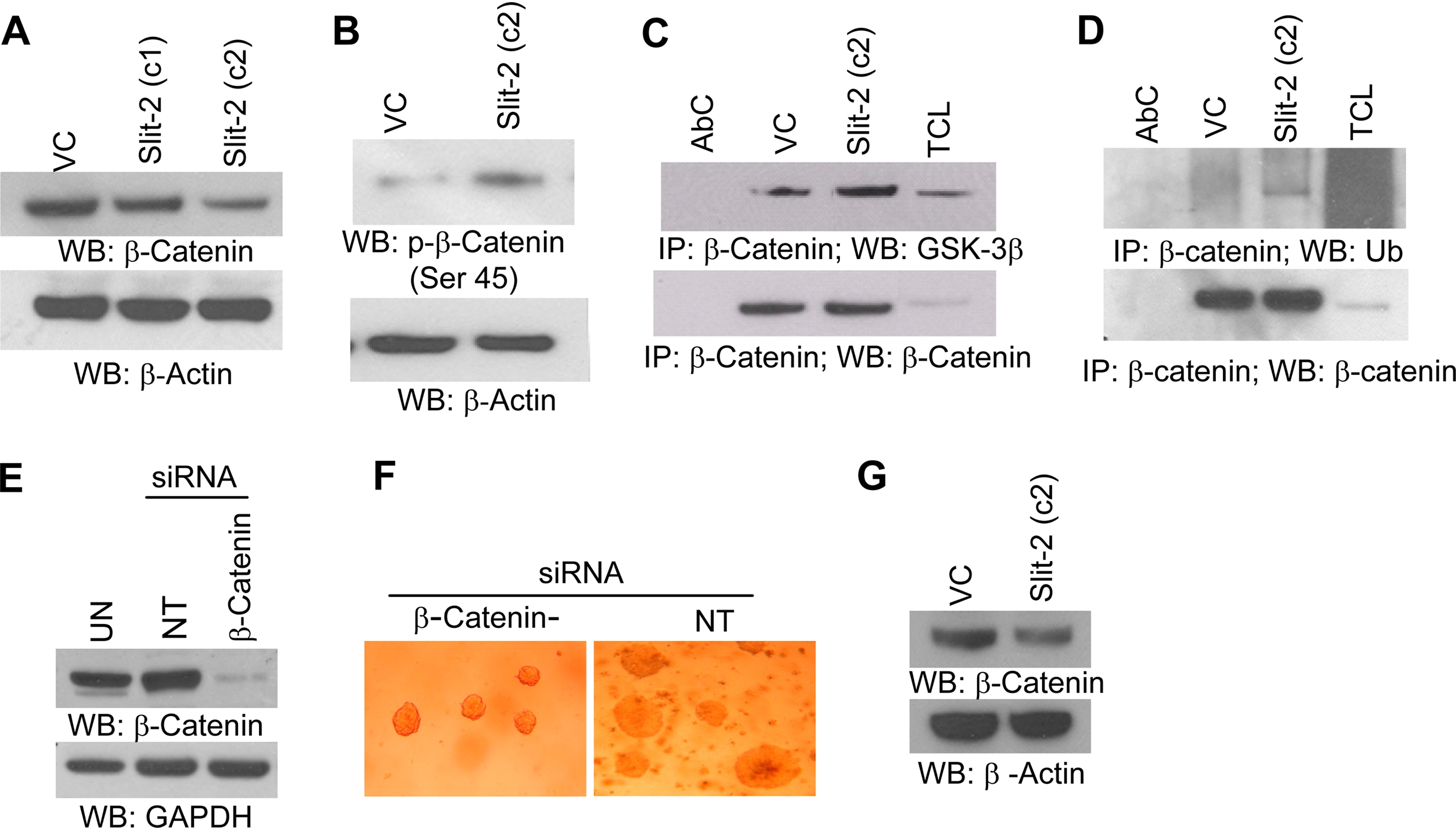
Increased β-catenin degradation was observed in MCF-7/Slit-2 cells compared with vector control (VC) cells. Both MCF-7/Slit-2 and MCF-7/VC cells were lysed and Western blotted with β-catenin (A, upper panel) or phospho-β-catenin (p-β-catenin) (serine 45) (B, upper panel) antibodies. Equal protein was confirmed in each sample by stripping and re-probing the blots with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or anti-β-actin antibodies (A and B, lower panels). The cell lysates were immunoprecipitated with anti-β-catenin antibody and Western blotted with anti-GSK-3β antibody (C, upper panel) or anti-ubiquitin (Ub) antibody (D, upper panel). Equal protein was confirmed in each sample by Western blotting with anti-β-catenin antibody (C and D, lower panels). AbC, antibody control; VC, vector control; TCL, total cell lysates. E, un-transfected (UN), control siRNA-transfected (NT), and β-catenin-siRNA-transfected MCF-7 cells lysed, and lysates were analyzed for β-catenin expression by Western blotting by using anti-β-catenin antibody. F, soft agar colony formation assay was preformed with control siRNA-transfected (NT) and β-catenin-siRNA transfected MCF-7 cells as mentioned above. G, tumors derived from MCF-7/VC and MCF-7/Slit-2 cells were excised, minced, and homogenized in modified radioimmune precipitation assay buffer. The lysates were analyzed for β-catenin expression by Western blotting by using anti-β-catenin antibody. All of the above experiments were repeated three times, and a representative one is shown.
To correlate that β-catenin down-regulation is responsible for inhibited soft agar colony formation, we knocked down β-catenin from MCF-7 cells by using an siRNA technique (Fig. 4E) and performed a soft agar colony formation assay. As shown in Fig. 4F, β-catenin knock-down MCF-7 cells exhibited decreased colony-forming ability compared with non-targeted siRNA-transfected cells. Further, we also analyzed β-catenin expression in lysates of tumor derived from MCF-7/VC- and MCF-7/Slit-2 (c2)-injected mice by Western blot analysis and observed that tumors derived from Slit-2-overexpressing cells show considerably decreased β-catenin expression as compared with tumors derived from MCF-7/VC cells (Fig. 4G). These results further confirm that down-regulation of β-catenin may be responsible for Slit-2-mediated tumor suppressor activity in breast cancer cells.
Slit-2-overexpressing MCF-7 Cells Exhibit Decreased β-Catenin Nuclear Translocation—Increased translocation of β-catenin to the nucleus leading to the enhanced expression of β-catenin target genes has been observed in many cancer cells (34–38, 47). In our study, we isolated the cytosolic fraction and nuclear fraction of MCF-7/Slit-2 and MCF-7/VC cells and checked the expression of β-catenin. We observed a decreased expression of β-catenin in both the cytosolic and nuclear fractions of MCF-7/Slit-2 cells as compared with vector control cells (Fig. 5A). Distribution analysis of β-catenin in subcellular compartments by confocal microscopy (as analyzed by using Volocity software) also revealed the decreased expression of β-catenin in both the cytosol and the nucleus of Slit-2-overexpressing cells compared with control cells (Fig. 5, B and C). However, β-catenin translocation to the membrane was increased in MCF-7/Slit-2 cells (Fig. 5B). These results confirmed the decreased nuclear translocation of β-catenin in the Slit-2-overexpressing cells. It is known that β-catenin controls E-cadherin-mediated cell adhesion at the plasma membrane and regulates the interaction of adherens junction molecules with the actin cytoskeletal system (32, 33). We also observed an increased association between β-catenin and E-cadherin in the Slit-2-overexpressing cells compared with vector control cells (Fig. 5D). However, there was no change observed in the association of E-cadherin with α-catenin (data not shown). The confocal microscopic analysis also revealed that there was an increased localization of E-cadherin in cell borders at sites of cell-cell interaction (Fig. 5E) and enhanced association between β-catenin and E-cadherin (Fig. 5F) in the MCF-7/Slit-2 cells compared with the MCF-7/VC cells. These data also clearly indicates overexpression of Slit-2 might enhance cell-cell adhesion by regulating β-catenin and E-cadherin in MCF-7 breast cancer cells.
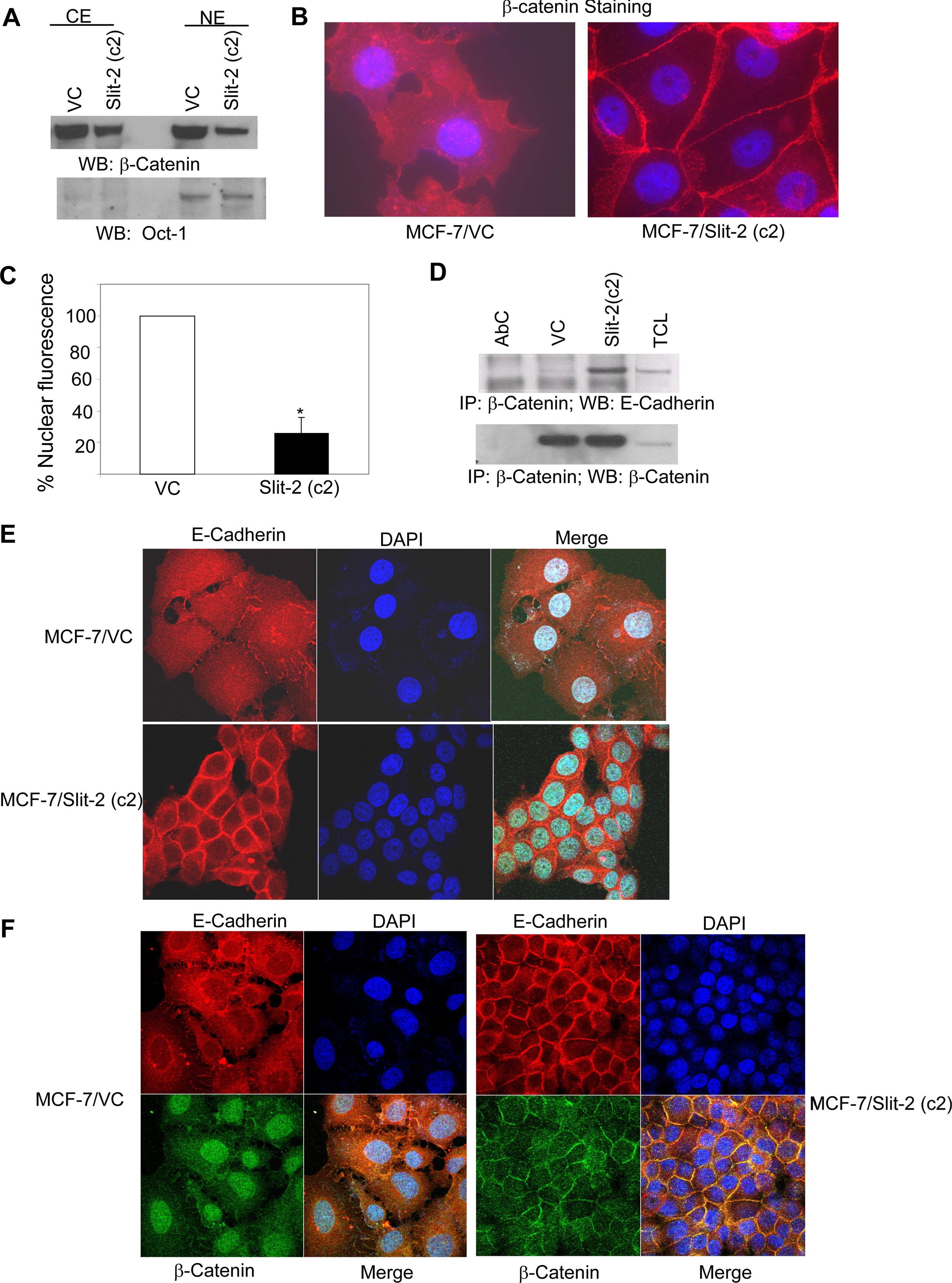
Slit-2-overexpressing cells show decreased nuclear translocation of β-catenin as well as increased expression of E-cadherin, enhanced intercellular adhesions, and association between β-catenin and E-cadherin. Nuclear extracts (NE) and cytoplasmic extracts (CE) were collected from both MCF-7/VC and MCF-7/Slit-2 cells by using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology) as per the manufacturer's protocol. The extracts were subjected to Western blotting using anti-β-catenin antibody (A, upper panel). The purity of fractionation and equal loading of protein in each lane were determined with Oct-1 antibody (A, lower panel). B, MCF-7/VC and MCF-7/Slit-2 cells were cultured in chamber slides. Cells were fixed and treated with rabbit anti-β-catenin antibody. After washing, cells were probed with Alexa 568-tagged anti-rabbit IgG antibody, and the slides were mounted by using Prolong Gold Antifade with DAPI (Invitrogen), and then examined under a Zeiss confocal microscope. The pictures were acquired by using LSM 510 software. C, nuclear fluorescent staining was quantitated by using Volocity software at the Beth Israel Deaconess Medical Center Microscopy Core Facility. The percentage of nuclear fluorescent staining was calculated by considering the fluorescence of the MCF-7/VC cells as 100%. Both MCF-7/Slit-2 and MCF-7/VC cells were lysed, and the cell lysates were immunoprecipitated with anti-β-catenin antibody and Western blotted with anti-E-cadherin antibody (D, upper panel). Equal protein was confirmed in each sample by stripping and re-probing the blot with anti-β-catenin antibody (D, lower panel). All of the above experiments were repeated three times, and a representative one is shown. *, p < 0.05 for all experiments. E and F, MCF-7/VC and MCF-7/Slit-2 cells were cultured in chamber slides. For panel E, cells were fixed and treated with mouse monoclonal anti-E-cadherin antibody. After washing, cells were probed with Alexa 568-tagged anti-mouse IgG secondary antibody. For panel F, cells were treated with mouse monoclonal anti-E-cadherin antibody and rabbit anti-β-catenin antibody, then cells were washed and probed with Alexa 568-tagged anti-mouse IgG and fluorescein isothiocyanate-labeled anti-rabbit IgG secondary antibodies. For both panels E and F, the slides were mounted by using Prolong Gold Antifade with DAPI (Invitrogen) and examined under a Zeiss confocal microscope. The pictures were acquired by using LSM510 software. All of the above experiments were repeated three times, and a representative one is shown.
Slit-2 Overexpression Inhibits β-Catenin Transcriptional Activity—Stabilized, hypophosphorylated β-catenin translocates to the nucleus where it interacts with transcription factors of the TCF/LEF-1 family, leading to the increased expression of genes, such as Cyclin D1, MMPs, and c-myc (34, 35, 39). Moreover, these genes also play an important role in tumor development and progression (40, 41). Because our initial microarray analysis revealed that the Slit-2-overexpressing MCF-7 cells exhibit decreased expression of TCF, we further investigated the transcriptional activity of β-catenin/TCF transcriptional factors by using a TCF reporter/LEF reporter assay (TOPFLASH). As shown in Fig. 6A, in the Slit-2-overexpressing cells, TOPFLASH activity was reduced by 47% compared with the vector control cells. It has been shown that EGF stimulation enhances β-catenin/TCF transcriptional activity in various cell types. Although we observed a significant increase in TOPFLASH activity in both the Slit-2-overexpressing and vector control cells upon EGF stimulation, the increase in activity in the MCF-7/Slit-2 cells was more modest as compared with the vector control cells.
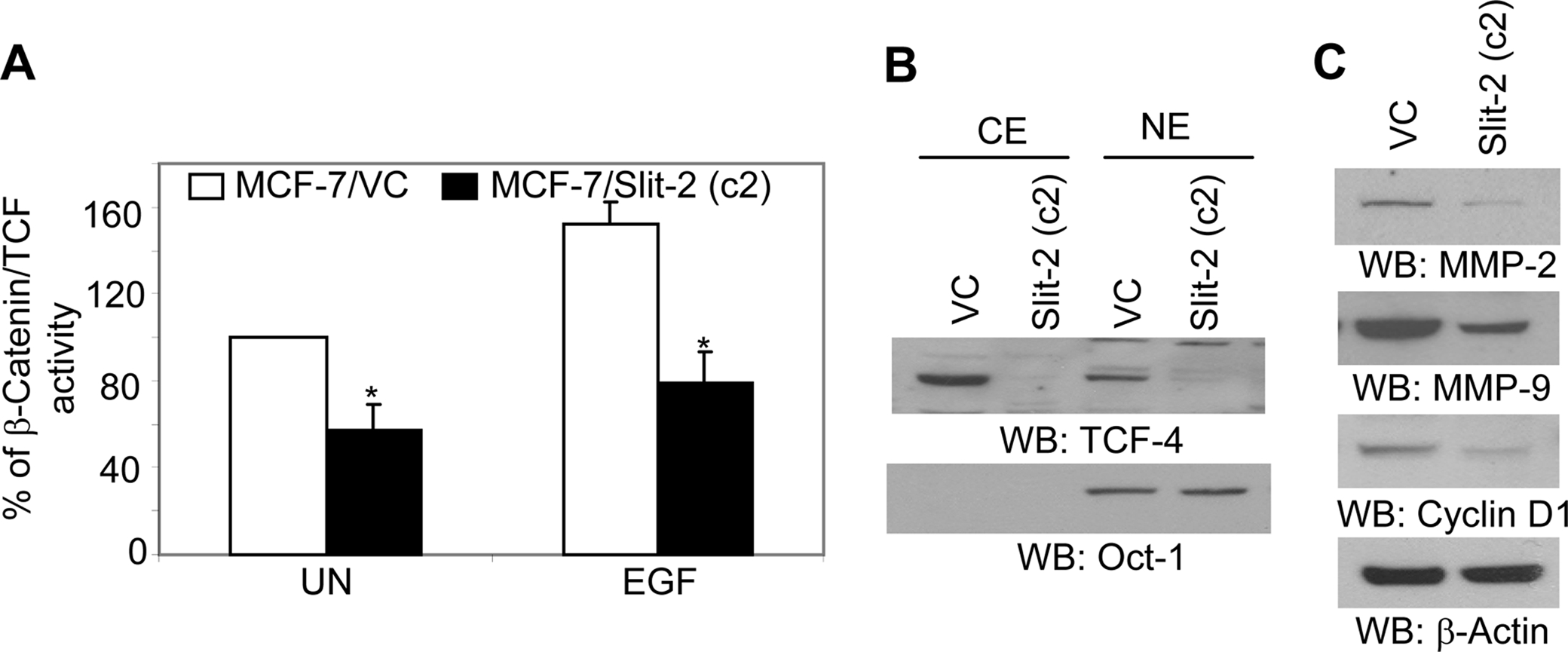
Slit-2-overexpressing cells exhibit decreased β-catenin/TCF transcriptional activity. A, TCF luciferase constructs (0.5 μg), containing the wild-type (pTOPFLASH) or mutant (pFOPFLASH, Upstate, Charlottesville, VA) TCF binding sites, were transfected into MCF-7/VC and MCF-7/Slit-2 cells (5 × 105 per well). Transfection experiments were carried out in triplicate using Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. The cells cotransfected with 0.1 μg of pRL-TK Renilla luciferase vector (Promega) were taken as an internal control. The cells were incubated for 48 h following the transfection and then untreated (UN) or treated with EGF (100 ng/ml) for 24 h. Cells were lysed and the activities of firefly and Renilla luciferase were assayed and measured in a Wallac Victor2 luminometer (PerkinElmer Life Sciences) using reagents from the Dual Luciferase kit (Promega), as mentioned under “Experimental Procedures.” The firefly (TOPFLASH or FOPFLASH) luciferase activity was corrected for Renilla luciferase activity (pRL-TK) to control for transfection efficiency. TOPFLASH activity was also normalized to the FOPFLASH activity. Data are expressed as the mean of triplicate values of the normalized TOPFLASH activity. Control, untreated value is taken as 100%, and other sample values were compared accordingly. All of the above experiments were repeated three times, and a representative one is shown. B, nuclear extracts (NE) and cytoplasmic extracts (CE) were collected from both MCF-7/VC and MCF-7/Slit-2 cells by using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology) as per the manufacturer's protocol. The extracts were subjected to Western blotting using anti-TCF-4 antibody (B, upper panel). The purity of fractionation and equal loading of protein in each lane was determined with Oct-1 antibody (B, lower panel). Both MCF-7/Slit-2 and MCF-7/VC cells were lysed, and the cell lysates were Western blotted with anti-MMP-2 (C, upper panel), anti-MMP-9 (C, second panel), and anti-cyclin D1 (C, third panel) antibodies. Equal protein was confirmed in each sample by stripping and re-probing the blot with anti-β-actin antibody (C, lower panel).
TCF-4, a member of the TCF transcription factor family, has been shown to be involved in cancer progression (51). We further analyzed the activity of TCF-4 and observed that its expression was significantly inhibited in the MCF-7/Slit-2 cells as compared with vector control cells (Fig. 6B). Moreover, we also analyzed the expression of different β-catenin/TCF target genes and found that MMP-2, MMP-9, and Cyclin D1 were inhibited in the MCF-7/Slit-2 cells compared with vector control cells (Fig. 6C).
In addition, we also transiently transfected Slit-2 gene in MDA-MB-231 cells (Fig. 7A) and analyzed for Slit-2-induced tumor-suppressive effects in these cells. We observed that Slit-2-expressing MDA-MB-231 cells also show decreased proliferation compared with vector control-transfected cells (Fig. 7B). These results confirm that Slit-2-induced tumor-suppressive effects are may not be due to clonal variation or heterogeneity in cancer cell lines. We further confirmed some of signaling results in Slit-2 transiently overexpressing MDA-MB-231 cells. Slit-2-transfected MDA-MB-231 cells showed decreased expression of β-catenin and cyclin D1 (Fig. 7C) and increased β-catenin/E-cadherin association (Fig. 7D) as compared with vector control-transfected cells.
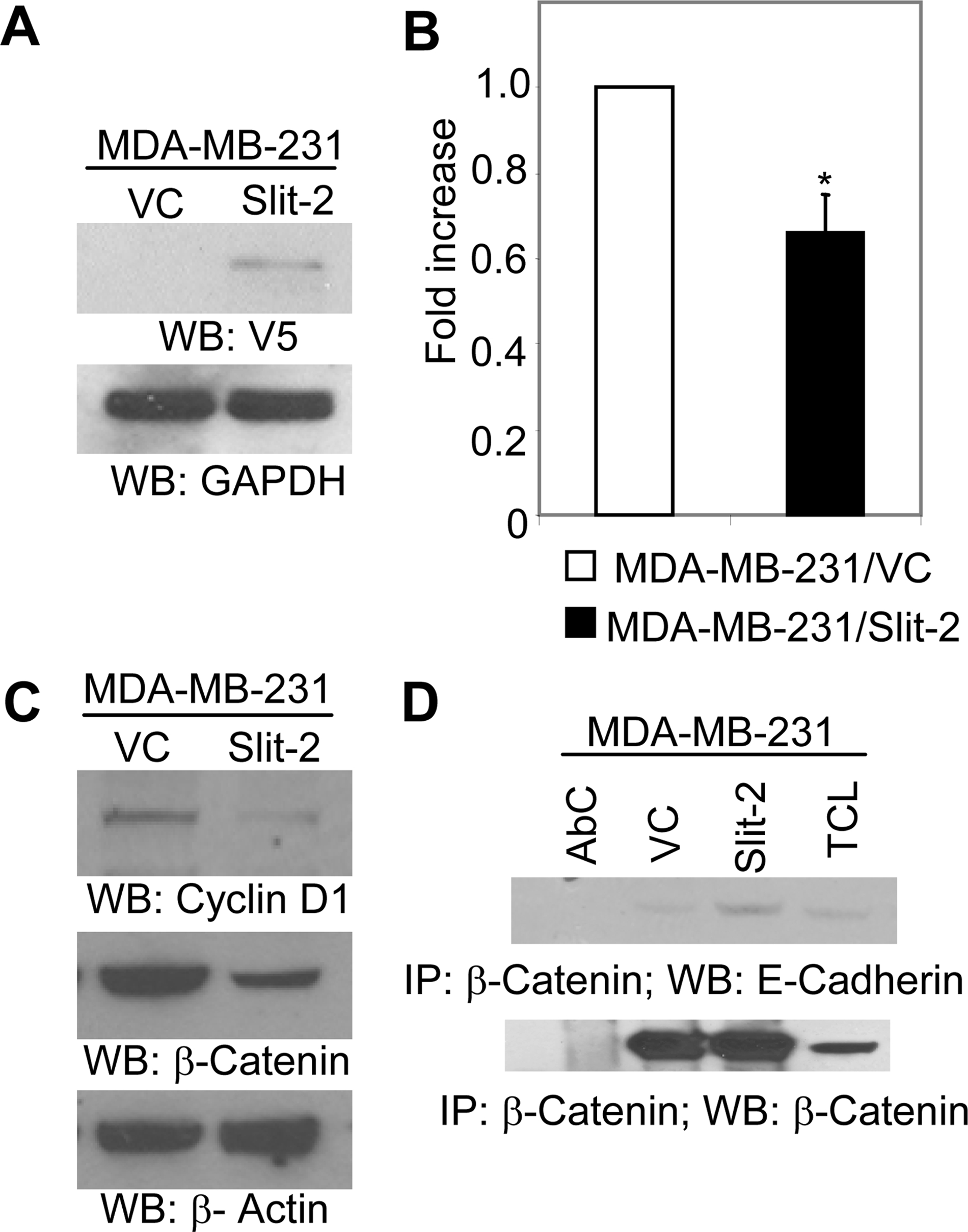
Slit-2 transiently transfected MDA-MB-231 cells show decreased proliferation, β-catenin, and cyclin D1 expression and increased β-catenin/E-cadherin association. pcDNA 3.1/V5-His-Slit-2 plasmid and vector control plasmids were transiently transfected to MDA-MB-231 cells as mentioned under “Experimental Procedures.” Cells were lysed and analyzed for Slit-2-V5 expression by Western blotting using anti-V5 antibody (A) or subjected to proliferation assay by using the CellTiter 96®Aqueous kit (Promega), as per the manufacturer's instructions (B). C, cells were lysed, and the cell lysates were Western blotted with anti-β-catenin antibody or anti-cyclin D1 antibody or (D) lysates were immunoprecipitated with anti-β-catenin antibody and Western blotted with anti-E-cadherin antibody (D, upper panel). Equal protein was confirmed in each sample by stripping and re-probing the blot with anti-β-catenin antibody or anti-β-actin antibody (C and D, lower panels). All of the above experiments were repeated three times, and a representative one is shown. *, p < 0.05 for all experiments.
Slit-2 Overexpression Also Inhibits Akt and GSK-3β Phosphorylation in Breast Cancer Cells—The coordination of β-catenin pathways and PI3K pathways has been reported in breast and lung cancer cells. Furthermore, the PI3K/Akt pathway also plays an important role in the stabilization of β-catenin by regulating GSK-3β activity (52, 53). In our previous study, we observed that exogenous Slit-2 stimulation inhibits PI3K activity (45). In the present study, we analyzed EGF-induced Akt phosphorylation and observed that the phosphorylation of Akt was significantly inhibited in the Slit-2-overexpressing cells compared with vector control cells (Fig. 8A). We also analyzed the phosphorylation of GSK-3β, which is a substrate of Akt, and found that its phosphorylation was decreased in the MCF-7/Slit-2 cells (Fig. 8B). Hence, it appears that, in Slit-2-overexpressing cells, the decreased activity of Akt results in the decreased phosphorylation of GSK-3β. Interestingly, the dephosphorylated form of GSK-3β has been shown to possess enhanced β-catenin phosphorylating activity (54).
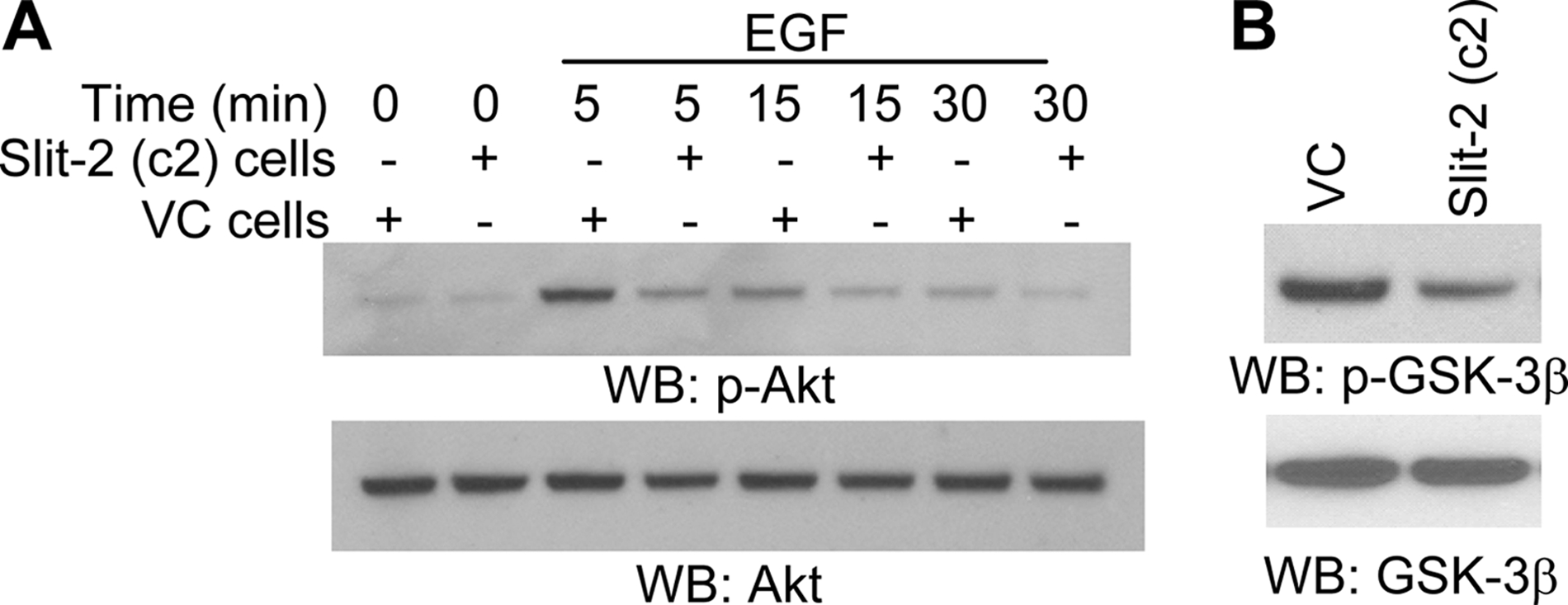
Slit-2-overexpressing cells show decreased phosphorylation of Akt and GSK-3β. MCF-7/Slit-2 and MCF-7/VC cells were lysed, and the cell lysates were Western blotted with anti-phospho-Akt (p-Akt) (A, upper panel) or anti-phospho-GSK-3β (p-GSK-3β) (B, upper panel) antibodies. In panel A, the MCF-7/Slit-2 (+) and MCF-7/VC (-) cells were untreated or treated with EGF (100 ng/ml) for various time points, as indicated. Equal protein was confirmed in each sample by stripping and re-probing the blots with anti-Akt antibody or anti-GSK-3β antibody (A and B, lower panels).
DISCUSSION
The Slit family of large extracellular matrix-secreted and membrane-associated glycoproteins are known to be crucial regulators of the repulsive cues on axons and growth cones during neural development (1, 3–7, 9, 14). Recently, Slit-2 has been proposed as a “tumor suppressor gene” because of its decreased expression and loss of heterozygosity in most of the human cancers, including breast cancer, due to hypermethylation at the CpG islands of its promoter region (Sundaresan et al. 55). In addition, the SLIT-2 gene is mapped to chromosome 4p15.2 and ~63% of breast tumors also show loss of heterozygosity at the 4p15.1–15.3 region (3–5). Recent studies have confirmed that epigenetic events such as hypermethylation of CpG sites in regulatory regions may be critical alternative mechanisms of tumor suppressor gene inactivation (56). These studies indicate that Slit-2 appears to function as a novel tumor suppressor gene. However, the exact mechanisms of its tumor suppressive function are not well defined.
In the present study, we characterized the tumor-suppressive effect of the SLIT-2 gene in Slit-2-overexpressing MCF-7 breast cancer cells and Slit-2 transiently expressing MDA-MB-231 cells. We observed a decreased rate of proliferation in Slit-2-overexpressing cells compared with control cells by using an in vitro proliferation assay. We also confirmed this phenomenon in a soft agar colony forming assay. This assay revealed that Slit-2-overexpressing cells almost lost their colony-forming ability. Dallol et al. (3) have demonstrated nearly similar results by showing that Slit-2-conditioned medium suppresses the growth of several breast cancer cell lines. Furthermore, our Robo-1 knockdown experiment suggests that Slit-2 may exert its function in an autocrine manner. We have also analyzed the tumorigenic effect of Slit-2-overexpressing cells in mouse model systems. Slit-2-overexpressing cells showed a remarkable decrease in their tumorigenic capability compared with control cells when xenografted into SCID mice. Several previous studies have demonstrated that continuous estrogen supplementation sustains and enhances the tumorigenic effect of MCF-7 cells in nude mice (57). Our study in both SCID and nude mice indicates that Slit-2 overexpression significantly overcomes the tumor-enhancing and -sustaining effects of estrogen by exhibiting a marked inhibition of tumor size even in the presence of estrogen. These results provide further evidence for the antitumor activities of the SLIT-2 gene.
Upon exploring the mechanisms of the tumor-suppressive function of the SLIT-2 gene, we observed the decreased expression of β-catenin in Slit-2-overexpressing MCF-7 cells. Elevated levels of β-catenin have been observed in various human cancers, including breast cancer. Moreover, β-catenin has been linked with a poor prognosis in human adenocarcinomas (58). Mammalian Slit proteins have been suggested as evolutionarily conserved targets of the wnt/β-catenin signaling pathway (30). Recently, it has become more evident that β-catenin plays a dual function within the cells. In addition to its structural role of associating with the E-cadherin/actin cytoskeletal system during the regulation of cell-cell adhesion, β-catenin can act as a transcription factor along with the TCF/LEF family of DNA-binding proteins (34, 35). Increased levels of β-catenin within the cytoplasm and/or nucleus in tumor cells are suggestive of stabilization of the β-catenin protein and can result in enhanced β-catenin-mediated transcription (36–38, 47).
In our study, we observed the increased phosphorylation of β-catenin at its Ser-45 phosphorylation site. It has been established that Ser-45 phosphorylation by casein kinase I initiates phosphorylation at Thr-41, Ser-37, and Ser-33 by GSK-3β, and these sites are recognized by the ubiquitin ligase complex that mediates β-catenin degradation (50). Furthermore, we observed an increased association of β-catenin with GSK-3β and enhanced ubiquitination in Slit-2-overexpressing MCF-7 cells. These results confirmed that there is an increased degradation of β-catenin in the Slit-2-overexpressing cells, resulting in the reduced cytosolic concentration and decreased nuclear translocation of β-catenin in these cells. Additionally, our luciferase gene reporter assay revealed inhibition of β-catenin/TCF transcriptional activity in the Slit-2-overexpressing cells. Further, upon analysis of the expression of various β-catenin/TCF genes, we found decreased expression of cyclin D1, MMP-2, and MMP-9 in the Slit-2-overexpressing cells. These genes have been identified as critical mediators of proliferation, invasiveness, and tumor progression (36–38, 47). Slit-2 also blocked the expression of the TCF-4 transcription factor, which is responsible for activation of vimentin, a crucial mediator of epithelial-mesenchymal transition (59).
In many types of human cancer, it has been reported that the transcriptional activity of β-catenin is regulated either through the wnt/wingless-dependent pathway or via independent signaling pathways (35, 60). In our previous studies, we showed that Slit-2 inhibits PI3K and Akt phosphorylation (45). Akt has been demonstrated to phosphorylate GSK-3β. Our present study indicates that Slit-2-mediated inhibition of Akt activation might result in the decreased phosphorylation of GSK-3β, a substrate of Akt. Dephosphorylated GSK-3β is an active form that phosphorylates β-catenin, leading to its degradation in the ubiquitin-dependent proteasome pathway. Our study suggests that Slit-2 may regulate β-catenin function through a coordinated regulation of the β-catenin and PI3K pathways.
Recently, the β-catenin and PI3K signaling pathways have also been implicated in the regulation of cell-cell adhesion (58). Cell-cell adhesion, which is mediated by cadherins and catenins, is fundamentally involved in the organization of epithelial tissues (61). Loss of intercellular adhesion has been identified as a key process in the development of an aggressive tumor cell phenotype (61). This alteration of tumor cell phenotype is mediated through the expression and regulation of E-cadherin. Decreased expression of E-cadherin signifies the transformation of epithelial cells to a more invasive mesenchymal phenotype (58). Moreover, the Slit/Robo complex is known to regulate β-catenin/N-cadherin-mediated cell-cell adhesion in neuronal cells (62, 63). In our study, we observed increased localization of E-cadherin in cell borders at sites of cell-cell adhesion and its enhanced association with β-catenin in Slit-2-overexpressing cells. Moreover, we also observed increased translocation of β-catenin to the membrane in Slit-2-overexpressing cells. Slit-2-mediated up-regulation of E-cadherin might also play an important role in the mechanism to sequester β-catenin at the plasma membrane and may halt β-catenin transport to the nucleus. Further confirmation of some of the functional and signaling data in Slit-2 transiently expressing MDA-MB-231 cells indicates that the tumor-suppressive effect of Slit-2 is due to Slit-2 overexpression rather than clonal variation or heterogeneity of breast cancer cell lines.
We demonstrate, in two mouse model systems, that Slit-2 overexpression induces tumor suppressor activity in breast cancer cells. Furthermore, we show that Slit-2 mediates its tumor-suppressive effect via a novel mechanism, through the coordinated regulation of the β-catenin/TCF and PI3K/Akt signaling pathways and by enhancing cell-cell adhesion.
Notes
*This work was supported, in whole or in part, by National Institutes of Health Grant CA109527. This work was also supported by the Susan G. Komen Breast Cancer Foundation (to R. K. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
3The abbreviations used are: EGF, epidermal growth factor; Robo-1, Roundabout-1; PI3K, phosphatidylinositol 3-kinase; DMEM, Dulbecco's modified Eagle' medium; siRNA, small interference RNA; PBS, phosphate-buffered saline; TCF, T-cell factor; LEF, lymphoid enhancer factor; VC, vector control; GSK-3β, glycogen synthase kinase-3β; SCID, severe combined immunodeficiency disease; DAPI, 4′,6-diamidino-2-phenylindole.
References
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m800679200
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2546548?pdf=render
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/283/39/26624
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/283/39/26624.pdf
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/283/39/26624
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1074/jbc.m800679200
Article citations
Phenotypic targeting using magnetic nanoparticles for rapid characterization of cellular proliferation regulators.
Sci Adv, 10(19):eadj1468, 08 May 2024
Cited by: 0 articles | PMID: 38718125 | PMCID: PMC11078187
Emerging roles of deubiquitinating enzymes in actin cytoskeleton and tumor metastasis.
Cell Oncol (Dordr), 47(4):1071-1089, 07 Feb 2024
Cited by: 0 articles | PMID: 38324230
Review
Slit2/Robo1 signaling inhibits small-cell lung cancer by targeting β-catenin signaling in tumor cells and macrophages.
Mol Oncol, 17(5):839-856, 10 Jan 2023
Cited by: 5 articles | PMID: 35838343 | PMCID: PMC10158774
The Role of Slit-2 in Gestational Diabetes Mellitus and Its Effect on Pregnancy Outcome.
Front Endocrinol (Lausanne), 13:889505, 23 Jun 2022
Cited by: 3 articles | PMID: 35813663 | PMCID: PMC9261261
Micro RNA-640 Targeting SLIT1 Enhances Glioma Radiosensitivity by Restraining the Activation of Wnt/β-Catenin Signaling Pathway.
Br J Biomed Sci, 79:10067, 07 Apr 2022
Cited by: 2 articles | PMID: 35996510 | PMCID: PMC9302537
Go to all (75) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Tumor-suppressive effects of psoriasin (S100A7) are mediated through the β-catenin/T cell factor 4 protein pathway in estrogen receptor-positive breast cancer cells.
J Biol Chem, 286(52):44845-44854, 20 Oct 2011
Cited by: 24 articles | PMID: 22016394 | PMCID: PMC3248020
Function of Slit/Robo signaling in breast cancer.
Front Med, 9(4):431-436, 05 Nov 2015
Cited by: 17 articles | PMID: 26542734
Review
MDA-7 negatively regulates the beta-catenin and PI3K signaling pathways in breast and lung tumor cells.
Mol Ther, 8(2):207-219, 01 Aug 2003
Cited by: 62 articles | PMID: 12907143
The tumor suppressor interferon regulatory factor 8 inhibits β-catenin signaling in breast cancers, but is frequently silenced by promoter methylation.
Oncotarget, 8(30):48875-48888, 01 Jul 2017
Cited by: 18 articles | PMID: 28388578 | PMCID: PMC5564732





